Background: IKKϵ can promote the ability of p52 to transactivate gene expression in a manner requiring p65.
Results: p52 is induced by TLR3 activation and regulates Sp1 transcription. Sp1 promotes the transcription of IL-15. Both events require the presence of IKKϵ and p52.
Conclusion: p52 is a target for IKKϵ in antiviral immunity.
Significance: This study reports a role for NFκB2 in the induction of antiviral gene expression.
Keywords: Cytokine Induction, Gene Regulation, Innate Immunity, Molecular Biology, NF-κB, Signal Transduction, Sp1, Toll-like Receptor (TLR), Transcription, Viral Immunology
Abstract
In this study we describe a previously unreported function for NFκB2, an NFκB family transcription factor, in antiviral immunity. NFκB2 is induced in response to poly(I:C), a mimic of viral dsRNA. Poly(I:C), acting via TLR3, induces p52-dependent transactivation of a reporter gene in a manner that requires the kinase activity of IκB kinase ϵ (IKKϵ) and the transactivating potential of RelA/p65. We identify a novel NFκB2 binding site in the promoter of the transcription factor Sp1 that is required for Sp1 gene transcription activated by poly(I:C). We show that Sp1 is required for IL-15 induction by both poly(I:C) and respiratory syncytial virus, a response that also requires NFκB2 and IKKϵ. Our study identifies NFκB2 as a target for IKKϵ in antiviral immunity and describes, for the first time, a role for NFκB2 in the regulation of gene expression in response to viral infection.
Introduction
NFκB2 (p100/p52), is a member of the NFκB family of transcription factors that comprises five mammalian members: Rel/c-Rel, RelA/p65, RelB, NFκB1 (p50 and its precursor p105), and NFκB2 (p52 and its precursor p100). These proteins exist in various homo- and heterodimeric complexes and control many biological processes, particularly in inflammation and immunity (1). There are two distinct NFκB activation pathways, termed the canonical and the alternative pathways. The canonical pathway is the better characterized NFκB pathway. It is activated by innate immune receptors, inflammatory cytokines, and stress pathways and leads to the phosphorylation of the inhibitory subunit IκBα by IKKα3 and IKKβ, leading to its degradation. The subsequent release and nuclear translocation of the p50/p65 dimer leads to the induction of a wide range of immune and inflammatory genes (1). The alternative pathway involves NFκB2. Known activators of this pathway are CD40, B-cell activating factor (BAFF) receptor, and lymphotoxin-β (LT-β) receptor. Activation of this pathway involves NFκB-inducing kinase. NFκB-inducing kinase activates IKKα, which phosphorylates p100, causing p100 to be partially processed to produce the active p52 subunit. Typically, p52 is described as part of a heterodimer with RelB. This complex is essential for the role of NFκB2 in humoral immunity and secondary lymphoid organogenesis.
A third IKK, termed IKKϵ, has been described. IKKϵ is activated downstream of the dsRNA receptors TLR3, RIG-I, and MDA5 and by TLR4 and IFN-β (2, 3) and, in turn, activates the transcription factors IRF3, IRF7, STAT1, and p65. IKKϵ has also been shown to be activated by respiratory syncytial virus (RSV) and influenza B virus (3, 4).
We identified p52 as a binding partner for IKKϵ in a yeast-two-hybrid screen and subsequently determined that overexpression of IKKϵ could promote the transactivating potential of p52 (5). However, the functional importance of this interaction remains elusive. Here we report the uncovering of a signaling pathway activated by TLR3, or RSV, that involves IKKϵ, NFκB2, and p65. We identify a conserved binding site for p52 on the Sp1 promoter and confirm IL-15, an antiviral cytokine, as a target for Sp1 on this pathway. Our study provides a previously undescribed function for both NFκB2 and Sp1 in antiviral immunity.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
LPS, (Alexis Corp.), poly(I:C) and poly(A:U) (Invivogen) were used. The antibodies used were p100/p52 (Cell Signaling Technology, catalog no. 4882), β-actin (Sigma-Aldrich, catalog no. A1978), and Sp1 (Millipore, catalog no. 07-645). Oligonucleotides were from Eurofins, and TaqMan probes were from Applied Biosystems. FLAG-IKKϵ and IKKϵ(K38A) were provided by Shizuo Akira (Osaka University, Japan). The TBK1-encoding plasmid was a gift from Dr. Makato Nakanishi (National Institute for Longevity Sciences, Japan). HA-p52 was a gift from Neil Perkins (University of Dundee, Scotland). The Gal-luciferase reporter gene was from Stratagene. The construction of p52-Gal4 has been described previously (5). The TRAF1, TRAF3, TRAF6, RIP1, and Nap1 plasmids were gifts from Andrew Bowie (Trinity College Dublin, Ireland). The HA-p65(S536A) plasmid was generated from the HA-p65 plasmid using the QuikChange XL site-directed mutagenesis kit (Stratagene).
Cell Culture and Isolation
WT and IKKϵ−/− MEFs obtained from Shizuo Akira (Osaka University, Japan), HEK293 cells, and HEK293 cells stably expressing either TLR3 (HEK293-TLR3) (Invivogen) were cultured in DMEM. WT, NFκB2−/−, and IKKϵ−/− (Kate Fitzgerald, University of Massachusetts) bone marrow was isolated from the tibias and femurs of C57/Bl6 mice, and the resulting cells were grown in macrophage colony stimulating factor-conditioned DMEM. Human peripheral blood mononuclear cells were isolated from whole blood using a Ficoll gradient and cultured in RPMI. In all cases, DMEM and RPMI medium were supplemented with 10% fetal calf serum, 2 mm l-glutamine, and 1% penicillin/streptomycin solution (v/v). Cells were plated at 1 × 105 cells/ml, and treated as described before isolation of RNA or lysate for quantitative PCR or Western blot analysis, respectively.
Sp1 Promoter Luciferase Construct
For construction of the Sp1 promoter luciferase reporter gene, we cloned its 5′ regulatory region of Sp1 from −1303 nt from the translational start site (ATG) between the NheI and XhoI sites of the reporter luciferase vector pGL3-enhancer (Promega). Progressive deletion constructs were generated using a common reverse primer and five different forward primers. The Sp1-specific sequences for these primers were taken from the EMBL-EBI AF261690 source (in uppercase, see below). For the forward primers, these specific sequences were preceded by an arbitrary sequence (in lowercase, see below), including a NheI restriction site (italics). The reverse primer followed a similar structure but contained a XhoI restriction site (italics) in the arbitrary sequence. The numbers indicated after the primer sequences correspond to the distance in nt from the 5′ end of the sequence in uppercase to the translational start site: forward, 5′-tcaagtcaggctagcTTGCTTTATGCATAGGCGGT-3′ (-1303); forward, 5′-tcaagtcaggctagcCGGATTCTGGTTGGCCGTTGT-3′ (-477); forward, 5′-tcaagtcaggctagcCTATCAAAGCTTTGCCTATCC-3′ (-443); forward, 5′-tcaagtcaggctagcGGGAGCCCGCCTGCCGGTTG-3′ (-415); forward 5′-tcaagtcaggctagcTCCTTCCAAGCCAATCATCTCC-3′ (-388); forward, 5′-tcaagtcaggctagcGCTCCCGCCCATCTTCACTTC-3′ (-365); and reverse, 5′-cagtgctgcctcgagGCTCAAGGGGGTCCTGTCCGG-3′ (-20).
Transfection-based Reporter Gene Assays
Cells were transfected with GeneJuice transfection reagent (Novagen, Madison, WI) with a total amount of 350–400 μg/well containing 150 ng of p-55UASGLuc and 50 ng of p52-Gal4 fusion construct (MEFs) or with a total of 250 ng of DNA containing 100 ng of p-55UASGLuc or 30 ng of p52-Gal4 (HEK293s). Assays also contained the plasmid DNA of interest, an empty vector, and 30 ng of Renilla reniformis luciferase construct. For Sp1-promoter-luciferase assays, HEK293 cells were transfected with a total amount of 220 ng of DNA/well comprising 80 ng of reporter construct, the plasmid DNA of interest, 40 ng of R. reniformis luciferase construct, and an empty vector. Cell extracts were monitored 24–36 h post-transfection for firefly luciferase activity following standard protocols with values normalized for transfection efficiency with R. reniformis luciferase.
RNA Extraction and PCR
MEFs and BMDM or Human peripheral blood mononuclear cells were set up at 5 × 105 or 1 × 106 cells/ml, respectively. Cells were stimulated with Poly(I:C). Total RNA was extracted using the RNeasy kit (Qiagen). For mRNA expression analysis, cDNA was prepared from 20 to 100 ng/ml total RNA using the High-Capacity cDNA archive kit (Applied Biosystems). Individual mRNAs were monitored with the following inventoried The AB7900FAST platform (Applied Biosystems) was used for all PCR, done in triplicate. Changes in expression were calculated by the change in threshold (ΔΔCT) method with Gapdh as an endogenous control for gene-expression analysis and were normalized to results obtained with untreated cells. TaqMan assays were from Applied Biosystems: mouse Sp1 assay (Mm00489039_m1), mouse IL-15 assay (Mm00434210_m1), mouse Gapdh (glyceraldehyde phosphate dehydrogenase) assay, human Sp1 assay (Hs00916521_m1), human IL-15 assay (Hs01003713), human Gapdh assay.
siRNA
The following RNA interference duplex was purchased from Qiagen Hs_NFκB2_1 FlexiTube siRNA SI00300965 and Allstars negative control siRNA (catalog no. 1027281) or Dharmacon ON-TARGET plus siRNA Sp1 (catalog no. L-026959). In all cases, 50 nm of siRNA was used. Human PBMCs were transfected with siRNA using an Amaxa electroporator and a Cell Line Nucleofector Kit V, program V-01 (PBMC). 1 × 106cells/ml PBMCs were used per point for nucleofection. Cells were harvested after 72 h and used for further analysis.
Immunoblotting
MEFs and BMDMs were seeded at 5 × 105 cells/ml, HEK293TLR3 cells were seeded at 1 × 105 cells/ml, and human peripheral blood mononuclear cells were set up 1 × 106 cells/ml 1 day prior to stimulation with 2% FCS. Cells were stimulated with poly(I:C) and lysed in 1 ml of low-stringency lysis buffer (50 mm HEPES, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors). Protein concentration was measured by Bradford, and equal amounts of protein were separated by SDS-gel electrophoresis, transferred to a PVDF membrane, incubated with antibody, and visualized by autoradiography.
Chromatin Immunoprecipitation
Genpathway, Inc. (CA) carried out an analysis of gene promoters that bound to p52 using samples prepared from WT and IKKϵ KO MEFs according to their instructions. Briefly, MEFs were set up at 5 × 105 cell/ml. A final volume of 1%, formaldehyde was added directly to the existing medium and incubated for 15 min. A 1/20 volume of 2.5 m glycine was then added to each flask and allowed to set at room temperature for 5 min. Cells were scraped, washed in PBS, and sent on dry ice to Genpathway, Inc. BMDMs were set up at 5 × 105 cell/ml, medium was removed, replaced with PBS, and fixed by adding a final concentration of 1% formaldehyde to each culture dish. Flasks were incubated for 10 min at room temperature. A 1/20 volume of 2.5 m glycine was then added to each flask and allowed to set at room temperature for 5 min. The primary antibodies anti-p52 (Abcam, catalog no. 7972), anti-p50 (Millipore, catalog no. 06-886), and anti-p65 (Santa Cruz Biotechnology, catalog no. (F-6) sc-8008) were determined to give the best ChIP results. Quantitative RT-PCR was carried out using primers for either the Sp1 promoter or β-actin promoter as indicated. Data are presented as percent of input.
Affinity Purification with Biotinylated Oligonucleotides
HEK293 cells were seeded at 1 × 105 cell/ml and incubated overnight. Cells were then transfected with either 2 μg of HA-p52 (five plates) or an empty vector control (five plates). 24 h later, cells were lysed in 100 μl of oligonucleotide buffer (25 mm Tris, 50 mm EDTA, 5% glycerol, 5 mm NaF, Nonidet P-40 1%, 1 mm DTT, 150 mm NaCl, and protease and phosphatase inhibitors), pooled, and snap-frozen. Samples were then thawed on ice and diluted with a further 4.5 ml of oligonucleotide buffer without NaCl. A 50-μl sample of lysate was kept, and the remainder was divided into five tubes and incubated for 2 h with streptavidin-agarose beads conjugated to biotinylated promoter regions, termed Seq 1–5, as depicted in Fig. 4F. Lysates were then centrifuged to pellet the beads, which were washed three times before 50 μl of 5× SDS sample buffer was added to the beads. Samples were then immunoblotted as indicated.
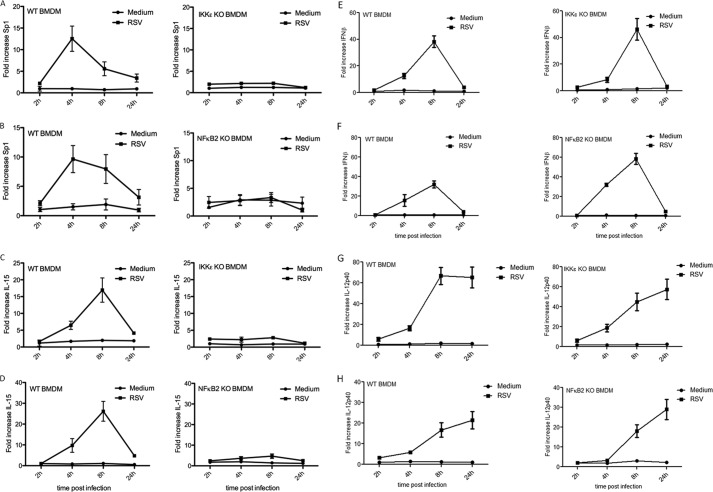
FIGURE 4.
RSV infection induces both Sp1 and IL-15 in an IKKϵ- and NFκB2-dependent manner. WT, IKKϵ −/−, and NFκB2−/− BMDMs were infected with the RSV long strain (group A) (multiplicity of infection = 2) or treated with medium for 2, 4, 8, or 24 h. Cells were lysed, and RNA was extracted for RT-PCR analysis with primers specific for Sp1 (A and B), IL-15 (C and D), IFNβ (E and F), or IL-12p40 (G and H). Expression was normalized to that of GAPDH and is presented relative to that of untreated controls. Data are representative of three separate experiments, with each point assayed in triplicate, with error bars representing S.D.
Viral Infection of BMDMs
RSV long strain (group A) was obtained from the ATCC and propagated in HEp-2 cells with serial plaque purifications to reduce defective interfering particles (6). WT and NFkB2-deficient BMDMs were plated in 6-well (3 × 106 cells/well) tissue culture plates. Macrophages were infected with RSV (multiplicity of infection = 2) or treated with medium alone and incubated at 37 °C for the indicated times.
RESULTS
Poly(I:C) Promotes p52-driven Transactivation in an IKKϵ- and p65-dependent Manner
Having previously identified p52 as a protein that interacts with IKKϵ and, furthermore, showing that IKKϵ promotes transactivation by p52 (5), we wished to probe the functional relevance of this interaction. Given the antiviral role of IKKϵ, we chose to test whether the dsRNA analog P(I:C) could promote p52-mediated transactivation. We cotransfected HEK293-TLR3 cells with a plasmid encoding full-length p52 fused to the DNA-binding domain of Gal4 (p52-Gal4) and a Gal4-driven luciferase construct. Fig. 1A shows that increasing concentrations of the dsRNA analogues P(I:C) and Poly(A:U) promote p52-driven transactivation. Because P(A:U) is a dsRNA analog only recognized by TLR3 (7) and P(I:C) and P(A:U) both promoted p52-driven transactivation to almost identical levels, the implication is that TLR3, and not the cytosolic RIG-1-like receptors, promote p52-driven transactivation. Having previously identified IKKϵ and p65 in a complex with p52 (5), we next examined whether IKKϵ and/or p65 mediate P(I:C)-inducible p52- driven transactivation. P(I:C) failed to promote p52-driven transactivation in IKKϵ−/− MEFs or p65−/− MEFs compared with WT MEFs (Fig. 1B). These MEF strains were responsive to TLR3 ligation, as demonstrated by comparable levels of P(I:C)-inducible phosphorylation of p38 (not shown). We next determined whether the kinase activity of IKKϵ was required for P(I:C)-inducible transactivation by p52. As shown in Fig. 1C, P(I:C) promoted p52-driven transactivation to ∼5-fold over the control, whereas a kinase-dead form of IKKϵ (IKKϵK38A) inhibited this induction, presumably acting as a dominant negative inhibitor.
FIGURE 1.
Poly(I:C) induces p100 and p52 expression and p52-dependent transactivation in an IKKϵ- and p65-dependent manner. A–J, p52-dependent transactivation was assayed in all cells using 150 ng of p-55UASGLuc and 50 ng of p52-Gal4 or Gal4-DBD. HEK293-TLR3 cells were stimulated with poly(A:U) or poly(I:C) 24 h post-transfection or left untreated (Control) and incubated for 6 h (A) or cotransfected with a plasmid encoding IKKϵKA (C), p65, p65S536A, or empty vector (EV) (D). B, WT, IKKϵ KO, or p65 KO MEFs were stimulated with P(I:C) 24 h post-transfection or left untreated (control) and incubated for 6 h. E, WT and IKKϵ KO MEFs were cotransfected with p65 or EV as indicated. F–I, HEK293 cells or wild-type and IKKϵ−/− MEFs (J) were cotransfected with plasmid encoding IRF3 (F); p65 (G); Trif (J); TRAF1, TRAF3, TRAF6, and RIP1 (H); IKKϵ, Nap1, and TBK1 (I) or with empty vector only, as indicated. Luciferase activity was determined 24–36 h after transfection. Data are the means of three measurements, with error bars representing S.D. K, HEK293-TLR3 cells were treated with increasing doses of P(I:C) for 24 h, lysed, and probed for p100/p52. Data are representative of three separate experiments.
Of the five NFκB family members, only c-Rel, p65, and RelB have transactivating potential (9). p50 and p52 are DNA binding subunits and are unable to transactivate gene expression on their own. Because we and others have shown that p52 interacts with p65 (5, 10), we next tested whether p65 was the transactivation partner for p52 downstream of P(I:C). Even small amounts of p65 could strongly drive p52-dependent transactivation, and this ability of p65 to induce p52 transactivation was substantially impaired when the serine residue at position 536 in p65 was mutated to an alanine (p65S536A) (Fig. 1D). Phosphorylation of p65 at position Ser-536 is known to be very important for the efficient transactivating potential of p65 in response to many ligands, and IKKϵ is known to phosphorylate Ser-536 in response to P(I:C) (8–11). We next investigated whether p65 could induce p52-dependent transactivation in IKKϵ−/− MEFs. As shown in Fig. 1E, p65 induced p52 transactivation in WT MEFs, but this induction was substantially impaired in IKKϵ−/− MEFs, indicating that p65 requires IKKϵ to confer its transactivation potential to p52. IRF3, another transcription factor with transactivating potential, is activated downstream of P(I:C) and phosphorylated by IKKϵ (10, 12). However, IRF3 is unable to mediate transactivation by p52 in HEK293-TLR3 cells (Fig. 1F). Together, these results imply that P(I:C) activates IKKϵ, which, in turn, mediates p65 transactivation of the p52-dependent reporter gene, likely by phosphorylating p65 at position Ser-536 (11–14).
We further investigated whether components of the TLR3 signaling pathway could promote p52-driven transactivation. Fig. 1, G–I demonstrates that TRIF, when overexpressed, can strongly induce p52-driven transactivation. TRAF3, TRAF6, RIP1, IKKϵ, and TBK1, all of which are known to be downstream of TRIF (13, 14), can promote p52-dependent transactivation in a dose-dependent manner, whereas TRAF1 and the IKKϵ/TBK1 adaptor NAP1 cannot. To determine whether TRIF-induced p52 transactivation is mediated by IKKϵ, we compared the ability of TRIF to drive p52 transactivation in WT and IKKϵ−/− MEFs (Fig. 1J). IKKϵ is required for TRIF-induced p52 transactivation because TRIF was unable to induce p52 transactivation in IKKϵ−/− MEFs (Fig. 1J). All of these data pointed to p52 as an important target for TLR3 signaling, and we further confirmed this by demonstrating that expression levels of both p100 and p52 were induced by P(I:C) in a dose-dependent manner (Fig. 1K).
p52 Binds to the Promoter of the Sp1 Gene to Activate Its Transcription
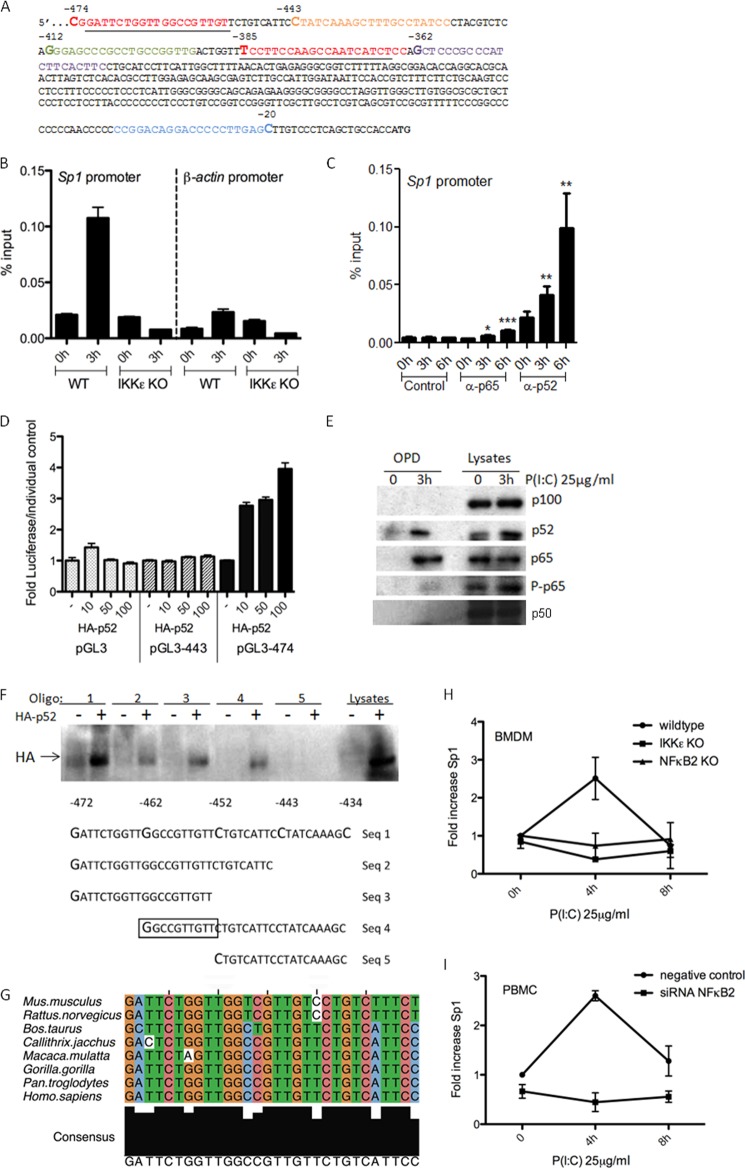
Having shown that p52 is induced and activated by P(I:C), we next interrogated the gene promoters that were bound by p52 in response to P(I:C). We carried out a ChIP analysis to determine genes to which p52 might bind. This was carried out commercially by Genpathway, Inc., who revealed that p52 binds to an enhancer sequence in the Sp1 gene promoter in the region shown in Fig. 2A. We next carried out a ChIP analysis comparing WT and IKKϵ−/− BMDMs treated with P(I:C) for 3 h. As shown in Fig. 2B, there was no difference between WT and IKKϵ−/− cells in the basal binding of p52 to the Sp1 promoter. However, P(I:C) induced a 5-fold increase in binding of p52 to the Sp1 promoter in WT BMDMs, and this was abrogated in IKKϵ−/− BMDMs. We also investigated the binding of the NFκB subunit p65 to the Sp1 promoter. p65 binding to the promoter was increased significantly, 2-fold, in response to P(I:C). Binding of p65 to the TNF promoter was measured as a positive control for p65 binding (not shown). p52 binding was again evident in the P(I:C)-treated cells (Fig. 2C).
FIGURE 2.
Poly(I:C) induces p52 and p65 to bind a previously undescribed site in the Sp1 promoter to drive transcription in an IKKϵ-, p65-, and NFκB2-dependent manner. A, a partial sequence of the Sp1 promoter region −474 nt to −362 nt 5′ from the start codon. The primers used for the ChIP assay are underlined. B, WT and IKKϵ KO BMDMs were treated with P(I:C) for 3 h or left untreated, after which a ChIP assay was performed using an anti-p52 antibody. Primers specific for promoters of Sp1 or β-actin were designed, and binding events of p52 were measured as percent of input. C, WT BMDMs were treated with P(I:C) for 0, 3, and 6 h, after which a ChIP assay was performed using antibodies against HA (control), p65, or p52. Binding events were measured as percent of input. Data are the means of three measurements, with error bars representing S.D. Statistical analysis was carried out using Student's t test. *, p < 0.05; **, p < 0.001; ***, p < 0.0001. Values are representative of three separate experiments. D, Sp1 promoter truncations were cloned into a pGL3 luciferase reporter vector, and Sp1 promoter activity was assayed in HEK293 cells transfected with 80 ng of pGL3 vector containing Sp1 promoter truncations −474 and −443 nt 5′ from the start site, respectively, or with pGL3 vector alone. Cells were cotransfected with a plasmid encoding HA-p52. Luciferase activity was determined 24 h after transfection and is represented as fold increase in luciferase over each individual pGL3-Sp1 promoter construct or empty vector control. Data are the means of three measurements, with error bars representing S.D. Values are representative of three independent experiments. E, HEK293-TLR3 cells were treated with P(I:C) for 0 and 3 h, lysed, and an oligo pull-down (OPD) assay was carried out with the −472 to −434 oligo sequence. Samples were probed for p100, p52, p65, P-p65S536, and p50. F, an OPD assay was carried out in HEK293 cells using the oligonucleotide sequence from −472 to −434 5′ from the Sp1 translational start codon (Seq 1) and truncations of this nucleotide sequence (Seq 2–5) as shown in the bottom panel. Cells were transfected with a plasmid encoding either HA-p52 (+) or empty vector (−). 24 h later, cells were lysed, incubated with oligos as indicated, and probed for HA (top panel). OPD assays are representative of two separate experiments. G, species sequence alignment of the site in the Sp1-promoter. Upstream regions were obtained via biomart taking the flanked regions 2500 base pairs upstream. An alignment was created using MUSCLE. The alignment was viewed, and an image was exported via Jalview. The 10 central base pairs in the alignment are the binding site. The binding site starts −364 upstream from the gene start site in the human sequence. H, WT, IKKϵ KO, and NFκB2 KO BMDMs or PBMCs (I) transfected with either siRNA targeting NFκB2 or a non-targeting control siRNA for 48 h were stimulated with P(I:C) for 0, 4, and 8 h, as indicated. H and I, quantitative RT-PCR analysis of RNA from these cells was carried out with primers specific for Sp1. Expression is normalized to that of GAPDH and is presented relative to that of untreated controls. Data are the mean of at least three separate experiments with each point assayed in triplicate. Error bars represent S.D.
To examine this region further and locate the DNA element important for the transcriptional regulation of the Sp1 gene by p52, a series of 5′-deletion promoter constructs was generated by PCR and cloned into the promoterless pGL3 enhancer luciferase reporter vector. The resulting constructs contain 5′ flanking regions from −20 to −474, −443, −412, −385, and −362 relative to the translational start codon. HEK293 cells were transfected with the deletion constructs in conjunction with increasing amounts of plasmid expressing HA-p52. A 4-fold increase in activity was induced by cotransfection of pGL3-474 with HA-p52 compared with that of pGL3-474 alone (Fig. 2D). Deletion to −443 abolished this activity, which was also abolished in all other constructs (not shown). These results demonstrate that the Sp1 promoter is activated by p52 in the region of sequence between −443 and −474 nt relative to the start site. We next employed an oligo pull-down assay to assess whether P(I:C) could induce the binding of p52 and p65 to this region of the promoter (−443 to −474). This is clearly the case because P(I:C) specifically induced the binding of both p52 (Fig. 2E, second panel) and p65 (third panel) but not p100 (first panel) or p50 (fourth panel) to the oligonucleotide sequence indentified from the Sp1 luciferase assay. Interestingly, the bound form of p65 appears to be in a phosphorylated state because we were also able to weakly detect P-p65S536 in the induced complex (Fig. 2E, fourth panel).
We further defined the p52 binding site using this assay. Overexpressed HA-p52 binds to the oligonucleotide consisting of the sequence from −434 to −472 (Fig. 2F, Seq 1, lane 2) and also to the sequences from −443 to −472 (Seq 2, lane 4), from −452 to −472 (Seq 3, lane 6), and from −434 to −462 (Seq 4, lane 8). However, HA-p52 does not bind to the oligonucleotide consisting of the sequence from −434 to −452 (Seq 5, lane 10) (Fig. 2F). Sequence 5 differs from the sequences 1–4 in that it lacks the sequence GGCCGTTGTT. Interestingly, this area in the promoter of Sp1 is conserved among species (Fig. 2G). This identifies, for the first time, GGCCGTTGTT as a binding site for p52.
Having demonstrated that p52 binds to the Sp1 promoter, we next tested the functional consequences of this response. As shown in Fig. 2H, P(I:C) induced the expression of Sp1 mRNA in WT BMDMs, whereas this effect was not observed in either IKKϵ−/− or NFκB2−/− BMDMs. To determine whether this effect could be seen in human cells, siRNA directed against NFκB2 or a non-targeting control were transfected into PBMCs, and Sp1 induction was measured. P(I:C) induced Sp1 expression in control cells. This induction was lost in NFκB2 knockdown cells (Fig. 2I).
Sp1, IKKϵ, and NFκB2 Regulate IL-15 Gene Transcription
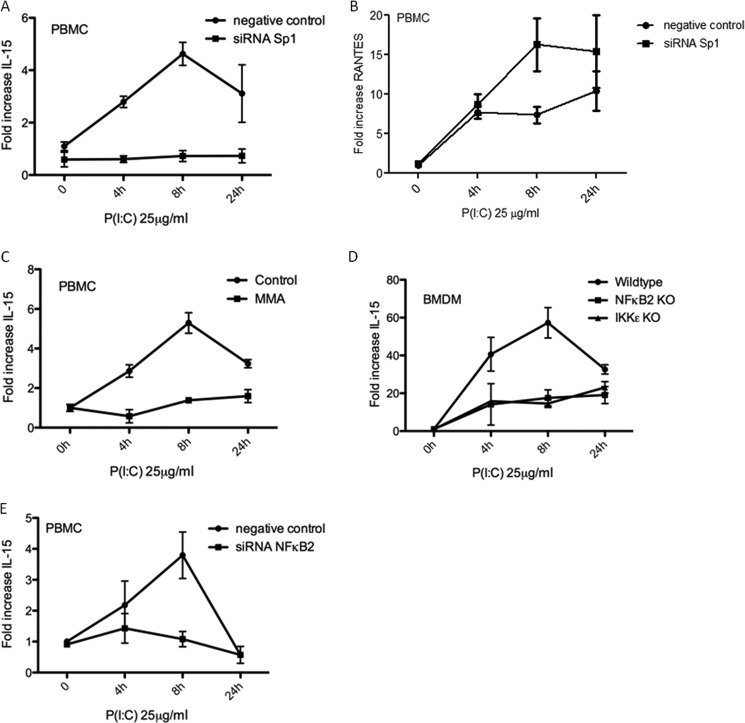
We next determined target genes for Sp1 that might be relevant to the antiviral response. The promoter of IL-15, a proinflammatory, antiviral cytokine, was strongly predicted to be regulated by Sp1. IL-15 is known to be induced by P(I:C) and by viral infection (16, 17). We hypothesized that a P(I:C)-inducible gene regulated by Sp1 should not be induced in either IKKϵ or NFκB2 knockout cells. To validate IL-15 as a Sp1 target gene, human PBMCs were transfected with siRNA directed against Sp1 or a non-targeting control, and IL-15 induction was measured. P(I:C) caused an increase in the level of IL-15 mRNA over time that was lost in cells deficient in Sp1 (Fig. 3A). Importantly, Sp1 knockdown did not reduce the ability of the PBMCs to induce IL-6 (not shown) or RANTES (regulated on activation, normal T cell expressed and secreted), which was, in fact, increased upon Sp1 knockdown (Fig. 3B). These are Sp1-independent, P(I:C)-inducible genes indicating specificity in the IL-15 observation. This effect was confirmed pharmacologically in PBMCs pretreated with 1 μm mithramycin A (MMA), an Sp1 inhibitor. P(I:C) caused an ∼5-fold increase in the level of IL-15 mRNA over basal levels after 8 h. This effect was inhibited by pretreatment with mithramycin A (Fig. 3C).
FIGURE 3.
IL-15 induction in response to poly(I:C) requires Sp1, IKKϵ, and NFκB2. A, B, and E, PBMCs were transfected with siRNA targeting Sp1 (A and B), NFκB2 (E), or a non-targeting control siRNA for 48 h. Cells were then stimulated with P(I:C) for 0, 4, and 8 h as indicated. C, PBMCs were pretreated for 1 h with 1 μm mithramycin A (MMA) or left untreated before stimulation with P(I:C). D, WT, IKKϵ−/−, and NFκB2−/− BMDMs were stimulated with P(I:C) for 4, 8, and 24 h or left untreated, as indicated. In each case, RT-PCR analysis of RNA was carried out with primers specific for IL-15 or RANTES, as indicated. Expression is normalized to that of GAPDH and is presented relative to that of untreated controls. Data are the mean of at least three separate experiments with each point assayed in triplicate, with error bars representing S.D.
Following this, we measured the IL-15 transcript in response to P(I:C) in WT, NFκB2−/−, and IKKϵ−/− BMDMs. IL-15 transcript levels increased 60-fold in WT BMDMs after 8 h of poly(I:C) treatment, whereas a marked inhibition of this response was observed in both NFκB2−/− and IKKϵ−/− BMDMs (Fig. 3D). This dependence of P(I:C)-induced IL-15 levels on NFκB2 was confirmed in human PBMC, whereas no increase was observed in PBMCs transfected with siRNA targeted against NFκB2 (Fig. 3E). These data, therefore, implicate IKKϵ, NFκB2, and Sp1 in the induction of IL-15 by P(I:C).
RSV Infection Fails to Up-regulate Sp1 and IL-15 Transcripts in Both IKKϵ and NFκB2 Knockout BMDMs
Finally, we examined whether this pathway was important for induction of IL-15 by a virus. RSV is recognized by TLR3 during infection (18, 19) and is a powerful inducer of IL-15 (16). We infected WT, IKKϵ−/−, and NFκB2−/− BMDMs with RSV (multiplicity of infection = 2) for 4, 8, or 24 h and measured Sp1 mRNA levels. As shown in Fig. 4A, left panel, the Sp1 transcript was induced 10-fold after 4 h of RSV infection in WT BMDMs. Conversely no induction of Sp1 was observed in IKKϵ−/− BMDMs after RSV infection (Fig. 4A, right panel). Similarly, no Sp1 was inducible by RSV infection in NFκB2−/− BMDMs (Fig. 4B). As shown in Fig. 4C, left panel, IL-15 is induced by RSV in WT BMDMs. However, this induction of IL-15 is completely abrogated in RSV-infected IKKϵ−/− BMDMs (Fig. 4C, right panel). Similarly, RSV-induced IL-15 is abrogated in NFκB2−/− BMDMs (Fig. 4D, right panel). To determine the specificity of the effect of NFκB2 and IKKϵ on RSV-inducible IL-15, we measured levels of IFNβ and IL-12p40, two further RSV-inducible genes, and found that neither NFκB2 or IKKϵ deficiency reduced the levels of these cytokines in response to RSV infection (Fig. 4, E–H). These results, therefore, indicate that, similar to P(I:C), RSV infection will trigger a pathway involving NFκB2 and activated by IKKϵ, leading to up-regulation of Sp1 and induction of IL-15, which could be critical for antiviral immunity.
DISCUSSION
In the NFκB field, the majority of studies concerned with infection and inflammation have centered on the canonical NFκB pathway, whereas NFκB2 is better known for its functions in lymphoid organogenesis and humoral immunity (20).
In this study, we present a novel inducer and activator of NFκB2 in the form of Poly(I:C) that acts via TLR3. A few studies have identified an indirect role for NFκB2 in host defense. With respect to viral immunity, RSV infection has been shown to induce the release of p65 from p100/p65 complexes (21). However, the elucidation of genes potentially regulated by NFκB2 in the host response to infection has been unexplored.
Sp1 was identified as a target gene for p52 in response to poly(I:C) through ChIP analysis. Sp1 is a transcription factor first identified on the basis of its ability to interact with the GC-rich motif of simian virus 40 regulatory sequences (22). Sp1 plays a critical role in many diverse cellular events, such as cell growth (23), differentiation (24), apoptosis (23), angiogenesis (25), and viral latency (26), by regulating the expression of other genes. Sp1 was once thought to serve mainly as a constitutive activator of housekeeping genes. However, growing evidence indicates that various posttranslational modifications can influence the transcriptional activity and stability of Sp1, making it a transcription factor responsive to extracellular signals.
Sp1 associates physically and cooperates functionally with several cellular transcriptional activators and also with several viral regulatory proteins, including the HIV-1 regulatory protein Tat (27) and the immediate early gene products of human cytomegalovirus (28). These associations determine the level of Sp1-mediated, viral, or host gene transcription (29, 30). Furthermore, Sp1-regulated elements are found in the promoters of various viruses such as HIV-1 (26), SV40 (22), HSV-1 (31), HCMV (32), and EBV (33). The fact that recognition elements of Sp1 are frequently found in the promoters of various viral genes and, furthermore, that viral regulatory proteins associate with Sp1 to affect its transactivating potential implies that viruses have hijacked the host response to infection in the form of increased Sp1 availability to their own advantage. This indicates that Sp1 induction upon viral infection is probably a common event during host defense. However, although there is an abundance of circumstantial evidence indicating a role for Sp1 in host defense against viral infection, only one study has demonstrated a functional role for Sp1 in the antiviral response in the skin to Vaccinia virus through its regulation of OAS2 expression (34). Therefore, we considered Sp1 a valid target for further investigation.
The identification of sequences to which NFκB dimers bind and effect gene expression in a dimer-specific manner is under intense investigation. Active NFκB dimers bind to specific DNA sites in the promoters of target genes that are collectively known as κB sites (35). The classical κB sites follow the 5′-GGGRNWYYCC-3′ consensus (r = purine, Y = pyrimidine, W = A or T, and n = any nucleotide). However, it has been reported that the p52/RelB heterodimer binds to and activates a unique class of κB site with the consensus 5′-RGGAGAYTTR-3′ (r = A or G and Y = C or T). This consensus sequence is present in the promoters of chemokines involved in lymphoid development and the maintenance of the splenic architecture (36). With increasing numbers of NFκB-regulated DNA target sites being discovered, the diversity of these κB-sequences is becoming more apparent, with even the stringency of the GGG and CC core sequences called into question (37). Our study has identified a previously undescribed κB site, 5′-GGCCGTTGTT-3′, targeted by p52/p65 in the promoter of the Sp1 gene in the region between −474 and −443 nt from the translational start site.
Functionally, we have demonstrated that Sp1 mRNA and protein levels increase upon P(I:C) treatment and that this increase is not observed in cells lacking either IKKϵ, p65, or NFκB2. We chose IL-15 as a possible candidate when considering which target genes might be regulated by Sp1. The role of IL-15 in host defense against viral infections is well documented, and it is known to be induced in response to numerous viruses, including RSV (16, 38). IL-15 is a potently proinflammatory cytokine with a diverse range of immunoregulatory functions (39). The IL-15 gene promoter is also predicted to have two Sp1 sites (40). We confirmed that IL-15 gene expression requires the presence of active Sp1 in PBMCs in response to P(I:C) and that its induction requires both IKKϵ and NFκB2.
Finally, we examined the role of IKKϵ and NFκB2 in a viral infection model. RSV is a major human respiratory pathogen and the leading cause of lower respiratory tract infection in infants worldwide (19). RSV is a single-stranded RNA virus. However, it makes dsRNA during its replication cycle, and it has been reported that TLR3 mediates inflammatory cytokine and chemokine production in response to RSV infection (19, 41).
Knocking out IKKϵ alone completely abrogates any Sp1 or IL-15 gene expression in response to RSV. It is known that IKKϵ phosphorylates both IRF3 and p65 in response to RSV to increase the transactivation potential of these transcription factors (4, 42). Because the IL-15 promoter has also been shown to have a virus-inducible region encompassing an interferon regulatory factor element and a consensus NFκB motif (43), the lack of IL-15 gene expression in response to RSV in IKKϵ-deficient BMDMs could conceivably be due to the insufficient transactivation of p65 and IRF3. However, we believe this is unlikely because other kinases can act in place of IKKϵ in this role, most notably TBK1 and IKKβ (8, 12). In addition, we observed that BMDMs that lack NFκB2 also fail to up-regulate IL-15 gene expression in response to either P(I:C) or RSV, suggesting that IKKϵ acts upstream of NFκB2 in our system. The mechanism of P(I:C)- and RSV-induced IL-15 expression is likely due to the ligation of TLR3, which both activates IKKϵ and induces p52. p52 then binds the promoter of Sp1 with p65, inducing its expression, and Sp1 then binds the promoter of IL-15, up-regulating its expression. We were unable to test RSV in vivo in NFκB2−/− mice because they are severely immunocompromised because of defective lymphoid organogenesis (20, 44).
The number of genes regulated by IKKϵ in a non-redundant manner are very few (3), so it is of interest that we report two new genes to add to this list, i.e. Sp1 and IL-15. Similarly, the number of genes known to be regulated by NFκB2 is small in number and relate only to lymphoid organogenesis, humoral immunity, and DNA damage (36, 45, 46). Considering the abundance of functions of IL-15 (41), we therefore present a role for NFκB2 as a key regulator of antiviral immunity. Furthermore, TLR3 signaling is activated by viral, bacterial, and parasite-derived dsRNA or by host-derived mRNA (47). Therefore, it is conceivable that NFκB2 alone or in conjunction with Sp1 will be found to play a role in host defense against a broader range of infectious agents and also in autoimmunity.
Acknowledgments
We thank Meghan E. Pennini for help in isolating BMDMs from the IKKϵ-deficient mice and Adrian Bracken for help in designing the ChIP assays.
This work was supported, in whole or in part, by National Institutes of Health Grant AI18797 (to S. N. V.). This work was also supported by Science Foundation Ireland Immunology Research Cluster Grant S.F.I. 07/SRC/B1144, by European Union FP7 INFLA-CARE Collaborative Research Program Contract 223151 (to J. H. C.), by Biotechnology and Biological Sciences Research Council Grant BB/D018234/1 (to J. H. C.), and by the School of Immunity and Infection of the College of Medicine of the University of Birmingham, United Kingdom.
- IKKα
- IκB kinase
- RSV
- respiratory syncytial virus
- MEF
- mouse embryonic fibroblast
- nt
- nucleotide(s)
- P(I:C)
- poly(I:C)
- BMDM
- bone marrow-derived macrophage
- TRIF
- TIR-domain containing adapter-inducing interferon β.
REFERENCES
- 1. Vallabhapurapu S., Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 2. Clément J. F., Meloche S., Servant M. J. (2008) The IKK-related kinases. From innate immunity to oncogenesis. Cell Res. 18, 889–899 [DOI] [PubMed] [Google Scholar]
- 3. Tenoever B. R., Ng S. L., Chua M. A., McWhirter S. M., García-Sastre A., Maniatis T. (2007) Multiple functions of the IKK-related kinase IKKϵ in interferon-mediated antiviral immunity. Science 315, 1274–1278 [DOI] [PubMed] [Google Scholar]
- 4. Bao X., Indukuri H., Liu T., Liao S. L., Tian B., Brasier A. R., Garofalo R. P., Casola A. (2010) IKKϵ modulates RSV-induced NF-κB-dependent gene transcription. Virology 408, 224–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wietek C., Cleaver C. S., Ludbrook V., Wilde J., White J., Bell D. J., Lee M., Dickson M., Ray K. P., O'Neill L. A. (2006) IκB kinase ϵ interacts with p52 and promotes transactivation via p65. J. Biol. Chem. 281, 34973–34981 [DOI] [PubMed] [Google Scholar]
- 6. Gupta C. K., Leszczynski J., Gupta R. K., Siber G. R. (1996) Stabilization of respiratory syncytial virus (RSV) against thermal inactivation and freeze-thaw cycles for development and control of RSV vaccines and immune globulin. Vaccine 14, 1417–1420 [DOI] [PubMed] [Google Scholar]
- 7. Wang L., Smith D., Bot S., Dellamary L., Bloom A., Bot A. (2002) Noncoding RNA danger motifs bridge innate and adaptive immunity and are potent adjuvants for vaccination. J. Clin. Invest. 110, 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buss H., Dörrie A., Schmitz M. L., Hoffmann E., Resch K., Kracht M. (2004) Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases including IκB kinase (IKK)-α, IKKβ, IKKϵ, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279, 55633–55643 [DOI] [PubMed] [Google Scholar]
- 9. Doyle S. L., Jefferies C. A., Feighery C., O'Neill L. A. (2007) Signaling by Toll-like receptors 8 and 9 requires Bruton's tyrosine kinase. J. Biol. Chem. 282, 36953–36960 [DOI] [PubMed] [Google Scholar]
- 10. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) IKKϵ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 11. Yang F., Tang E., Guan K., Wang C. Y. (2003) IKK β plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J. Immunol. 170, 5630–5635 [DOI] [PubMed] [Google Scholar]
- 12. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 13. Gauzzi M. C., Del Cornò M., Gessani S. (2010) Dissecting TLR3 signalling in dendritic cells. Immunobiology 215, 713–723 [DOI] [PubMed] [Google Scholar]
- 14. Su X., Li S., Meng M., Qian W., Xie W., Chen D., Zhai Z., Shu H. B. (2006) TNF receptor-associated factor-1 (TRAF1) negatively regulates Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF)-mediated signaling. Eur. J. Immunol. 36, 199–206 [DOI] [PubMed] [Google Scholar]
- 15.Deleted in proof
- 16. Ennaciri J., Ahmad R., Menezes J. (2007) Interaction of monocytic cells with respiratory syncytial virus results in activation of NF-κB and PKC-α/β leading to up-regulation of IL-15 gene expression. J. Leukocyte Biol. 81, 625–631 [DOI] [PubMed] [Google Scholar]
- 17. Mattei F., Schiavoni G., Belardelli F., Tough D. F. (2001) IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J. Immunol. 167, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 18. Liu P., Jamaluddin M., Li K., Garofalo R. P., Casola A., Brasier A. R. (2007) Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 81, 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rudd B. D., Burstein E., Duckett C. S., Li X., Lukacs N. W. (2005) Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J. Virol. 79, 3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caamaño J. H., Rizzo C. A., Durham S. K., Barton D. S., Raventós-Suárez C., Snapper C. M., Bravo R. (1998) Nuclear factor (NF)-κ B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J. Exp. Med. 187, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu P., Li K., Garofalo R. P., Brasier A. R. (2008) Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-κ B2 complexes via a novel retinoic acid-inducible gene-I·NF-κB-inducing kinase signaling pathway. J. Biol. Chem. 283, 23169–23178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dynan W. S., Tjian R. (1983) The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35, 79–87 [DOI] [PubMed] [Google Scholar]
- 23. Kaczynski J., Cook T., Urrutia R. (2003) Sp1- and Krüppel-like transcription factors. Genome Biol. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Opitz O. G., Rustgi A. K. (2000) Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 60, 2825–2830 [PubMed] [Google Scholar]
- 25. Mazure N. M., Brahimi-Horn M. C., Pouysségur J. (2003) Protein kinases and the hypoxia-inducible factor-1, two switches in angiogenesis. Curr. Pharm. Des. 9, 531–541 [DOI] [PubMed] [Google Scholar]
- 26. Jones K. A., Kadonaga J. T., Luciw P. A., Tjian R. (1986) Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 232, 755–759 [DOI] [PubMed] [Google Scholar]
- 27. Loregian A., Bortolozzo K., Boso S., Sapino B., Betti M., Biasolo M. A., Caputo A., Palú G. (2003) The Sp1 transcription factor does not directly interact with the HIV-1 Tat protein. J. Cell. Physiol. 196, 251–257 [DOI] [PubMed] [Google Scholar]
- 28. Xu J., Ye L. (2002) Human cytomegalovirus IE2 protein interacts with transcription activating factors. Sci. China C. Life Sci. 45, 604–612 [DOI] [PubMed] [Google Scholar]
- 29. Rossi A., Mukerjee R., Ferrante P., Khalili K., Amini S., Sawaya B. E. (2006) Human immunodeficiency virus type 1 Tat prevents dephosphorylation of Sp1 by TCF-4 in astrocytes. J. Gen. Virol. 87, 1613–1623 [DOI] [PubMed] [Google Scholar]
- 30. Yurochko A. D., Mayo M. W., Poma E. E., Baldwin A. S., Jr., Huang E. S. (1997) Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates up-regulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 71, 4638–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones K. A., Tjian R. (1985) Sp1 binds to promoter sequences and activates herpes simplex virus “immediate-early” gene transcription in vitro. Nature 317, 179–182 [DOI] [PubMed] [Google Scholar]
- 32. Walker S., Hagemeier C., Sissons J. G., Sinclair J. H. (1992) A 10-base-pair element of the human immunodeficiency virus type 1 long terminal repeat (LTR) is an absolute requirement for transactivation by the human cytomegalovirus 72-kilodalton IE1 protein but can be compensated for by other LTR regions in transactivation by the 80-kilodalton IE2 protein. J. Virol. 66, 1543–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Howe J. G., Shu M. D. (1989) Epstein-Barr virus small RNA (EBER) genes. Unique transcription units that combine RNA polymerase II and III promoter elements. Cell 57, 825–834 [DOI] [PubMed] [Google Scholar]
- 34. Bin L., Howell M. D., Kim B. E., Streib J. E., Hall C. F., Leung D. Y. (2011) Specificity protein 1 is pivotal in the skin's antiviral response. J. Allergy Clin. Immunol. 127, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen F. E., Huang D. B., Chen Y. Q., Ghosh G. (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391, 410–413 [DOI] [PubMed] [Google Scholar]
- 36. Bonizzi G., Bebien M., Otero D. C., Johnson-Vroom K. E., Cao Y., Vu D., Jegga A. G., Aronow B. J., Ghosh G., Rickert R. C., Karin M. (2004) Activation of IKKα target genes depends on recognition of specific κB binding sites by RelB:p52 dimers. EMBO J. 23, 4202–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fusco A. J., Huang D. B., Miller D., Wang V. Y., Vu D., Ghosh G. (2009) NF-κB p52:RelB heterodimer recognizes two classes of κB sites with two distinct modes. EMBO Rep 10, 152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fawaz L. M., Sharif-Askari E., Menezes J. (1999) Up-regulation of NK cytotoxic activity via IL-15 induction by different viruses. A comparative study. J. Immunol. 163, 4473–4480 [PubMed] [Google Scholar]
- 39. Carroll H. P., Paunovic V., Gadina M. (2008) Signalling, inflammation and arthritis. Crossed signals. The role of interleukin-15 and -18 in autoimmunity. Rheumatology 47, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 40. Washizu J., Nishimura H., Nakamura N., Nimura Y., Yoshikai Y. (1998) The NF-κB binding site is essential for transcriptional activation of the IL-15 gene. Immunogenetics 48, 1–7 [DOI] [PubMed] [Google Scholar]
- 41. Huang S., Wei W., Yun Y. (2009) Upregulation of TLR7 and TLR3 gene expression in the lung of respiratory syncytial virus infected mice. Wei Sheng Wu Xue Bao 49, 239–245 [PubMed] [Google Scholar]
- 42. Indukuri H., Castro S. M., Liao S. M., Feeney L. A., Dorsch M., Coyle A. J., Garofalo R. P., Brasier A. R., Casola A. (2006) IKKϵ regulates viral-induced interferon regulatory factor-3 activation via a redox-sensitive pathway. Virology 353, 155–165 [DOI] [PubMed] [Google Scholar]
- 43. Azimi N., Shiramizu K. M., Tagaya Y., Mariner J., Waldmann T. A. (2000) Viral activation of interleukin-15 (IL-15). Characterization of a virus-inducible element in the IL-15 promoter region. J. Virol. 74, 7338–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franzoso G., Carlson L., Poljak L., Shores E. W., Epstein S., Leonardi A., Grinberg A., Tran T., Scharton-Kersten T., Anver M., Love P., Brown K., Siebenlist U. (1998) Mice deficient in nuclear factor (NF)-κ B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J. Exp. Med. 187, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schneider G., Saur D., Siveke J. T., Fritsch R., Greten F. R., Schmid R. M. (2006) IKKα controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 25, 3801–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schumm K., Rocha S., Caamano J., Perkins N. D. (2006) Regulation of p53 tumour suppressor target gene expression by the p52 NF-κB subunit. EMBO J. 25, 4820–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. (2004) mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 279, 12542–12550 [DOI] [PubMed] [Google Scholar]