Background: dGTP allosterically activates SAMHD1.
Results: GTP can also efficiently activate SAMHD1.
Conclusion: The cellular concentration of GTP is much higher than that of dGTP, and GTP can activate SAMHD1 to the same extent. Therefore, GTP may serve as a primary activator of SAMHD1.
Significance: Activator availability does not limit SAMHD1 phosphohydrolase activity because, unlike dGTP, GTP is not hydrolyzed by SAMHD1.
Keywords: Allosteric Regulation, HIV, Hydrolases, Macrophages, Reverse Transcription, SAMHD1, dNTP
Abstract
SAMHD1 (SAM domain- and HD domain-containing protein 1) is a dGTP-dependent dNTP triphosphohydrolase that converts dNTPs into deoxyribonucleosides and triphosphates. Therefore, SAMHD1 expression, particularly in non-dividing cells, can restrict retroviral infections such as HIV and simian immunodeficiency virus by limiting cellular dNTPs, which are essential for reverse transcription. It has previously been established that dGTP acts as both an activator and a substrate of this enzyme, suggesting that phosphohydrolase activity of SAMHD1 is regulated by dGTP availability in the cell. However, we now demonstrate biochemically that the NTP GTP is equally capable of activating SAMHD1, but GTP is not hydrolyzed by the enzyme. Activation of SAMHD1 phosphohydrolase activity was tested under physiological concentrations of dGTP or GTP found in either dividing or non-dividing cells. Because GTP is 1000-fold more abundant than dGTP in cells, GTP was able to activate the enzyme to a greater extent than dGTP, suggesting that GTP is the primary activator of SAMHD1. Finally, we show that SAMHD1 has the ability to hydrolyze base-modified nucleotides, indicating that the active site of SAMHD1 is not restrictive to such modifications, and is capable of regulating the levels of non-canonical dNTPs such as dUTP. This study provides further insights into the regulation of SAMHD1 with regard to allosteric activation and active site specificity.
Introduction
The concentration of dNTPs in dividing cells is tightly regulated by the cell cycle (1). During G1/S phase, dNTP biosynthesis is up-regulated such that replicative DNA polymerases have sufficient dNTPs to synthesize genomic DNA (1). In cells that are terminally differentiated/non-dividing such as macrophages, dNTPs are maintained at a much lower concentration (20–70 nm) compared with dividing cells (1–15 μm) (2). Although dNTPs are not needed for replication in these cells, dNTPs are still conditionally required for DNA repair. Unlike dNTPs, the concentration of NTPs, which have a 2′-OH on the ribose moiety, is not significantly impacted by the cell cycle and remains in the millimolar range (2). NTPs are essential energy carriers in the cell (i.e. ATP) and are also required for RNA synthesis.
Enzymes such as 5′-nucleotidases, deaminases, and nucleoside phosphorylases carry out the degradation of nucleotides (3). Additionally, SAMHD1 (SAM domain- and HD domain-containing protein 1), which was recently characterized as the first dNTP triphosphohydrolase in mammalian cells, can also degrade dNTPs. SAMHD1 harbors both an allosteric site and an active site. The binding of dGTP to the allosteric site results in a conformational change such that the active site is able to bind and hydrolyze dNTPs into deoxyribonucleosides and triphosphates (4, 5). Several studies have demonstrated that SAMHD1 plays a role in restricting infection of lentiviruses such as HIV-1, HIV-2, and simian immunodeficiency virus in non-dividing cells (6–9). The phosphohydrolase activity of SAMHD1 maintains dNTP concentrations in the low nanomolar range in macrophages, dendritic cells, and resting CD4+ T cells, thus limiting the substrate available for viral DNA synthesis (6–9). To overcome this restriction, some lentiviruses such as HIV-2 and several simian immunodeficiency viruses express the accessory protein Vpx to target SAMHD1 for proteasomal degradation (6, 7). This leads to an elevation in cellular dNTP concentrations and rapid completion of reverse transcription in non-dividing target cells (10, 11).
Beloglazova et al. (12) have recently revealed that SAMHD1 also acts as a nonspecific single-stranded DNA (ssDNA)2 and RNA (ssRNA) exonuclease and that several mutations in SAMHD1 associated with Aicardi-Goutières syndrome (AGS) reduce this activity. AGS is an immune-mediated neurological genetic disorder that is caused by mutations in genes encoding TREX1 (three prime repair exonuclease 1), ADAR1 (adenosine deaminase acting on RNA 1), RNase H2, and SAMHD1 (13). This disease is associated with increased levels of IFN-α and has clinically been described as a mimic of congenital viral infection (13). Because these AGS-related proteins are involved in nucleic acid metabolism, it is proposed that mutations in these proteins cause an accumulation of nucleic acids that trigger an autoimmune response (14).
As mentioned above, dGTP can allosterically activate SAMHD1 and is also hydrolyzed to dG by the active site of the enzyme (4, 5). Therefore, it has been thought that SAMHD1 is regulated by the availability of dGTP in the cell. In this study, we tested SAMHD1 allosteric activation by other deoxyguanosine and guanosine derivatives to determine the specificity of the allosteric site. Surprisingly, GTP activates SAMHD1 as well as dGTP; however, in contrast to dGTP, GTP is not hydrolyzed by the active site (5). Furthermore, because the concentration of GTP is 1000-fold higher than that of dGTP (2), we show that, at physiological concentrations, GTP acts as the primary activator of SAMHD1. We also demonstrate that SAMHD1 is able to hydrolyze base-modified nucleotides, suggesting that the active site specificity is defined to a greater extent by interactions with the ribose moiety rather than the base. In this study, we identified novel activators and substrates of SAMHD1, providing further insights into the regulation and substrate specificity of this enzyme.
EXPERIMENTAL PROCEDURES
SAMHD1 Protein Preparations
Recombinant human GST-SAMHD1 (R-SAMHD1) was overexpressed in and purified from Escherichia coli as described by Amie et al. (15). Immunoprecipitated human HA-SAMHD1 (IP-SAMHD1) was prepared by transfecting 293FT cells with 60 μg of pLVX-SAMHD1 vector containing an N-terminal HA tag (Clontech Laboratories, Inc.). Cells were lysed in radioimmune precipitation assay lysis buffer (Millipore), and SAMHD1 was immunoprecipitated from the lysate using the HA-Tag IP/Co-IP application set (Thermo Scientific).
HPLC-based SAMHD1 Phosphohydrolase Assay
In all SAMHD1 reactions, 1 μm R-SAMHD1 was incubated with 1 mm substrate (as indicated), 500 μm activator (as indicated; unless stated otherwise in the figure legends), and reaction buffer (50 mm Tris-HCl (pH 8), 50 mm KCl, 5 mm MgCl2, and 0.1% Triton X-100). To test for hydrolysis of base-modified nucleotides, 100 μm dGTP as activator was added to the reactions. Reactions were incubated for 30 min at 37 °C and terminated by incubation for 10 min at 75 °C. Reactions were then separated and quantified by anion exchange HPLC as described by White et al. (16).
TLC-based SAMHD1 Phosphohydrolase Assay
R-SAMHD1 (1 μm) or IP-SAMHD1 (740 nm) was incubated with 0.125 μCi/μl [α-32P]dATP and 250 μm unlabeled dATP with or without 250 μm activator (as indicated) and reaction buffer. The reactions shown in Fig. 2 (D and E) contained 0.125 μCi/μl [α-32P]dTTP, 250 μm unlabeled dTTP, and 1 μm ssDNA or ssRNA (5′-GGGGUCCUAAGCCAGUGCCAGAAGAGCCAAGGA-3′) (17). Reactions were incubated for 30 min at 37 °C and heat-killed for 10 min at 70 °C. Analysis and quantification of TLC reactions were completed as described by White et al. (16).
FIGURE 2.
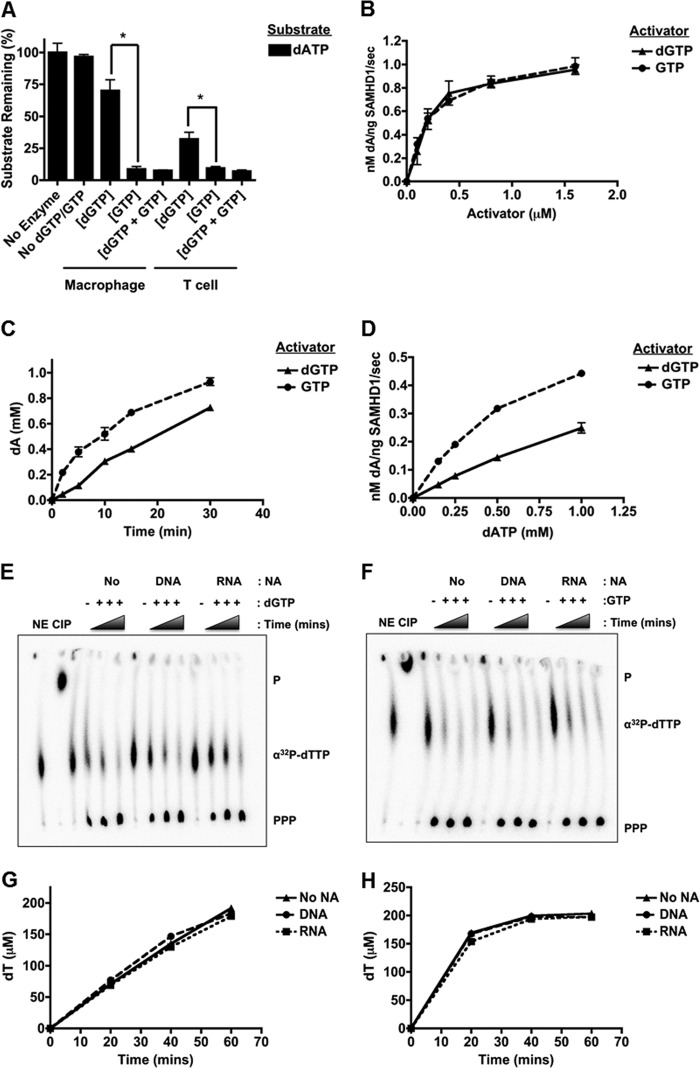
SAMHD1 allosteric activation by dGTP and GTP at physiological concentrations. A, R-SAMHD1 (1 μm) was incubated with 1 mm dATP (substrate) and physiological concentrations of either dGTP or GTP: 70 nm dGTP or 0.32 mm GTP for macrophage reactions (*, p = 0.0143) and 1.5 μm dGTP or 1.75 mm GTP for T cell reactions (*, p = 0.0451). SAMHD1 was also incubated with physiological concentrations of both nucleotides (dGTP + GTP). B, R-SAMHD1 was incubated with 1 mm dATP and increasing concentrations of dGTP or GTP. C, R-SAMHD1 was incubated with 1 mm dATP and 500 μm dGTP or GTP for the indicated time points. D, R-SAMHD1 was incubated with 250 μm dGTP or GTP and increasing concentrations of dATP. E–H, R-SAMHD1 was incubated with 250 μm dGTP (E) or GTP (D), 250 μm dTTP, and 0.125 μCi/μl [α-32P]dTTP in the presence or absence (No) of 33-mer ssDNA or ssRNA (1 μm). Reaction products were separated by TLC to detect [α-32P]dTTP hydrolysis (PPP), and densitometry was used to quantify product formation in the presence of dGTP (G) or GTP (H). NE, no-enzyme control; CIP, calf intestinal phosphatase control; P, monophosphates; NA, nucleic acids.
SAMHD1 Western Blot Analysis
1 μm R-SAMHD1 and the same volume of IP-SAMHD1 used in the TLC reactions (740 nm) were separated by SDS-PAGE and transferred to a membrane. SAMHD1 was detected using an anti-SAMHD1 primary antibody (Abcam) and a HRP-conjugated anti-mouse IgG secondary antibody (GE Healthcare).
Statistics
All error bars represent S.E. (n = 3). Significance was determined using an unpaired t test with the Welch correction.
RESULTS
SAMHD1 Allosteric Activation by dGTP Derivatives
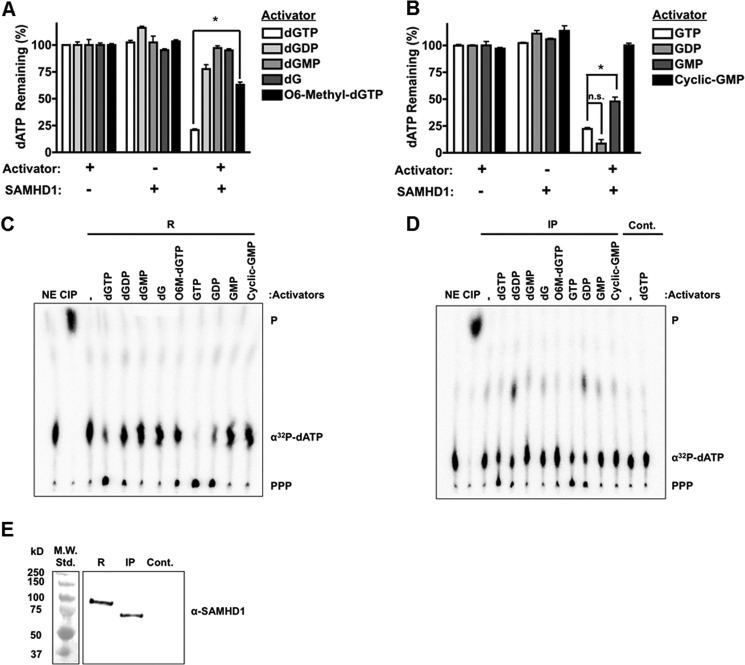
To test whether dGTP derivatives can activate SAMHD1, we utilized an assay in vitro that employs HPLC. Purified R-SAMHD1 (1 μm) was incubated with 1 mm dATP (substrate) with or without 500 μm activator (dGTP, dGDP, dGMP, dG, or O6-methyl-dGTP) (Fig. 1A). No-enzyme controls were used to determine the input amount of dATP in each reaction (100%), and reactions with SAMHD1 but without activator were used to control for any phosphatase contaminants in the purified preparation. Samples were injected onto an anion exchange column, and the remaining amount of dATP in the presence of each activator was quantified and normalized to the no-enzyme controls. In Fig. 1A, dATP was hydrolyzed in the presence of the known activator dGTP, but it was not hydrolyzed in the no-activator control, indicating that our purified preparation is active, specific, and free of phosphatase contaminants. The addition of dGDP minimally activated SAMHD1 (78% dATP remaining compared with 20% with dGTP); however, nucleotides with fewer phosphates did not efficiently activate the enzyme. Interestingly, O6-methyl-dGTP activated SAMHD1, but not to the same extent as dGTP (*, p = 0.0008), suggesting that the allosteric site can accommodate this base modification.
FIGURE 1.
SAMHD1 allosteric activation by guanosine derivatives. A and B, R-SAMHD1 (1 μm) was incubated with 1 mm dATP (substrate) with or without the indicated activators (500 μm). The amount of non-hydrolyzed dATP remaining after incubation was measured by HPLC and normalized to the no-enzyme controls. Statistical significance was determined using an unpaired t test with the Welch correction. *, p = 0.0008 (A); *, p = 0.0179 (B). n.s., not significant. C and D, R-SAMHD1 (R), IP-SAMHD1 (IP), or immunoprecipitate from an empty vector control (Cont.) was incubated with 250 μm dATP and 0.125 μCi/μl [α-32P]dATP with or without 250 μm activator (as indicated), and reaction products were separated by TLC. No-enzyme (NE) and calf intestinal phosphatase (CIP) controls were used for detection of [α-32P]dATP and monophosphates (P), respectively. SAMHD1 hydrolysis of [α-32P]dATP is indicated (PPP). E, Western blot analysis of SAMHD1 levels used in the above TLC reactions. Std.,, standard.
SAMHD1 Allosteric Activation by GTP Derivatives
We next tested whether the ribonucleotide GTP and its derivatives (GDP, GMP, and cGMP) can activate SAMHD1. As shown in Fig. 1B, 500 μm GTP, GDP, and even GMP (although not to the same extent as GTP; *, p = 0.0179) activated SAMHD1, whereas rG (data not shown) and cGMP failed to activate the protein. This suggests that a minimum of two phosphate groups for deoxyguanosine and one phosphate group for guanosine derivatives are required for efficient activation. Additionally, the allosteric site can accommodate the additional 2′-OH on the ribose moiety, but it cannot accommodate a cyclic phosphate group.
Allosteric Activation of IP-SAMHD1
Next, allosteric activation with each deoxyguanosine or guanosine derivative was confirmed with TLC using either R-SAMHD1 (Fig. 1C) or IP-SAMHD1 (Fig. 1D). IP-SAMHD1 was prepared by overexpressing SAMHD1 with an N-terminal HA tag in 293FT cells, a human embryonic kidney cell line, and by immunoprecipitation with an anti-HA antibody. Utilization of IP-SAMHD1 in these assays ensured that there were no post-translational modifications on SAMHD1 that could alter allosteric site specificity. In these reactions, SAMHD1 was incubated with [α-32P]dATP and unlabeled dATP with or without the indicated activators. Reactions were spotted on a PEI-cellulose TLC plate and separated with a LiCl-containing solvent. Production of the labeled triphosphate (Fig. 1, C and D, PPP) from dATP can be visualized if the indicated activator activated SAMHD1. A no-enzyme control (NE) and a calf intestinal phosphatase control (CIP) were used to detect [α-32P]dATP and monophosphates (P), respectively. As shown in Fig. 1 (C and D), the activators dGTP, dGDP, O6-methyl-dGTP, GTP, and GDP were able to activate both R-SAMHD1 and IP-SAMHD1, consistent with the data generated with the HPLC-based assay (Fig. 1, A and B). However, no significant hydrolysis was detected by the TLC-based assay with the addition of GMP. This is due to the difference in activator concentration used in the two assays (500 μm in the HPLC-based assay and 250 μm in the TLC-based assay; data not shown). Western blot analysis with an anti-SAMHD1 antibody was used to verify that HA-tagged SAMHD1 was indeed pulled down from 293FT cells and that it was not present in the empty vector control pulldown (Fig. 1E).
Comparison of SAMHD1 Allosteric Activation by GTP and dGTP at Physiological Concentrations
The concentration of GTP is 1000-fold higher than that of dGTP in both dividing and non-dividing cells (2). Although GDP and GMP activated SAMHD1 (Fig. 1B), they were also found at much lower concentrations compared with GTP (18). Therefore, we tested activation of SAMHD1 under the physiological dGTP and GTP concentrations found in macrophages. As shown in Fig. 2A, SAMHD1 hydrolyzed dATP in the presence of GTP by 8-fold more compared with dGTP (*, p = 0.0143). A similar trend was observed under the physiological concentrations found in T cells (*, p = 0.0451). When the two nucleotides were mixed at physiological concentrations, dATP was hydrolyzed to the same extent as with the addition of GTP alone. These biochemical studies suggest that GTP serves as the primary activator of SAMHD1 in cells and that the concentration of available activator does not limit SAMHD1 activation because GTP is not hydrolyzed by SAMHD1 (5). This was also confirmed with our full-length R-SAMHD1 (data not shown).
We next wanted to determine whether dGTP and GTP activate SAMHD1 to similar extents. R-SAMHD1 was first incubated with increasing concentrations of each activator, and hydrolysis of dATP was measured. As shown in Fig. 2B, dGTP and GTP had similar activation potentials at each concentration tested, resulting in the same apparent Km (240 nm), Vmax (1.1 nm dA/ng of SAMHD1/s), and kcat (0.003 s−1) values. However, when a time course with 1 mm dATP and 500 μm each activator was conducted, GTP allowed for more hydrolysis of dATP at each time point (Fig. 2C). To determine whether these activators alter the apparent Km for dATP, titration with dATP in the presence of a fixed concentration of each activator (250 μm) demonstrated that the Km for dATP was lower with GTP (0.74 mm) than with dGTP (2.7 mm), whereas the kcat values were similar (Fig. 2D). This could be caused by GTP altering the conformation of SAMHD1 such that dATP binds more efficiently and/or by dGTP competing with dATP for the active site of SAMHD1. Additionally, the Km for dATP was in the millimolar range, suggesting that the protein would not be active in non-dividing cells, where the dNTP concentration is in the nanomolar range, but perhaps post-translational modifications on SAMHD1 that increase substrate binding affinity occur in non-dividing cells.
Recently, SAMHD1 was characterized as a single-stranded nucleic acid exonuclease (12). Exonuclease and phosphohydrolase activities are associated with the HD domain (12, 16). Therefore, we wanted to confirm that the presence of 33-mer ssDNA or ssRNA does not affect SAMHD1 activation by GTP and subsequent phosphohydrolase activity. R-SAMHD1 was incubated with [α-32P]dTTP and unlabeled dTTP (phosphohydrolase substrate) with or without the nucleic acid substrates (1 μm) and with or without dGTP (Fig. 2E) or GTP (Fig. 2F) as activators. We chose dTTP as the substrate in these reactions to demonstrate that GTP can activate SAMHD1 and allow hydrolysis of a pyrimidine nucleotide in addition to the previously tested purine nucleotide (dATP). Product formation (PPP) for each reaction condition was monitored throughout a 60-min time course. As shown in Fig. 2 (G and H), the presence of ssDNA or ssRNA did not affect activation by dGTP or GTP, suggesting that GTP can still activate SAMHD1 even if these exonuclease substrates are present in the cell. This was additionally confirmed at different concentrations of dGTP and GTP (1 μm to 1 mm), and at each concentration tested, the nucleic acids did not have an effect (data not shown).
SAMHD1 Hydrolysis of Base-modified Nucleotides
We next tested the substrate specificity of the active site of SAMHD1. Although there is a crystal structure of the catalytic core of SAMHD1 (5), there has been no co-crystal of SAMHD1 bound to a nucleotide in the active site. Additionally, the crystallized SAMHD1 was most likely in the inactive state because dGTP was absent, making it difficult to distinguish the exact moieties of the nucleotide substrates necessary to interact with the active site. However, because SAMHD1 efficiently hydrolyzes all four canonical dNTPs, we predicted that SAMHD1 likely hydrolyzes base-modified dNTPs. For this test, R-SAMHD1 was incubated with or without dGTP (activator), and the remaining amount of non-hydrolyzed nucleotides was quantified by HPLC. As shown in Fig. 3 (A and B), SAMHD1 was able to hydrolyze 2-amino-2′-dATP, O6-methyl-dGTP, 5-methyl-2′-dCTP, and 2-thio-dTTP to the same extent as the canonical nucleotides (dATP, dGTP, dCTP, and dTTP, respectively). Interestingly, SAMHD1 hydrolyzed dUTP to a greater extent than dTTP (*, p = 0.0057). dUTP is a non-canonical nucleotide formed in the cell as a side product of dTTP formation. dUTPase is an essential enzyme expressed by the cell to decrease the concentration of dUTP, thus preventing incorporation of dUMP into genomic DNA (19). This result suggests that SAMHD1 also contributes to maintenance of the cellular dUTP concentration. Overall, these data indicate that the active site of SAMHD1 can accommodate various base modifications and that the active site specificity of SAMHD1 is maintained mainly by interactions with the ribose moiety rather than the base moiety of a nucleotide substrate.
FIGURE 3.
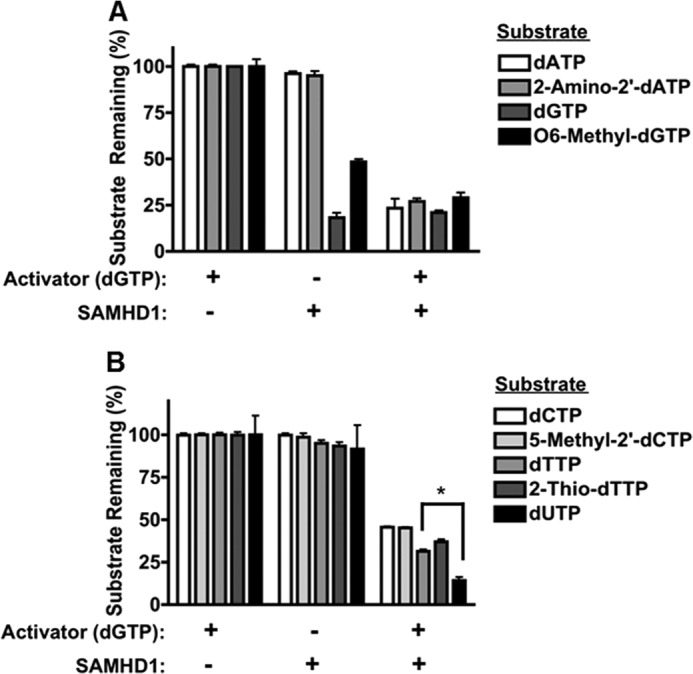
SAMHD1 hydrolysis of base-modified nucleotides. R-SAMHD1 (1 μm) was incubated with the indicated purine (A) and pyrimidine (B) nucleotide substrates (1 mm) with or without 100 μm dGTP for activation. The amount of substrate remaining in the triphosphate form was quantified by HPLC. *, p = 0.0057.
DISCUSSION
SAMHD1 has been characterized as the first mammalian dGTP-dependent dNTP triphosphohydrolase and more recently as a single-stranded nucleic acid exonuclease (4, 5, 12). The phosphohydrolase activity is thought to be responsible for restricting retroviral infections in non-dividing cells, and loss of both the phosphohydrolase and exonuclease activities may be involved in the autoimmune disorder AGS (13). The role of SAMHD1 as an antiviral restriction factor has been well established; however, the exact role of SAMHD1 in cellular processes and the regulation of enzymatic activities is still being investigated.
Here, we sought to further define the specificity of the allosteric and active sites of SAMHD1. First, we identified GTP as an allosteric activator of SAMHD1 that can activate SAMHD1 to the same extent as the previously reported activator, dGTP. Because the cellular concentration of GTP is much higher than that of dGTP (18), in this study, we have demonstrated that GTP likely serves as the primary activator of SAMHD1 in the cell. Unlike dGTP, GTP is not hydrolyzed by SAMHD1 (5), suggesting that the phosphohydrolase activity of SAMHD1 is either constitutively active or is regulated by other mechanisms such as protein expression and/or post-translational modifications. A recent study has shown that SAMHD1 expression is high at G1 phase and low at S phase to ensure that the cellular dNTP concentration is at the proper level for DNA replication (20). Additionally, two groups have demonstrated that the ability of SAMHD1 to restrict retroviral infections is regulated by cyclin A2/Cdk1 phosphorylation of Thr-592 (21, 22). In dividing cells, SAMHD1 is phosphorylated at this residue, thereby preventing restriction of HIV-1. Although SAMHD1 restriction of HIV-1 has previously been linked to phosphohydrolase activity, White et al. (21) suggested that Thr-592 phosphorylation does not alter the ability of SAMHD1 to hydrolyze dNTPs. This corroborates the results generated from our assays in vitro using SAMHD1 immunoprecipitated from dividing 293FT cells because IP-SAMHD1 is likely phosphorylated at Thr-592 yet has similar specific activity as R-SAMHD1 (1.7 and 2.0 nm dA/ng of SAMHD1/min, respectively) (Fig. 1, C and D). However, it is still possible that other post-translational modifications and/or protein-protein interactions regulate SAMHD1 activity, but further work regarding both possibilities must be done to determine the exact mechanisms of regulation.
Finally, we have shown that SAMHD1 can accommodate and hydrolyze base-modified nucleotides. Interestingly, SAMHD1 efficiently hydrolyzes dUTP, suggesting that SAMHD1 may also control the levels of mutagenic non-canonical dNTPs, particularly in non-dividing cells. This demonstrates that the active site is not very restrictive to such base modifications but is restrictive to the presence of a 2′-OH group on the ribose moiety because ribonucleotides are not substrates of the enzyme (5). These data may be helpful in future drug design because nucleoside analogues are utilized as antiviral therapies and chemotherapeutic agents (23).
This work was supported, in whole or in part, by National Institutes of Health Grants AI077401 and AI049781 (to B. K.) and by Training Grants GM068411 (Cellular, Biochemical, and Molecular Sciences) and AI049815 (HIV-1 Replication and Pathogenesis) (to S. M. A.).
- ssDNA
- single-stranded DNA
- ssRNA
- single-stranded RNA
- AGS
- Aicardi-Goutières syndrome
- R-SAMHD1
- recombinant human GST-SAMHD1
- IP-SAMHD1
- immunoprecipitated human HA-SAMHD1.
REFERENCES
- 1. Engström Y., Eriksson S., Jildevik I., Skog S., Thelander L., Tribukait B. (1985) Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J. Biol. Chem. 260, 9114–9116 [PubMed] [Google Scholar]
- 2. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fox I. H. (1981) Metabolic basis for disorders of purine nucleotide degradation. Metabolism 30, 616–634 [DOI] [PubMed] [Google Scholar]
- 4. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutières syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 6. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. St Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim B., Nguyen L. A., Daddacha W., Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 level, cellular dNTP levels and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beloglazova N., Flick R., Tchigvintsev A., Brown G., Popovic A., Nocek B., Yakunin A. F. (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutières syndrome and HIV-1 restriction. J. Biol. Chem. 288, 8101–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crow Y. J. (2013) Aicardi-Goutières syndrome. Handb. Clin. Neurol. 113, 1629–1635 [DOI] [PubMed] [Google Scholar]
- 14. Rigby R. E., Leitch A., Jackson A. P. (2008) Nucleic acid-mediated inflammatory diseases. BioEssays 30, 833–842 [DOI] [PubMed] [Google Scholar]
- 15. Amie S. M., Daly M. B., Noble E., Schinazi R. F., Bambara R. A., Kim B. (2013) Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J. Biol. Chem. 288, 20683–20691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L., Kim B., Brojatsch J., Diaz-Griffero F. (2013) Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noble E., Cox A., Deval J., Kim B. (2012) Endonuclease substrate selectivity characterized with full-length PA of influenza A virus polymerase. Virology 433, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 19. Tchigvintsev A., Singer A. U., Flick R., Petit P., Brown G., Evdokimova E., Savchenko A., Yakunin A. F. (2011) Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme. Biochem. J. 437, 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., Bianchi V. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1312033110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L. A., Kim B., Tuzova M., Diaz-Griffero F. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cribier A., Descours B., Valadão A. L. C., Laguette N., Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 23. McKenna C. E., Kashemirov B. A., Peterson L. W., Goodman M. F. (2010) Modifications to the dNTP triphosphate moiety: from mechanistic probes for DNA polymerases to antiviral and anti-cancer drug design. Biochim. Biophys. Acta 1804, 1223–1230 [DOI] [PubMed] [Google Scholar]