Background: The antibody 6C2 exhibited an unusually potent neutralizing ability against ricin.
Results: We determined the crystal structure of 6C2 Fab in complex with RTA and mapped the epitope on RTA.
Conclusion: The binding of 6C2 hinders the interaction between RTA and the ribosome, thus inhibiting the activities of RTA.
Significance: Our findings further confirm the role of ribosomal elements in ricin activity and specificity.
Keywords: Antibodies, Ribosomal RNA Processing, Ribosomes, RIP, Structural Biology
Abstract
Ricin belongs to the type II ribosome-inactivating proteins that depurinate the universally conserved α-sarcin loop of rRNA. The RNA N-glycosidase activity of ricin also largely depends on the ribosomal proteins that play an important role during the process of rRNA depurination. Therefore, the study of the interaction between ricin and the ribosomal elements will be better to understand the catalysis mechanism of ricin. The antibody 6C2 is a mouse monoclonal antibody exhibiting unusually potent neutralizing ability against ricin, but the neutralization mechanism remains unknown. Here, we report the 2.8 Å crystal structure of 6C2 Fab in complex with the A-chain of ricin (RTA), which reveals an extensive antigen-antibody interface that contains both hydrogen bonds and van der Waals contacts. The complementarity-determining region loops H1, H2, H3, and L3 form a pocket to accommodate the epitope on the RTA (residues Asp96–Thr116). ELISA results show that Gln98, Glu99, Glu102, and Thr105 (RTA) are the key residues that play an important role in recognizing 6C2. With the perturbation of the 6C2 Fab-RTA interface, 6C2 loses its neutralization ability, measured based on the inhibition of protein synthesis in a cell-free system. Finally, we propose that the neutralization mechanism of 6C2 against ricin is that the binding of 6C2 hinders the interaction between RTA and the ribosome and the surface plasmon resonance and pulldown results confirm our hypothesis. In short, our data explain the neutralization mechanism of mAb 6C2 against ricin and provide a structural basis for the development of improved antibody drugs with better specificity and higher affinity.
Introduction
Ribosome-inactivating proteins (RIPs),4 mostly from plants, are N-glycosidases (E.C. 3.2.2.22) that damage ribosomes irreversibly by removing one or more adenine residues from the rRNA (1). Damaged ribosomes are rendered unable to bind elongation factor 2, thus losing the ability to synthesize proteins (2). RIPs are usually divided into two classes (3). Type I RIPs are monomers, strongly basic proteins of ∼30 kDa with enzymatic activity. Type II RIPs are heterodimers with an approximate molecular mass of 60 kDa, in which one polypeptide of ∼30 kDa with enzymatic activity (A-chain) is linked by a disulfide bridge to another polypeptide of ∼35 kDa with lectin properties (B-chain).
Some type 2 RIPs are potent toxins, the best known of these is ricin, which is a potential biological weapon. Surprisingly, ricin also can be harnessed for biomedicine. Scientists have used a portion of the ricin toxin to develop a new class of therapeutics called “immunotoxins.” Over the past few decades, immunotoxins have been taken from discovery to clinical trials in patients with a variety of cancers. From another perspective, ricin is a good prototype to investigate the N-glycosidase mechanism of RIPs because of its powerful catalytic ability for mammalian ribosomes. Endo and colleagues (4) discovered that the ricin A-chain (RTA) catalyzes the depurination of an invariant adenosine A4324, which is located within a GAGA sequence in a universally conserved sarcin-ricin loop of eukaryotic 28S rRNA. The x-ray crystallographic structure of the heterodimer plant toxin ricin has been determined at 2.8 Å resolution by Montfort and colleagues (39), revealing that RTA has a prominent cleft that recognizes the target rRNA stem loop. Site-directed mutagenesis has identified five residues (Tyr80, Tyr123, Glu177, Arg180, and Trp211) within or near the active site cleft that are critical for the enzymatic activity of RTA (5, 6). Recently, the structure of ricin was solved with transition state analogues bound, confirming that these invariant residues of the RIPs in the catalytic active site of ricin were essential for efficient catalysis by RTA (7). Although the biochemical properties of RIPs have been extensively studied, the enzymatic mechanism of RIPs remains elusive.
Exploiting the extraordinary capacity of ricin to provoke an immune response, Erhlich (40) and others (8) were the first to demonstrate the potential of Abs to completely inactivate ricin in the late 1880s. Since those early studies, dozens of polyclonal antibodies derived from different animal species (e.g. mouse and rabbit) have been tested on a diversity of cell types (e.g. human, non-human primate, and mouse) and animal models (e.g. mice, rats, and rabbits) (9–19). These results have confirmed that Abs directed against the holotoxin are generally sufficient to neutralize ricin in vitro and to confer passive immunity in vivo.
To better protect against ricin, we have developed a potent neutralizing antibody called 6C2 that has more effective therapeutic immunotoxins (19). The antibody 6C2 is obtained from individual mice immunized with RTA. Our results indicate that 6C2 can efficiently inhibit the enzymatic activity of ricin through the cell-free translation assay in vitro and the ability of 6C2 to protect against the cytotoxic effect of ricin in vivo is also substantial. One of the most striking findings is that the epitope is distant from the previously reported catalytic active site of ricin, but 6C2 exhibits more marked neutralizing ability than the Ab binding to the enzymatic active site of RTA.
To map the epitope accurately on RTA and to elucidate the neutralization mechanism of 6C2 against ricin, we determined the crystal structure of 6C2 Fab-RTA. The structure presents the first detailed atomic level description of the RTA complex with a monoclonal antibody and reveals that the complementarity-determining region (CDR) loops H1, H2, H3, and L3 form a pocket to accommodate the epitope on the RTA (residues Asp96–Thr116). We further confirm the residues on the epitope that play an important role in interacting with 6C2 by ELISA. The inhibition of protein synthesis experiment also indicates that perturbation of the 6C2 Fab-RTA interface will eliminate the protective efficacy of 6C2. Combining the surface plasmon resonance (SPR) and pulldown results, we propose that the binding of 6C2 hinders the interaction between RTA and the ribosome, thus inhibiting the activities of RTA.
EXPERIMENTAL PROCEDURES
Construction, Expression, and Purification of Recombinant RTA
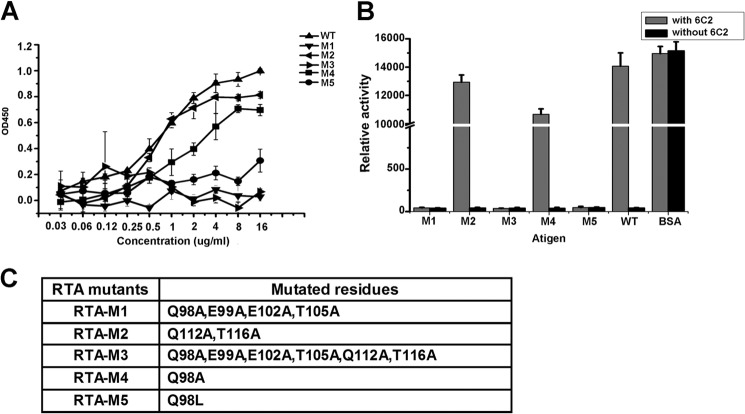
The gene encoding RTA was chemically synthesized by Sangon (Shanghai, China) and then subcloned into expression vector pGEX-4T-2. The recombinant RTA was expressed in Escherichia coli BL21(DE3) under induction by isopropyl-β-d-thiogalactopyranoside. The expressed product was purified using a B-PER GST fusion protein purification kit (Pierce, Rockford, IL). Then, the GST fusion sequence was digested by enterokinase (Sino Biological, Inc., Beijing, China). The digested proteins were dialyzed against 10 mm Tris-HCl, pH 7.5, 200 mm NaCl, and then applied to a Superdex-75 column (2.6 × 100 cm). The mutant RTA-M1 represented the mutations of residues Gln98, Glu99, Glu102, and Thr105 to alanine; the mutant RTA-M2 represented the mutations of residues Gln112 and Thr116 to alanine; the mutant RTA-M3 represented the mutations of residues Gln98, Glu99, Glu102, Thr105, Gln112, and Thr116 to alanine; the mutant RTA-M4 represented the mutation of residue Gln98 to alanine and the mutant RTA-M5 represented the mutation of residue Gln98 to leucine (see Fig. 2C). All of these RTA mutations were produced by the MutanBEST kit (TaKaRa) and purified in the same way as that of the wild-type proteins.
FIGURE 2.
Validation of the RTA-6C2 Fab interface. A, binding ability of 6C2 with RTA and RTA mutants are measured by ELISA. B, inhibition of RTA and RTA mutants enzymatic activities by 6C2. After RTA or RTA mutants were preincubated with (or without) 6C2 antibody, luciferase template mRNA was added to the rabbit reticulocyte lysates containing intact ribosomes. The relative luciferase activity, which was linear with the concentration of luciferase, was measured by GloMaxTM 96 Microplate Luminometer. C, table showing the RTA mutants.
Preparation of 6C2 Fab Fragment
The mAb 6C2 IgG was produced according to our previous method (14, 20). The 6C2 IgG was proteolysed for 4 h at 37 °C with immobilized papain (Roche Applied Science) in buffer A (0.05 m Bis-Tris, pH 6.3). After this reaction was stopped with E-64, the proteolysed 6C2 was dialyzed overnight at 4 °C in buffer B (0.025 m sodium acetate, pH 5.0), and the Fab fragments were separated from the Fc fragments by protein A affinity chromatography. Further purification was achieved through cationic exchange chromatography and gel filtration on a Superdex 200 column (GE Healthcare).
Crystallization and Diffraction Data Collection
The purified 6C2 Fab and the RTA were mixed in a molar ratio of 1:1 at 4 °C for 6 h. The mixed protein was purified with HiLoad 16/60 Superdex 200 (GE Healthcare). The purified protein was dialyzed into buffer A (50 mm Tris-HCl, pH 7.5, 80 mm NaCl) and concentrated to 20–30 mg/ml. Co-crystallization was performed using the hanging drop vapor diffusion method by mixing equal volumes of the RTA-6C2 Fab mixture solution and a reservoir solution at 16 °C. After 5–7 days, crystals in 0.1 m sodium citrate, pH 5.0, and 8% PEG 8000 grew to a size suitable for x-ray diffraction. The crystals were then soaked in a cryoprotection solution (0.1 m sodium citrate, pH 5.0, 8% PEG 8000, 25% glycerol) prior to data collection. Unfortunately, these crystals diffracted x-rays only weakly. We attempted dehydration to help improve the diffraction. Finally, x-ray diffraction data of the crystal were collected on beamline 17U1 of the Shanghai Synchrotron Radiation Facility. The data were processed and scaled with HKL2000 (21) and programs in the CCP4 package (22). The statistics for the diffraction data are summarized in Table 1.
TABLE 1.
Statistics of x-ray diffraction data collection and structure refinement
Values in parentheses are for last resolution shell.
| Data collection | |
| Space group | I222 |
| Unit cell parameters (Å) | a = 108.52, b = 151.80, c = 208.42 |
| Wavelength (Å) | 0.9792 |
| Resolution limits (Å ) | 50.00–2.80 (2.85–2.80) |
| No. of unique reflections | 40,594 (1188) |
| Completeness (%) | 93.6 (55.6) |
| Redundancy | 7.1 (4.9) |
| Rmerge (%)a | 9.4 (34.5) |
| Mean I/σ(I) | 19.0 (3.2) |
| Refinement statistics | |
| Resolution limits (Å ) | 50.00–2.80 |
| Rworkb/Rfreec (%) | 23.84/28.89 |
| r.m.s.d. for bonds (Å) | 0.009 |
| r.m.s.d. for angles | 1.217° |
| Mean B factor (Å2) | 49.98 |
| No. of non-hydrogen protein atoms | 8287 |
| Ramachandran plot (%) | |
| Most favored regions | 87.4 |
| Additional allowed regions | 11.9 |
| Generously allowed regions | 0.7 |
a Rmerge = ΣhΣl|Ihl − 〈Ih〉|/ΣhΣl 〈Ih〉, where Ihl is the lth observation of reflection h and 〈Ih〉 is the weighted average intensity for all observations l of reflection h.
b Rwork factor = Σh|Fobs(h)| − |Fcal(h) ‖/Σh|Fobs(h)|, where Fobs(h) and Fcal(h) are the observed and calculated structure factors for reflection h, respectively.
c Rfree factor was calculated similar to the Rwork factor using 5% of the reflections, which were selected randomly and omitted from refinement.
Structure Determination and Refinement
The structure of the 6C2 Fab in complex with RTA was solved using the molecular replacement method in Molrep (23). The structures of the ricin A-chain (Protein Data Bank code 3HIO) and Fab antibody (Protein Data Bank code 3EOA) were used as the search models. The initial model was refined by using the maximum likelihood method implemented in Refmac5 (24) as part of the CCP4i program suite and refined interactively using the σA-weighted electron density maps with the coefficients 2Fo − Fc and Fo − Fc in the program COOT (25). During the latter stage, the restrained positional and B-factor refinement was performed using the program PHENIX.refine (26) during the refinement. The final models were evaluated with the programs MOLPROBITY (27) and PROCHECK (28). The final coordinates and structure factors were refinement converged to an R-factor of 23.84% and R-free of 28.89% at a resolution of 2.8 Å. The final coordinates and structure factors were deposited in the Protein Data Bank under the accession code of 4KUC. The data collection and structure refinement statistics are listed in Table 1. All structure figures were created using the program PyMOL.
ELISA
The microtiter wells were coated with 100 μl of anti-RTA antibody 13G6 (1 μg/ml) in PBS (pH 7.4) overnight at 4 °C, washed three times with PBST (PBS-Tween 20; 0.1%v/v), and blocked with 10% skim milk in PBST for 2 h at 37 °C before the addition of RTA and its mutants in 10 concentration scales (0.031–16.00 μg/ml). The wells were washed six times in PBST, and chimeric 6C2 consisting of mouse antibody variable domain and human antibody conservation domain was added and incubated for 3 h at room temperature. The plates were again washed, and HRP-conjugated goat anti-human IgG antibody was added. The wells were again washed sixtimes, and the colorimetric HRP substrate was added. The absorbance at 450 nm was read 5 min later. In a standard ELISA, each bar is the average of three wells. The results are the means±S.E. for three independent experiments, p < 0.05.
Inhibition of Ricin Enzymatic Activity
The ability of 6C2 to inhibit the enzymatic activity of RTA and its mutants was measured in a cell-free in vitro translation system using rabbit reticulocyte lysates (Promega) as the source of both ribosomes and luciferase mRNA. The protein translation efficiency was determined by measuring the relative activity of luciferase. After preincubating 0.5 μg of RTA or RTA mutants with (or without) 5 μg of 6C2, 0.5 μg of luciferase template mRNA was added to the rabbit reticulocyte lysates containing intact ribosomes and incubated for 90 min. The relative luciferase activity was measured by GloMaxTM 96 microplate luminometer (Promega). BSA, which could not bind to the ribosomes and impact the translation activity of ribosomes, was used as a control.
SPR
The Biacore T200 (GE Healthcare) was used to analyze the change in the interaction between the ribosomes and the RTA in the presence and absence of the antibody 6C2. The GST-RTA was used as the ligand, and the rabbit reticulocyte 80S ribosomes purified from untreated rabbit reticulocyte lysate as described (29) were used as the analyte under conditions both with and without 6C2. The GST-RTA was diluted in the running buffer at 50 nm and injected at a flow rate of 20 μl/min to generate a resonance signal of 150 resonance units. Ribosomes at the concentrations of 25, 16.67, 11.11, 7.41, and 4.94 nm were passed through both the target and the reference surfaces at a flow rate of 30 μl/min for 3 min to monitor the association. The dissociation was monitored at the same flow rate in the running buffer for another 3 min. The signal from the reference surface was subtracted for nonspecific binding. After each ribosome binding, regeneration was executed using a 60-s injection of 10 mm glycine/HCl (pH 2.0). For the antibody-blocking experiments, the 6C2 antibody was passed through at an abundance of 200 resonance units before the ribosomes were used. Then ribosomes at 40, 26.67, 17.78, 11.85, and 7.90 nm were passed through both the target and the reference surfaces as described above. The data were analyzed using the Biacore T200 Evaluation software. The curves were modeled assuming a simple 1:1 interaction to generate the kinetic data.
Pulldown Assays
After preincubation with 0.5 μg of RTA or RTA mutants, rabbit reticulocyte 80S ribosomes were purified from untreated rabbit reticulocyte lysate (Promega). A volume of 100 μl of lysate was layered onto 50 μl of sucrose cushion (1 m sucrose, 20 mm Tris-HCl pH 7.5, 500 mm KCl, 2.5 mm MgCl2, 0.1 mm EDTA, and 0.5 mm DTT) and centrifuged at 100,000 rpm in a TLA 120.2 rotor for 2 h at 4 °C. The glassy pellet was washed twice on ice with ribosome storage buffer (20 mm Hepes-KOH, pH 7.7, 100 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 250 mm sucrose, and 1 mm DTT) and suspended in 10 μl of the same buffer. The solution was analyzed by Western blot using the anti-RTA antibody 13G6 as a recognizing antibody and the HRP-anti-mouse-Fc antibody as a detecting antibody. As controls, 10 μg of extra 6C2 or BSA was added to the untreated rabbit reticulocyte lysate (Promega) when the samples were preincubated.
RESULTS
Overall Structure of the 6C2 Fab-RTA Complex
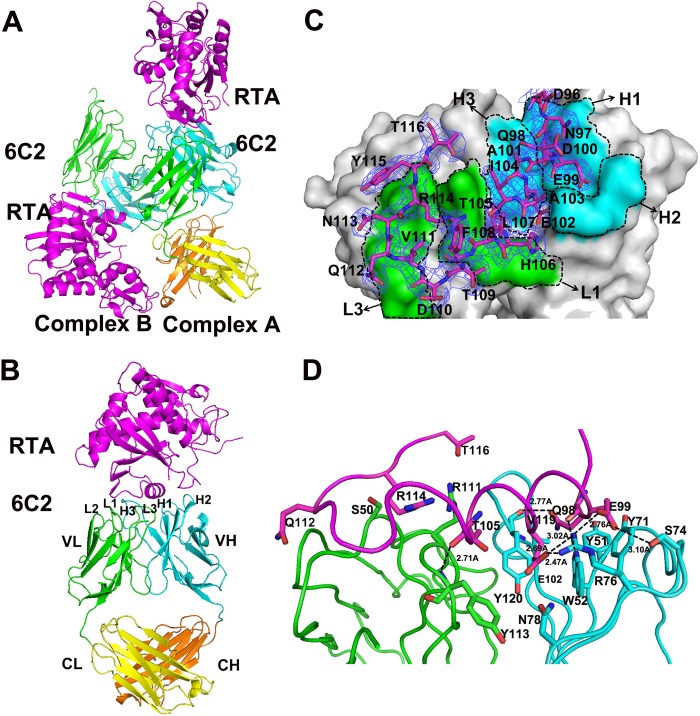
RTA was co-crystallized in complex with the 6C2 Fab fragment. The dataset was severely anisotropic, and the high-resolution diffraction spots were very weak. The structure of the complex was solved using molecular replacement and refined to a resolution of 2.8 Å with an Rfactor of 23.84% and Rfree of 28.89% (Table 1). There are two Fab-RTA complexes (A and B) in the crystallographic asymmetric unit (Fig. 1A). Due to the better electron density, the structure model of complex A will be used for further structural analysis and discussion. The length of the complex is ∼110 Å, and the width of the complex is ∼50 Å. The complex contains one molecular RTA (chain A) and one molecular 6C2 Fab (chain D and H) (Fig. 1B). The RTA contains residues 5–263, adopting a similar globular fold as in a previously described structure (PDB code 2AAI). Due to their flexibility, four residues at the N terminus and four residues at the C terminus are not observed (Table 2). The 6C2 Fab has a canonical immunoglobin fold consisting of 4β-barrel domains, a VL (variable domain) and a CL (constant domain) consisting of residues 21–228 from the light chain (chain D), and a VH (variable domain) and CH (constant domain) consisting of residues 24–237 from the heavy chain (chain H) (Fig. 1B). The heavy chain and light chain are connected by four pairs of disulfide bonds. Due to their flexibility, the N-terminal 20 residues in the VL domain and 23 residues in the VH domain could not be traced, and the loops (residues 154–161, 196–199) in the CH domain are also not observed (Table 2). Note that as the electron density quality of complex B is poor, the CL domain (residues 130–231) in chain E and CH domain (residues 143–237) in chain F could not be observed (Fig. 1A, Table 2, and supplemental Fig. S1). The details of the refinement statistics and model quality of the final model are summarized in Table 1.
FIGURE 1.
Overall structure of the 6C2 Fab-RTA complex. A, crystal structure of 6C2 Fab-RTA complex in an asymmetric unit. B, crystal structure of 6C2 Fab-RTA (complex A). The RTA is colored magenta, the light chain (VL) and the heavy chain (VH) of 6C2 Fab are colored green and cyan, the light chain (CL) and the heavy chain (CH) of 6C2 Fab are colored yellow and orange, respectively, and the CDRs of 6C2 Fab are also labeled. C, surface of the 6C2 Fab-RTA complex and the relative role of each CDR loop in the interaction with the RTA. The residues on RTA are shown as sticks and colored magenta, the light chain (VL) and the heavy chain (VH) of 6C2 Fab are labeled and colored green and cyan, respectively. The 2Fo − Fc electron density map (contoured at 1σ) for the bound epitope peptide is shown as blue. D, binding interface between the 6C2 Fab and the epitope of the RTA. The important residues on the interface are shown as sticks, and hydrogen bonds are indicated by dashed lines. RTA and the CDRs are colored similarly on Fig. 1B.
TABLE 2.
Summary of the missing residues of the RTA-6C2 complex
| Complex A | |
| RTA (chain A) | Residues 1–4, 264–267 |
| Light chain (chain D) | Residues 1–20, 229–231 |
| Heavy chain (chain H) | Residues 1–23, 154–161, 196–199 |
| Complex B | |
| RTA (chain I) | Residues 1–4, 264–267 |
| Light chain (chain E) | Residues 1–20, 130–231(CL domain) |
| Heavy chain (chain F) | Residues 1–23, 143–237(CH domain) |
Overview of the Interface of 6C2 Fab and RTA
The 6C2 Fab-RTA complex structure reveals that the CDRs of the heavy chain (H) and light chain (L) of 6C2 Fab form a pocket to interact with the epitope on the RTA (residues Asp96-Thr116) (Fig. 1, C and D). The interaction interface buries 828 Å2 of 6C2 Fab and 783 Å2 of the surface area of RTA, which represent ∼6.7% of the total accessible surface area (21,234.1 Å2) of 6C2 Fab and 9.7% of total accessible surface area of RTA (11,770.7 Å2), respectively. The value is also within the range for other antibody-antigen interactions. The CDR loops H1, H2, H3, L1, and L3 contribute 61.5 Å2 (7.4%), 240.3 Å2 (29.0%), 113.4 Å2 (13.7%), 139.6 Å2 (16.9%), and 254.7 Å2 (30.8%) of the buried surface area, respectively. The data are very consistent with the structure: as shown in Fig. 1C, the CDR loops H1, H2, H3, and L3 form a pocket to accommodate the epitope on the RTA, and L1 also makes direct contact with RTA. The heavy chain of 6C2 Fab contributes more interface area for RTA binding than the light chain. The L2 loop of the light chain does not protrude from the 6C2 Fab surface and makes no contact with RTA. A limited role of L2 in antigen-antibody interaction is frequently observed in many antigen-antibody complexes (30).
Interactions between 6C2 Fab and RTA
In total, the interactions between 6C2 Fab and RTA consist of seven hydrogen bonds and 106 van der Waals contacts (<4 Å) and can be analyzed in two parts. First, H1, H2, H3 and L3 (6C2 Fab) form a pocket to interact with residues Asp96–His106 (RTA). As shown in Fig. 1D, Gln98 (RTA) is a key residue of RTA, embedding its side chain in a hydrophobic pocket formed by residues Tyr51 (H1), Trp52 (H1), Tyr71 (H2), Tyr120 (H3), and Val119 (H3). Gln98 contributes two hydrogen bonds and 17 van der Waals contacts. The main chain carbonyl of Gln98 forms a hydrogen bond with the side chain atoms NH1 of Wrp52 (H1), and the side chain atoms OE1 form hydrogen bond with the main chain carbonyl of Tyr120 (H3). Glu99 (RTA) forms two hydrogen bonds with the side chain of Arg76 (H2) and Ser74 (H2), respectively. Glu102 is another important residue that embeds its side chain in a cleft formed by residues Trp52 (H1), Arg76 (H2), Asn78 (H2), and Tyr120 (H3). It contributes 20 van der Waals contacts and forms two hydrogen bonds. The side chain atoms OE2 of Glu102 form two hydrogen bonds with the side chain of Arg76 (H2). Near Glu102, the main chain carbonyl of Thr105 forms a hydrogen bond with the main chain amide of Tyr113 (L3) and contributes 15 van der Waals contacts. Second, the interactions between L1 and the β-sheet on the RTA (Val111–Thr116), Gln112, Arg114, and Thr116 contribute another 15 van der Waals contacts (Fig. 1D).
Perturbation of the 6C2 Fab-RTA Interface Affects the Protective Efficacy of 6C2
To further validate the interface and to understand the determinant interaction elements, we generated three mutants, RTA-M1 (Q98A, E99A, E102A, T105A), RTA-M2 (Q112A, T116A), and RTA-M3 (Q98A, E99A, E102A, T105A, Q112A, T116A) (Fig. 2C). The ELISA results showed that RTA-M1 and RTA-M3 fail to bind 6C2, but the binding capacity of RTA-M2 decreased only slightly compared with that of the wild type RTA (Fig. 2A). The data indicated that the interaction between RTA and 6C2 was mainly mediated by the first binding pocket, and Gln98, Glu99, Glu102, and Thr105 (RTA) were the key residues in recognizing 6C2. As discussed above, Gln98 (RTA) embeded its side chain in a hydrophobic pocket formed by residues Tyr51 (H1), Trp52 (H1), Tyr71 (H2), Tyr120 (H3) and Val119 (H3). To further validate the role of residue Gln98 in the interface, we mutated Gln to Ala (RTA-M4) and Leu (RTA-M5) (Fig. 2C). The ELISA results showed that the binding capacity of RTA-M4 decreased moderately, but the binding capacity of RTA-M5 decreased substantially compared with that of the WT-RTA (Fig. 2A), indicating that the interaction was not dependent on the hydrophobic properties of the side chain. This result was consistent with our analysis that the interaction between 6C2 and RTA mainly relied on hydrogen bonds.
Studies have shown that 6C2 exhibits potent neutralizing ability against RTA. To investigate whether perturbation of the 6C2 Fab-RTA interface will affect the protective efficacy of 6C2, we tested the ability of 6C2 to inhibit the enzymatic activity of RTA and its mutants, which was measured as the inhibition of protein synthesis in a cell-free system using rabbit reticulocyte lysates. When the ribosomes and luciferase mRNA were mixed, the ribosome would translate the mRNA to luciferase in the absence of RTA, resulting in a high luciferase signal (Fig. 2B). However, in the presence of RTA (and no 6C2), ribosome was inactivated by RTA and could not translate the mRNA to luciferase, leading to a low luciferase signal (Fig. 2B). The presence of 6C2 antibody would suppress the enzymatic activity of RTA, leading to an active ribosome to translate luciferase and a high luciferase signal (Fig. 2B). However, 6C2 almost lost the neutralizing ability against RTA-M1, RTA-M3, and RTA-M5 (Fig. 2B). The neutralizing ability against RTA-M2 decreased slightly compared with that against the wild type RTA, and the neutralizing ability against RTA-M4 was weaker than that against the RTA-M2 (Fig. 2B). These results were very consistent with the ELISA data, indicating that the specific interaction between 6C2 and RTA was vital for the protective efficacy of 6C2.
Structural Comparison of RTA before and after 6C2 Binding
As mentioned above, we identified the exact epitope on the RTA that was recognized by 6C2 Fab. The epitope mainly contains the residues Asp96–Thr116, which forms the substrate binding pocket together with the residues Gly120–Asn136 and residues Asp201–Ser221 (7). To investigate whether 6C2 binding to RTA will change its structure near the enzyme activity center and eliminate the rRNA depurination activity of RTA, we compared the apo-form RTA (PDB code 2AAI) with the RTA in complex with 6C2 and a substrate analog (PDB codes 4KUC and 3HIO). The structural superposition indicates that there are no obvious changes in the main chain, especially the enzyme activity center (root mean square deviation of 0.397 Å for 232 comparable Cα atoms), but there is a small difference at the C terminus because of the flexibility of three tandem Pro residues (Fig. 3). The changes are mainly focused on the side chains of the residues on the epitope, which participate in the interaction with 6C2 Fab (Fig. 3). However, the side chains of these residues neither participate in recognizing the substrate nor are necessary for the enzymatic activity of ricin (Fig. 2B). In particular, changing these residues does not affect the residues that are important for substrate binding. Therefore, we conclude that the loss of the rRNA depurination activity of RTA does not result from the structural rearrangements caused by 6C2 binding.
FIGURE 3.
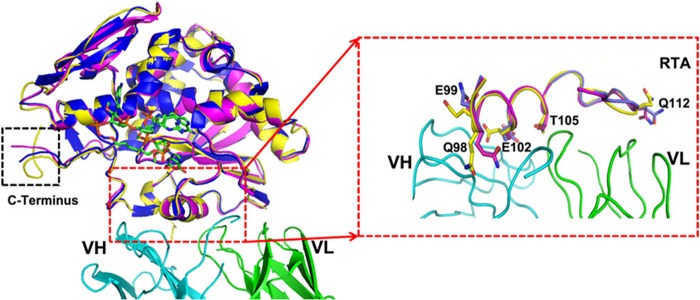
Superposition of RTA in the free form (PDB code 2AAI, yellow), in the complex with 6C2 (PDB code 4KUC, magenta) and in the complex with cyclic G (9-DA) GA2′-OMe (PDB code 3HIO, blue). Cyclic G (9-DA) GA2′-OMe (which contains the tetranucleotide sequence of the GAGA sarcin-ricin loop but with the ricin-susceptible adenosine replaced with 4′-deaza-1′-aza-2′-deoxy-1′-(9-methylene)-immucillin-A (9-DA), a transition state mimic) serves to mimic the substrate and is shown as sticks (green). 6C2 is colored the same as described in the legend to Fig. 1A. The overall structures are almost identical. Changing of the residues on the epitope is highlighted by the red dashed lines.
6C2 Hinders the Interaction between RTA and Ribosome
Although the sarcin-ricin loop has been established as a common substrate for all RIPs, several studies have suggested that ribosomal proteins proximal to the sarcin-ricin loop also played an important role in promoting ribosome depurination. It was observed that although the Km values of RTA for the rat ribosome and naked 28S rRNA were similar, RTA exhibited 105-fold higher catalytic activity (Kcat) on the rat ribosome than on naked 28S rRNA (31). A subsequent study revealed that RTA could interact with the C-terminal region of the ribosomal P stalk proteins (P0, P1, and P2), and yeast ribosome with the P stalk protein mutants were depurinated less than the wild-type ribosomes (32, 33). To examine whether 6C2 interfered with the interaction between RTA and ribosome, and thus inhibited the activities of RTA, we performed an SPR experiment. Briefly, the purified GST-RTA was immobilized onto the surface of a CM5 chip. Then, purified ribosomes were passed over the surface of the chip at a range of concentrations, and the binding kinetics was determined using the Biacore T200 Evaluation software. In another situation, 6C2 antibody was passed over the GST-RTA connecting CM5 chip, obtaining a reaction of ∼200 resonance units before the injections of ribosomes were carried out (Fig. 4, A and B). The results showed an affinity that is ∼500-fold less than that in the direct interaction situation (Fig. 4C), indicating that the binding of 6C2 to RTA substantially influenced the interaction between RTA and the ribosome, at least in terms of kinetics.
FIGURE 4.
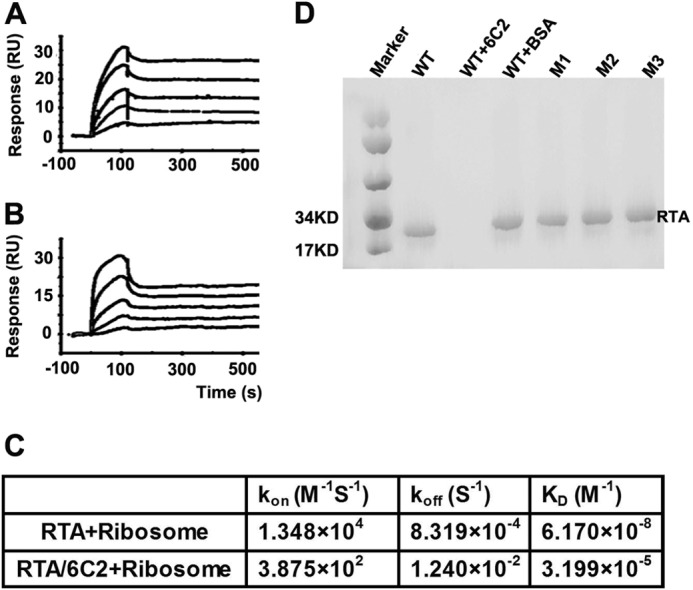
6C2 hinders the interaction between RTA and ribosome. SPR analysis of ribosome binding to RTA in the presence (A) and absence of the antibody 6C2 (B). The signal obtained from the reference surface is subtracted to avoid the binding of non-specificity. C, table showing the binding kinetic parameters. D, pulldown analysis of ribosome binding to RTA in the presence and absence of the antibody 6C2. Mutants disrupting RTA-6C2 binding retain ribosome binding ability. RU, resonance units.
To further confirm the SPR results, we performed pulldown assays. Briefly, the WT-RTA, 6C2 Fab-RTA complex, and WT-RTA+BSA were mixed separately with untreated rabbit reticulocyte lysates (Promega). Then, the rabbit reticulocyte 80S ribosomes were purified, and the solution was analyzed by Western blot. The results showed that WT-RTA and WT-RTA+BSA could bind 80S ribosomes normally, but RTA was not detected after preincubating with 6C2 Fab (Fig. 4D). This result was very consistent with the SPR results, indicating that 6C2 disrupted the interaction between RTA and the ribosome. Next, to determine whether the residues on the interface between RTA and 6C2 were also involved in interacting with the ribosome, we performed pulldown assays to test the interactions of the mutants RTA-M1, RTA-M2, and RTA-M3 with the ribosome. The results showed that these mutants could bind the ribosome similarly to WT-RTA, indicating that these residues may not directly interact with the ribosome (Fig. 4D).
DISCUSSION
To date, 27 crystal structures of RTA have been reported in the PDB database, which were mainly complexes of RTA with small molecule antagonists. The structure of 6C2 Fab-RTA determined and refined at 2.8 Å resolution presented the first detailed atomic level description of RTA in complex with a monoclonal antibody. Based on the structure, we determined the interface between 6C2 Fab and RTA, and we further mapped the residues on RTA that played an important role in interacting with 6C2 Fab by an ELISA experiment. The results of the inhibition of protein synthesis also showed that disruption of the 6C2 Fab-RTA interface eliminated the protective efficacy of 6C2. To elucidate the neutralization mechanism of 6C2 against ricin, we compared the structure of apo-form RTA (PDB code 2AAI) with RTA in the complex with 6C2 and with a substrate analog (PDB codes 4KUC and 3HIO). The structural superposition indicated that the residue conformation changes caused by 6C2 binding did not affect the residues that were vital for substrate binding and catalysis.
Molecular Mechanism of Inhibition of Ricin Enzymatic Activity by 6C2
To our knowledge, the catalytic subunit of ricin was an N-glycosidase that depurinated a universally conserved adenine residue within the sarcin-ricin loop of the 28S rRNA. However, several studies had established the importance of the ribosomal proteins in facilitating the depurination activity and ribosome specificity of ricin (34, 35). Thus, targeting ricin accurately to the ribosome was a prerequisite for the RNA N-glycosidase activity. As discussed above, the complex structure showed that the 6C2 binding site was very close to the substrate binding pocket of RTA, and the length of the complex was ∼110 Å, whereas the width of 6C2 was ∼50 Å. According to the structure of a eukaryotic ribosome that was determined and refined at 2.8 Å resolution (36), we speculated that there was not enough space for the sarcin-ricin loop region of the ribosome to accommodate the RTA-6C2 complex, in contrast to RTA alone. The SPR and pulldown results also confirmed this hypothesis. Based on our data, we proposed that the binding of 6C2 sterically hindered the interaction between RTA and the ribosome, thus inhibiting the activities of RTA. However, a detailed model for the interaction of RTA with the ribosome remained elusive. RTA mutants with defects in binding 6C2 retained the ability to bind the ribosome, indicating that these residues may not directly participate in interacting with the ribosome. The interaction of ricin with the ribosomal elements may represent a promising target for protection against ricin and further experimental approaches were still needed to understand the relationship between the ribosomal elements and the relative activity of ricin.
Significance of the 6C2 Antibody for Further Clinical Drug Development
First, the structure of the 6C2 Fab-RTA complex showed the detailed interface between the antibody and the epitope on the RTA. Based on this valuable structural data, we could develop improved antibodies with much higher binding affinity and specificity against RTA. Second, the epitope on the RTA that 6C2 Fab recognized was different from the recognition epitopes for other antibodies. For instance, murine IgG1 mAb GD12 recognized the residues Leu161–Ile175 on the RTA (37); mAb FGA12 recognized the residues Asp37–Arg48 on the RTA; BD7 recognized the residues Cys259–Phe267 on the RTA and some others recognized the peptides on the RTB (38), so we could use 6C2 in combination with these antibodies to achieve an improved therapeutic effect. Third, the neutralization mechanism of mAb 6C2 against ricin was also different from that of the small molecule antagonists, which depended on blocking the enzyme activity center. Therefore, 6C2 could also be used in combination with the small molecule antagonists.
In short, ricin cytotoxicity is a multistep event. Although a number of antibodies have been reported to bind specifically to the ricin and inhibit the rRNA depurination activity, most of the neutralization mechanism remains unknown. Here, we are the first to reveal the mechanism of mAb 6C2 against ricin through structural biology method, the results indicate that 6C2 binding to RTA greatly affects the interaction between ribosome and RTA via steric hindrance. Our findings also confirm the role of ribosomal elements in ricin activity and specificity.
Acknowledgments
We thank the staff at Shanghai Synchrotron Radiation Facility beamline BL17U for assistance with synchrotron data collection.
This work was supported by Chinese Ministry of Science and Technology Grants 2012CB917200 and 2009CB825500; Chinese National Natural Science Foundation Grants 31270014, 31130018, 31170726, 30900224, and 10979039; and Ministry of Science and Technology 973 Project Grants 2010CB833600 and 2010CB735605.

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 4KUC) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- RIP
- ribosome-inactivating protein
- PDB
- Protein Data Bank
- Ab
- antibody
- CDR
- complementarity-determining region.
REFERENCES
- 1. Stirpe F., Battelli M. G. (2006) Ribosome-inactivating proteins: progress and problems. Cell Mol. Life Sci. 63, 1850–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartley M. R., Lord J. M. (2004) Genetics of ribosome-inactivating proteins. Mini. Rev. Med. Chem. 4, 487–492 [DOI] [PubMed] [Google Scholar]
- 3. Montanaro L., Sperti S., Mattioli A., Testoni G., Stirpe F. (1975) Inhibition by ricin of protein synthesis in vitro. Inhibition of the binding of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 to ribosomes. Biochem. J. 146, 127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Endo Y., Tsurugi K. (1988) The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 263, 8735–8739 [PubMed] [Google Scholar]
- 5. Kim Y., Mlsna D., Monzingo A. F., Ready M. P., Frankel A., Robertus J. D. (1992) Structure of a ricin mutant showing rescue of activity by a noncatalytic residue. Biochemistry 31, 3294–3296 [DOI] [PubMed] [Google Scholar]
- 6. Morris K. N., Wool I. G. (1992) Determination by systematic deletion of the amino acids essential for catalysis by ricin A chain. Proc. Natl. Acad. Sci. U.S.A. 89, 4869–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho M. C., Sturm M. B., Almo S. C., Schramm V. L. (2009) Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 20276–20281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olsnes S. (2004) The history of ricin, abrin and related toxins. Toxicon 44, 361–370 [DOI] [PubMed] [Google Scholar]
- 9. Chanh T. C., Romanowski M. J., Hewetson J. F. (1993) Monoclonal antibody prophylaxis against the in vivo toxicity of ricin in mice. Immunol. Invest. 22, 63–72 [DOI] [PubMed] [Google Scholar]
- 10. Houston L. L. (1982) Protection of mice from ricin poisoning by treatment with antibodies directed against ricin. J. Toxicol. Clin. Toxicol. 19, 385–389 [DOI] [PubMed] [Google Scholar]
- 11. Godal A., Fodstad O., Pihl A. (1983) Antibody formation against the cytotoxic proteins abrin and ricin in humans and mice. Int. J. Cancer 32, 515–521 [DOI] [PubMed] [Google Scholar]
- 12. Hewetson J. F., Rivera V. R., Creasia D. A., Lemley P. V., Rippy M. K., Poli M. A. (1993) Protection of mice from inhaled ricin by vaccination with ricin or by passive treatment with heterologous antibody. Vaccine 11, 743–746 [DOI] [PubMed] [Google Scholar]
- 13. Griffiths G. D., Lindsay C. D., Allenby A. C., Bailey S. C., Scawin J. W., Rice P., Upshall D. G. (1995) Protection against inhalation toxicity of ricin and abrin by immunisation. Hum. Exp. Toxicol. 14, 155–164 [DOI] [PubMed] [Google Scholar]
- 14. Maddaloni M., Cooke C., Wilkinson R., Stout A. V., Eng L., Pincus S. H. (2004) Immunological characteristics associated with the protective efficacy of antibodies to ricin. J. Immunol. 172, 6221–6228 [DOI] [PubMed] [Google Scholar]
- 15. Smallshaw J. E., Richardson J. A., Pincus S., Schindler J., Vitetta E. S. (2005) Preclinical toxicity and efficacy testing of RiVax, a recombinant protein vaccine against ricin. Vaccine 23, 4775–4784 [DOI] [PubMed] [Google Scholar]
- 16. Smallshaw J. E., Richardson J. A., Vitetta E. S. (2007) RiVax, a recombinant ricin subunit vaccine, protects mice against ricin delivered by gavage or aerosol. Vaccine 25, 7459–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smallshaw J. E., Vitetta E. S. (2012) Ricin vaccine development. Curr Top Microbiol. Immunol. 357, 259–272 [DOI] [PubMed] [Google Scholar]
- 18. Yermakova A., Mantis N. J. (2011) Protective immunity to ricin toxin conferred by antibodies against the toxin's binding subunit (RTB). Vaccine 29, 7925–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai J., Zhao L., Yang H., Guo H., Fan K., Wang H., Qian W., Zhang D., Li B., Wang H., Guo Y. (2011) Identification of a novel functional domain of ricin responsible for its potent toxicity. J. Biol. Chem. 286, 12166–12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemley P. V., Amanatides P., Wright D. C. (1994) Identification and characterization of a monoclonal antibody that neutralizes ricin toxicity in vitro and in vivo. Hybridoma 13, 417–421 [DOI] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 23. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) Procheck–a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 29. Sturm M. B., Schramm V. L. (2009) Detecting ricin: sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem 81, 2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson I. A., Stanfield R. L., Rini J. M., Arevalo J. H., Schulze-Gahmen U., Fremont D. H., Stura E. A. (1991) Structural aspects of antibodies and antibody-antigen complexes. Ciba Found. Symp. 159, 13–28; discussion 28–39 [DOI] [PubMed] [Google Scholar]
- 31. Endo Y., Tsurugi K. (1988) The RNA N-glycosidase activity of ricin A-chain. Nucleic Acids Symp. Ser. 139–142 [PubMed] [Google Scholar]
- 32. McCluskey A. J., Poon G. M., Bolewska-Pedyczak E., Srikumar T., Jeram S. M., Raught B., Gariépy J. (2008) The catalytic subunit of shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J. Mol. Biol. 378, 375–386 [DOI] [PubMed] [Google Scholar]
- 33. Chiou J. C., Li X. P., Remacha M., Ballesta J. P., Tumer N. E. (2008) The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol. Microbiol. 70, 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harley S. M., Beevers H. (1982) Ricin inhibition of in vitro protein synthesis by plant ribosomes. Proc. Natl. Acad. Sci. U.S.A. 79, 5935–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor S., Massiah A., Lomonossoff G., Roberts L. M., Lord J. M., Hartley M. (1994) Correlation between the activities of five ribosome-inactivating proteins in depurination of tobacco ribosomes and inhibition of tobacco mosaic virus infection. Plant J 5, 827–835 [DOI] [PubMed] [Google Scholar]
- 36. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 [DOI] [PubMed] [Google Scholar]
- 37. Neal L. M., O'Hara J., Brey R. N., 3rd, Mantis N. J. (2010) A monoclonal immunoglobulin G antibody directed against an immunodominant linear epitope on the ricin A chain confers systemic and mucosal immunity to ricin. Infect Immun 78, 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Hara J. M., Neal L. M., McCarthy E. A., Kasten-Jolly J. A., Brey R. N., 3rd, Mantis N. J. (2010) Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine 28, 7035–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Montfort W., Villafranca J. E., Monzingo A. F., Ernst S. R., Katzin B., Rutenber E., Xuong N. H., Hamlin R., Robertus J. D. (1987) The three-dimensional structure of ricin at 2.8 A. J. Biol. Chem. 262, 5398–5403 [PubMed] [Google Scholar]
- 40. Ehrlich P., et al. (1891) Experimentelle Untersuchungen über Immunität. I. Ueber Ricin. Deut. Med. Wochenschr. 17, 976–979 [Google Scholar]