Background: The regulatory mechanisms that control skeletal muscle bioenergetics are not fully understood.
Results: The muscle-specific protein Perm1 is induced by exercise and exercise-activated regulators and controls the expression of energy homeostasis genes.
Conclusion: Perm1 is a regulator of mitochondrial oxidative capacity and bioenergetics.
Significance: Novel muscle regulators of bioenergetics may facilitate approaches for preserving/enhancing muscle function.
Keywords: Bioenergetics, Energy Metabolism, Nuclear Receptors, Skeletal Muscle, Transcription Regulation, Estrogen-related Receptor, PGC-1α, Mitochondrial Biogenesis
Abstract
Mitochondrial oxidative metabolism and energy transduction pathways are critical for skeletal and cardiac muscle function. The expression of genes important for mitochondrial biogenesis and oxidative metabolism are under the control of members of the peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) family of transcriptional coactivators and the estrogen-related receptor (ERR) subfamily of nuclear receptors. Perturbations in PGC-1 and/or ERR activities have been associated with alterations in capacity for endurance exercise, rates of muscle atrophy, and cardiac function. The mechanism(s) by which PGC-1 and ERR proteins regulate muscle-specific transcriptional programs is not fully understood. We show here that PGC-1α and ERRs induce the expression of a so far uncharacterized muscle-specific protein, PGC-1- and ERR-induced regulator in muscle 1 (Perm1), which regulates the expression of selective PGC-1/ERR target genes. Perm1 is required for the basal as well as PGC-1α-enhanced expression of genes with roles in glucose and lipid metabolism, energy transfer, and contractile function. Silencing of Perm1 in cultured myotubes compromises respiratory capacity and diminishes PGC-1α-induced mitochondrial biogenesis. Our findings support a role for Perm1 acting downstream of PGC-1α and ERRs to regulate muscle-specific pathways important for energy metabolism and contractile function. Elucidating the function of Perm1 may enable novel approaches for the treatment of disorders with compromised skeletal muscle bioenergetics, such as mitochondrial myopathies and age-related/disease-associated muscle atrophies.
Introduction
Cellular energy homeostasis is central to skeletal and cardiac muscle function. Impaired capacity for energy production as seen in patients with mutations in mitochondrial DNA or in nuclear genes encoding lipid oxidation enzymes and respiratory chain components can lead to myopathies and cardiac dysfunction (1–3). An inability to match energy supply to energy needs is also seen in heart failure (4, 5). Thus, a better understanding of the signaling and transcriptional networks that regulate the expression of genes important for mitochondrial biogenesis, fueling, and function may identify novel therapeutic targets for the treatment of diseases associated with muscle bioenergetic defects.
Peroxisome proliferator-activated receptor (PPAR) γ coactivator 1α (PGC-1α)2 and PGC-1β are two members of the PGC-1 family and important regulators of mitochondrial biogenesis and function in cardiac and skeletal muscle (5–8). They are expressed at high levels in heart and skeletal muscle and can induce mitochondrial biogenesis and oxidative capacity when expressed ectopically in myotubes in vitro and muscle tissue in vivo (9–14). Mice lacking PGC-1α or PGC-1β show defects in cardiac and skeletal muscle energetics and decreased exercise capacity (15–20). PGC-1α KO mice in particular develop cardiomyopathy when stressed (16). PGC-1α and PGC-1β act complementarily to each other in maintaining muscle bioenergetics; deletion of both PGC-1s leads to prominent mitochondrial dysfunction accompanied by death after birth (in heart-specific double KOs) or a dramatic loss in exercise capacity (in skeletal muscle-specific KOs) (19, 20). Interestingly, PGC-1α and/or PGC-1β expression in muscle is decreased in states associated with mitochondrial dysfunction, such as diabetes, denervation, cancer cachexia, and amyotrophic lateral sclerosis (ALS), suggesting that decreases in PGC-1 activity may contribute to the pathology of these states (21–24). Supporting this notion, transgenic expression of PGC-1α in skeletal muscle protects from age-related or denervation-induced muscle atrophy and delays the onset of mitochondrial myopathies, whereas transgenic expression of PGC-1β in the heart protects from sepsis-induced cardiomyopathy (23, 25–27). However, transgenic expression of PGC-1α can also promote pathophysiological changes in a tissue-, developmental stage-, and environment-dependent manner, leading to heart failure or skeletal muscle insulin resistance, and underlying the need for a better understanding of the mechanisms by which PGC-1α/β control muscle energy homeostasis pathways (28, 29).
PGC-1α and PGC-1β exert their regulatory functions by interacting with DNA-binding transcription factors, such as estrogen-related receptor α (ERRα), nuclear respiratory factor 1, and GA-binding protein, and activating the expression of genes targeted by these factors (6). ERRα and the related receptors ERRβ and ERRγ are orphan nuclear receptors that act both downstream and parallel to PGC-1 coactivators to control the expression of a broad set of genes important for energy homeostasis, including genes for mitochondrial biogenesis and oxidative function (30, 31). Genome-wide studies show that ERRα and ERRγ bind and regulate the same target genes, suggesting that they act in a redundant and/or complementary fashion (32). Mice lacking ERRα show similarities to mice lacking PGC-1α; they have decreased mitochondrial gene expression, are unable to defend their body temperature when exposed to cold, and develop heart failure when challenged with cardiac pressure overload (33, 34). ERRγ-null mice also show defects in PGC-1-controlled pathways, such as cardiac oxidative capacity, and die shortly after birth (35). Skeletal muscle-specific knock-out of ERRγ and ERRβ leads to decreased capacity for exercise (36). Conversely, mice overexpressing ERRγ in skeletal muscle show increased oxidative capacity and exercise performance (37, 38).
The transcriptional programs induced by PGC-1α/β and ERRs include genes that encode components of cellular energy pathways (e.g. tricarboxylic acid cycle, OxPhos, and mitochondrial biogenesis genes) as well as genes that encode additional gene expression regulators, which act with or downstream of PGC-1/ERRs to amplify and/or specify the induced program. For example, PGC-1α acts with ERRα in myotubes to induce nuclear respiratory factor 1, GA-binding protein α chain, PPARα, and striated muscle activator of Rho signaling, each of which further contributes to the activation of subsets of PGC-1/ERR targets (39–41). Similarly, PGC-1α, ERRα, and ERRγ in heart induce Lpin1, which can specify and amplify the PGC-1α-induced program (42, 43). To identify new regulators that may act in the PGC-1/ERR pathway and enable the execution of muscle energy programs, we searched for uncharacterized genes that are induced by PGC-1α in differentiated myotubes, are expressed selectively in muscle, and are required for the induction of PGC-1/ERR-regulated genes. We show here that PGC-1/ERR-induced regulator in muscle 1 (Perm1) is a muscle-specific gene that is induced by PGC-1α/β and ERRs and required for the expression of a subset of genes important for muscle mitochondrial biogenesis and oxidative capacity.
EXPERIMENTAL PROCEDURES
Identification and Cloning of Perm1
For the identification of PGC-1α-induced genes, C2C12 myotubes were infected on day 6 of differentiation with adenoviruses expressing LacZ (control) or PGC-1α at an m.o.i. of 50 and in triplicates. RNA was isolated 20 h later and profiled using Illumina MouseWG-6 v1.1 Expression Beadchip microarrays (48,318 probes). Raw data were normalized using Illumina software. Significantly differentially regulated genes were determined by one-way analysis of variance. Genes of unknown function that were strongly induced by PGC-1α were next screened in silico for tissue-specific expression patterns using BioGPS (44) and the Attie laboratory genome database (45). Expression patterns for genes of interest were validated by RT-qPCR as described for Fig. 2C. Perm1 (2310042D19Rik) was among the 15 highest induced genes. The mouse Perm1 (2310042D19Rik) and human PERM1 (C1orf170) cDNAs were amplified using RT-PCR with gene-specific primers (5′-TATAGAATTCATGGACAACTTCCAGTACAGCGT-3′ and TATATCTAGACTAGCAGCTGGGGTTTGAGCTG for Perm1 and 5′-TATAAAGCTTATGCCGACCCAGGACGGGCAG-3′ and 5′-TATATCTAGAGCGCCTGTGGTCGTCTC-3′ for PERM1) and RNA isolated from mouse and human skeletal muscles, respectively. The amplified PCR products were cloned into pDrive vector (Qiagen), sequenced, and subcloned into expression vectors.
FIGURE 2.
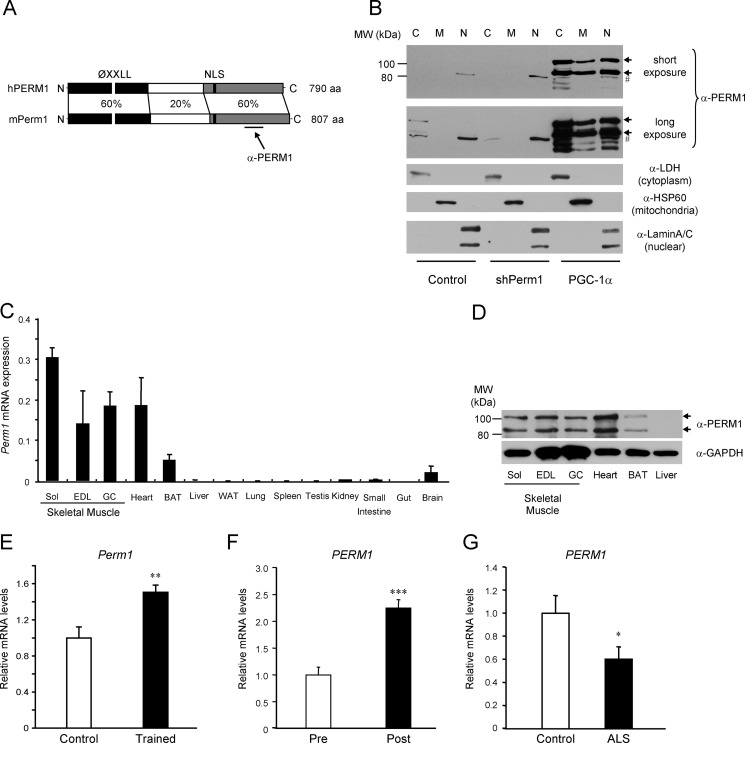
Perm1 encodes a protein found in multiple cellular compartments, is selectively expressed in muscle, and is regulated by physical activity. A, schematic representation of the human (h) and mouse (m) PERM1 proteins, showing the percent identity at the amino acid level and highlighting the conserved motifs (ØXXLL, protein interaction motif; NLS, nuclear localization signal; aa, amino acids) and the region recognized by the anti-PERM1 serum. B, detection of endogenous Perm1 in C2C12 myotubes infected with adenoviruses expressing shGFP (control), shPerm1, or PGC-1α on day 6 of differentiation. Cells were harvested on day 8, cell lysates were subjected to subcellular fractionation, and Perm1 protein was detected using the PERM1 antibody (α-PERM1). Endogenous Perm1 in control cells was detectable only after long exposure (second panel from top). Antibodies against cytoplasmic lactate dehydrogenase (LDH), mitochondrial HSP60, and nuclear lamins (three bottom panels) were used to assess the purity of the cytoplasmic (C), mitochondrial (M), and nuclear (N) fractions. Arrows indicate the two major protein isoforms encoded by Perm1; # indicates a nonspecific protein reacting with the antibody and enriched in the nuclear fraction. C, Perm1 mRNA levels in the indicated tissues of 10-week-old male mice (n = 3) were determined by RT-qPCR and normalized to levels of 36B4 in each tissue (Sol, soleus; EDL, extensor digitorum longus; GC, gastrocnemius). D, Perm1 protein in lysates from skeletal muscles, heart, brown adipose tissue (BAT), and liver of 10-week-old male mice was detected by SDS-PAGE and Western blot using the PERM1 antibody (upper panel); an antibody against GAPDH was used as a loading control (lower panel). E, Perm1 mRNA levels in the quadriceps muscles of 19-week-old female mice that had access to electronically monitored in-cage running wheels for 5 weeks (Trained; n = 7) and control sedentary littermate females (Control; n = 7) were determined by RT-qPCR, normalized to levels of GAPDH, and expressed relative to Perm1 levels in the control sedentary mice. F, PERM1 mRNA levels in muscle biopsies taken from male subjects (n = 7) before (Pre) or 3 h after (Post) an acute cycling bout (60 min at ∼70% of their VO2 peak) were quantified by RT-qPCR as described (41). G, PERM1 mRNA levels in biopsies taken from male control subjects or ALS patients (n = 9) were quantified by RT-qPCR as described (24). Error bars represent S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Generation of Adenoviruses
Adenoviruses expressing LacZ, FLAG-tagged PGC-1α, FLAG-tagged PGC-1β, FLAG-tagged ERRα, FLAG-tagged ERRγ, and short hairpin RNA (shRNA) for ERRα have been described (46, 47). Adenoviruses expressing mouse FLAG-tagged Perm1 and shRNA for Perm1 and GFP were generated as described previously (46) using pAdlox (for FLAG-Perm1) and pAdlox-SUPER (for shPerm1 and shGFP). The shPerm1 sequences are shown in supplemental Table 1. All plasmids were sequenced to verify the sequence of inserts.
Cell Culture and Adenoviral Infections
C2C12 myoblasts were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37 °C with 5% CO2. Confluent myoblasts were differentiated into myotubes by the addition of DMEM with 2% horse serum. For experiments using shRNA, myotubes were infected on day 4 after differentiation with adenoviruses expressing shRNAs (at an m.o.i. of 50) and on day 6 after differentiation with adenoviruses expressing LacZ (control), FLAG-PGC-1α, FLAG-PGC-1β, or FLAG-ERRγ (at an m.o.i. of 50) together with a second dose of shRNA-expressing adenoviruses (at an m.o.i. of 25). The cells were assayed or harvested 24 h later. For overexpression of Perm1 and ERRs, differentiated myotubes were infected on day 4 or 5 (as specified in the figure legends) with adenoviruses expressing LacZ (control), FLAG-Perm1, FLAG-ERRα, FLAG-ERRβ, or FLAG-ERRγ at an m.o.i. of 50. Cells were harvested on day 6.
Reverse Transcription and Quantitative Real Time PCR Analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen), and cDNA was synthesized using the Superscript II reverse transcription system (Invitrogen) as published (47). Relative mRNA levels were determined by quantitative PCR using cDNA, gene-specific primers (supplemental Table 1), and SYBR Green reagent (USB Corp.) and normalized to levels of 36B4 as described (34).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed with adenovirus-infected C2C12 myotubes as described previously (48). Briefly, C2C12 myotubes were cross-linked for 10 min at 37 °C in 1% formaldehyde in PBS. After quenching, sonication to ∼500-bp fragments, and preclearing by treatment with protein A/G-Sepharose, soluble chromatin was immunoprecipitated with the following antibodies: anti-GFP, anti-FLAG (Clone M2, Sigma) for the detection of FLAG-tagged PGC-1α, and anti-ERRα (2131-1S1353, Epitomics). Genomic DNA from each immunoprecipitation was quantified by quantitative PCR using primers flanking an ERR response element (ERRE) in the first intron of Perm1 as well as primers specific to the ERR response element-containing region of the Esrra gene (positive control) and a region that lacks ERREs and is distal to the Esrra promoter (negative control) (primers are described in Supplemental Table 1). Data were first normalized against total genomic input and then expressed relative to levels of immunoprecipitated DNA in anti-GFP control samples.
DNA Isolation and Quantification
Total DNA was prepared according to standard procedures (49) and digested with 100 μg/ml RNase A for 30 min at 37 °C. The relative copy numbers of mitochondrial and nuclear DNAs were determined by quantitative PCR with primers specific to the CoxII (mitochondrial) and Nrip1 (nuclear) genes (Supplemental Table 1).
Oxygen Consumption
Oxygen consumption rates were measured using a Clark-type electrode as described previously (34) and normalized to total protein levels in each sample.
Subcellular Fractionation
The mitochondrial fraction was obtained according to Sun et al. (50), and the nuclear fraction was obtained according to Dignam et al. (51). Briefly, C2C12 myotubes were suspended in homogenizing buffer (10 mm Tris-HCl, pH 8.0, 300 mm sucrose, 1 mm EDTA, 1 mm EGTA) supplemented with protease inhibitors (1 mm PMSF, 2 μg/ml aprotinin, 5 μg/ml pepstatin A, 10 μg/ml leupeptin), homogenized using a Dounce homogenizer, and centrifuged at 600 × g for 10 min to generate a crude nuclear pellet and a supernatant containing the mitochondria and cytosol. The supernatant was then centrifuged at 5,000 × g for 10 min, giving rise to a pellet (crude mitochondrial fraction) and a supernatant with the crude cytosolic fraction. The crude mitochondrial fraction was washed with homogenizing buffer and centrifuged again at 7,000 × g for 10 min; the pellet was designated as the mitochondrial fraction. The supernatant from the 5,000 × g centrifugation was centrifuged again at 14,000 × g for 20 min; the resulting supernatant was designated as the cytosolic fraction. The first crude nuclear fraction pellet was further washed with Buffer A (20 mm Tris-HCl, pH 8.0, 1.5 mm MgCl2, 10 mm KCl) containing 0.5% (v/v) Nonidet P-40 and centrifuged at 1,000 × g for 10 min. Nuclear proteins were extracted from the pellet by incubation with Buffer B (20 mm Tris-HCl, pH 8.0, 10% (v/v) glycerol, 0.5 m NaCl, 1.5 mm MgCl2, I mm EDTA, 1 mm EGTA) supplemented with protease inhibitors. The extracted proteins were cleared by centrifugation at 14,000 × g for 20 min; the final supernatant was designated as the nuclear fraction. All procedures were conducted at 4 °C.
Western Blot and Antibodies
Protein samples were separated by SDS-PAGE and transferred onto nitrocellulose membrane (Hybond C Extra, Amersham Biosciences). Western blot was performed using the following antibodies: anti-FLAG (Clone M2, Sigma), anti-PERM1 (anti-C1orf170, Sigma), anti-GAPDH (MAB374, Chemicon), Anti-Rt/Ms Total OxPhos Complex kit (Invitrogen), anti-lactate dehydrogenase (sc-33781, Santa Cruz Biotechnology), anti-HSP60 (Clone LK-1, Cayman Chemical), anti-lamin A/C (2032, Cell Signaling Technology). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies were purchased from Bio-Rad. The blots were developed using an enhanced chemiluminescence reagent (Pierce).
Animal Studies
C57BL/6J mice were housed under a 12:12-h light-dark cycle at constant temperature and given food and water ad libitum. To determine the expression pattern of Perm1, tissues were collected from 10-week-old male mice. To determine the effect of exercise training on Perm1 expression, 14-week-old C57Bl/6 female mice were housed individually with access to electronically monitored in-cage running wheels for 5 weeks. Control littermates were housed individually in similar cages and for the same time but without access to wheels. All procedures were performed in compliance with standard principles and guidelines for the care and use of laboratory animals at The Scripps Research Institute. Tissues were dissected immediately after euthanization and stored at −80 °C until processing.
Muscle Biopsies
Human skeletal muscle samples were from the belly of the vastus lateralis muscle and have been described previously (24, 41).
Statistics
For cell culture experiments, the data presented are the mean ± S.D. of three to four experimental replicates from one of at least two representative experiments. For animal experiments, values are shown as mean ± S.E. The unpaired two-group Student's t test was performed to assess significant differences.
RESULTS
PGC-1α, PGC-1β, and ERRs Induce Perm1 in C2C12 Myotubes
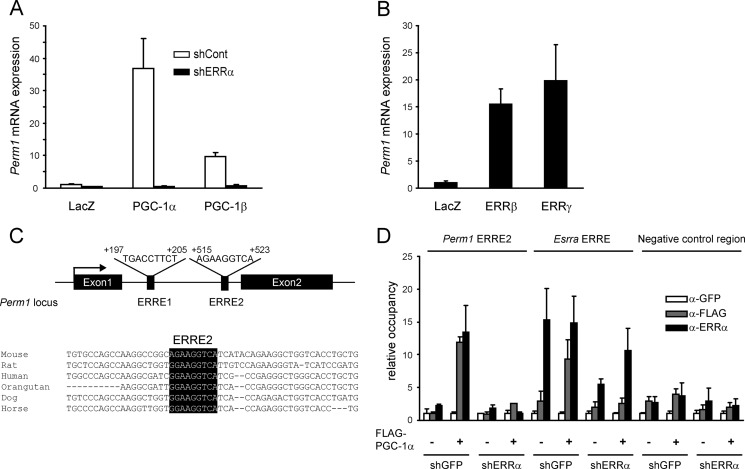
To identify novel proteins that may cooperate with PGC-1α and/or mediate the effects of PGC-1α in skeletal muscle, we screened for genes that are induced by PGC-1α in differentiated C2C12 cells, are expressed selectively in muscle, and encode for so far uncharacterized proteins. Based on these criteria, we focused our studies on the gene 2310042D19Rik, which we name here Perm1. As shown in Fig. 1A, Perm1 was strongly induced by PGC-1α and to a lesser extent by PGC-1β in C2C12 myotubes. The induction by PGC-1α and PGC-1β was lost when endogenous ERRα expression was suppressed (Fig. 1A), suggesting that Perm1 is regulated by PGC-1α/β via ERRα, the predominant ERR isoform in C2C12 cells. Consistent with Perm1 being an ERR target gene, Perm1 was also strongly induced by the introduction of ERRβ and ERRγ whose endogenous levels are low in C2C12 myotubes (Fig. 1B). The presence of two putative ERREs in the first intron of Perm1 coupled to the conservation of one of them, ERRE2, among mammalian species (Fig. 1C) suggested that Perm1 is a direct transcriptional target of ERRα. Indeed, ChIP assays showed PGC-1α and ERRα occupancy at the DNA region that includes ERRE2 and is dependent on the expression of endogenous ERRα (Fig. 1D). Notably, PGC-1α and ERRα occupancy at the Perm1 ERRE2 in PGC-1α-expressing cells was comparable with that seen at the Esrra proximal promoter region, one of the best characterized ERREs (31). Neither PGC-1α nor ERRα binding were detected at significant levels in a control region that lacks ERREs (Fig. 1D). These results suggest that PGC-1α/β and ERRs induce Perm1 expression directly via binding to the intronic ERRE2.
FIGURE 1.
PGC-1α, PGC-1β, and ERRs regulate Perm1 expression in myotubes. A, C2C12 myotubes were infected with adenoviruses expressing shGFP (shCont) or shERRα on day 4 of differentiation and LacZ, PGC-1α, or PGC-1β on day 6. RNA was harvested on day 7. Perm1 mRNA levels were determined by RT-qPCR, normalized to 36B4 levels in each sample, and expressed relative to Perm1 levels in control cells (LacZ/shCont). B, C2C12 myotubes were infected with adenoviruses expressing LacZ, ERRβ, or ERRγ on day 5. RNA was harvested on day 6, and Perm1 levels were quantified as in A. C, graphic representation of the Perm1 locus showing the location of the ERREs relative to the transcriptional start site and an alignment of the ERRE2 sequence (highlighted in black) across species. D, ChIPs were performed using antibodies against GFP (α-GFP; control), FLAG-tagged PGC-1α (α-FLAG), or ERRα (α-ERRα), and C2C12 myotubes infected with adenoviruses as described in A. The abundance of the Perm1 ERRE2, the Esrra ERRE (positive control for comparison), and a negative control genomic region in the ChIPs was quantified by qPCR, normalized to input signal, and expressed relative to the levels of each region in the control α-GFP samples. In A, B, and D, data are the mean of three to four experimental replicates from one of at least two representative experiments. Error bars represent S.D.
Perm1 Is Expressed in a Muscle-selective Manner and Regulated by Physical Activity
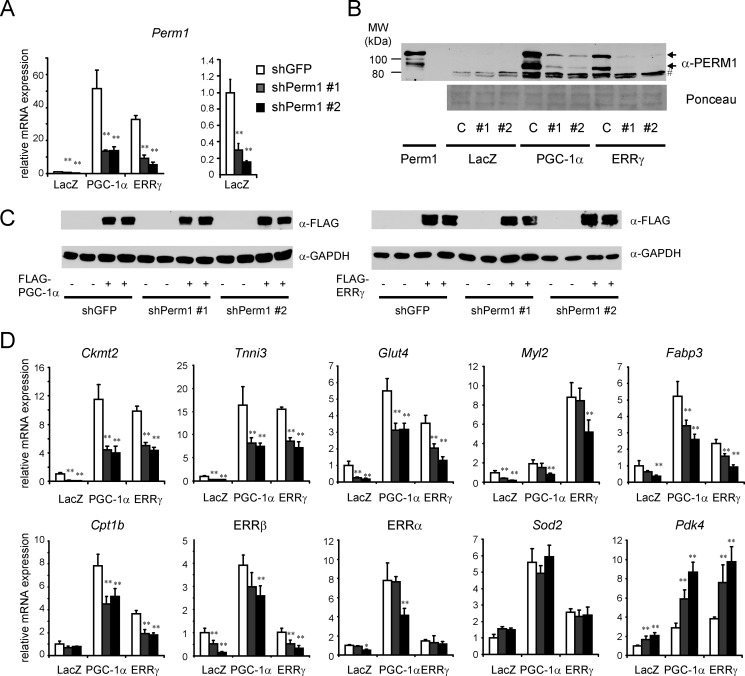
The Perm1 protein has not been described so far. The mouse and human loci have been predicted to give rise to both mRNAs and non-coding RNAs. To gain insights into the encoded products, we amplified, subcloned, and sequenced cDNAs corresponding to the human (C1orf170) and mouse (2310042D19Rik) genes using human and mouse skeletal muscle RNA, respectively (GenBankTM accession numbers KF150175 and KF150176). The predicted protein sequences (790 and 807 amino acids) contain a conserved putative nuclear localization signal (NLS), nuclear export signals, and a ØXXLL-type motif (where Ø is a hydrophobic amino acid, L is a leucine, and X is any amino acid) that is often involved in protein-protein interactions (52) but no other obvious protein motifs suggestive of a molecular function. The identity between the mouse and human proteins is highest in the N- and C-terminal regions, which include the putative ØXXLL, nuclear localization signal, and nuclear export signal motifs (Fig. 2A). Consistent with the presence of the nuclear localization signal and nuclear export signals, the majority of the PGC-1α-induced Perm1 protein was in the cytoplasmic and nuclear fractions of C2C12 myotubes (Fig. 2B). In the absence of PGC-1α, basal Perm1 protein was barely detectable and mostly in the cytoplasm. Interestingly, the anti-PERM1 serum (raised against the human amino acids 555–658) detected two major Perm1 protein isoforms in C2C12 cells, both of which were induced by PGC-1α and silenced by shRNA specific for Perm1 (Figs. 2B and 4B). The upper isoform co-migrated with the predominant protein species expressed from the cloned cDNA (Fig. 4B). The molecular events underlying the presence of two Perm1 isoforms are currently unclear and may not involve alternative splicing as we have so far identified just one cDNA species in skeletal muscle.
FIGURE 4.
Endogenous Perm1 is required for the expression and/or induction of selective PGC-1α/ERR target genes. C2C12 myotubes were infected on day 4 of differentiation with adenoviruses expressing control (shGFP) or shPerm1 and on day 6 with adenoviruses expressing LacZ, PGC-1α, or ERRγ. RNA and protein were harvested on day 7. A and D, mRNA levels of Perm1 and the indicated PGC-1/ERR-regulated genes were measured by RT-qPCR, normalized to 36B4 levels, and expressed relative to levels of each gene in control cells (shGFP/LacZ). Perm1 mRNA levels in LacZ (control) cells are shown twice: in the same scale as in PGC-1α/ERRγ expressing cells (A, left panel) and in a smaller scale for better view of the mRNA levels (A, right panel). Data are the mean of three to four experimental replicates from one of at least two representative experiments. Error bars represent S.D. *, p < 0.05; **, p < 0.01. B, Perm1 protein levels were determined by Western blot using the PERM1 antibody. C, shGFP; #1, shPerm1-1; #2, shPerm1-2. The two arrows indicate the Perm1 protein isoforms. As a control, the first lane shows the Perm1 protein expressed from the cloned cDNA (myotubes infected with adenovirus expressing Perm1); the major protein species co-migrates with the upper isoform of the doublet seen for the endogenous protein. Even when expressed from the cloned cDNA, a faster migrating species is also detectable. Protein loading was assessed by Ponceau staining of the membrane (lower panel); lane 1 (lysates of C2C12 infected with Ad/Perm1) has 8-fold less protein than all other lanes). # indicates nonspecific bands. D, the levels of FLAG-tagged PGC-1α and FLAG-tagged ERRγ were determined by Western blot analysis using an anti-FLAG antibody (upper panel). As a control for protein loading, the same blots were probed with an anti-GAPDH antibody (lower panel).
We next determined the tissue distribution of Perm1 in mice. Perm1 mRNA was highly expressed in skeletal muscles and heart with lower but detectable levels in brown adipose tissue. Consistent with the mRNA levels, Perm1 protein was also enriched in skeletal muscle and heart and to a lesser extent in brown adipose tissue (Fig. 2D). Similar to C2C12 myotubes, two major Perm1 protein isoforms were seen in muscle in vivo.
The expression and activity of skeletal muscle PGC-1α and ERRα are increased by endurance exercise, which promotes mitochondrial biogenesis and function (48, 53, 54). Thus, we asked whether Perm1 expression is also changing in response to exercise, as expected for a PGC-1/ERRα-regulated gene. Indeed, Perm1 mRNA was significantly higher in the quadriceps muscle of voluntary wheel-trained mice compared with control sedentary littermates (Fig. 2E). Similar regulation was seen in human subjects where PERM1 expression was increased 2.3× by an acute endurance exercise bout (Fig. 2F). Conversely, PGC-1α and ERRα muscle levels are decreased in states of physical inactivity or in patients with ALS who present muscle atrophy and mitochondrial dysfunction (24). PERM1 levels were also significantly lower in muscle biopsies from ALS patients compared with control subjects (Fig. 2G) (24). These findings suggest that Perm1 is a muscle-selective protein that can cycle between the cytoplasm and nucleus and whose levels are regulated by physical activity.
Perm1 Regulates the Expression of Selective PGC-1 and ERR Targets
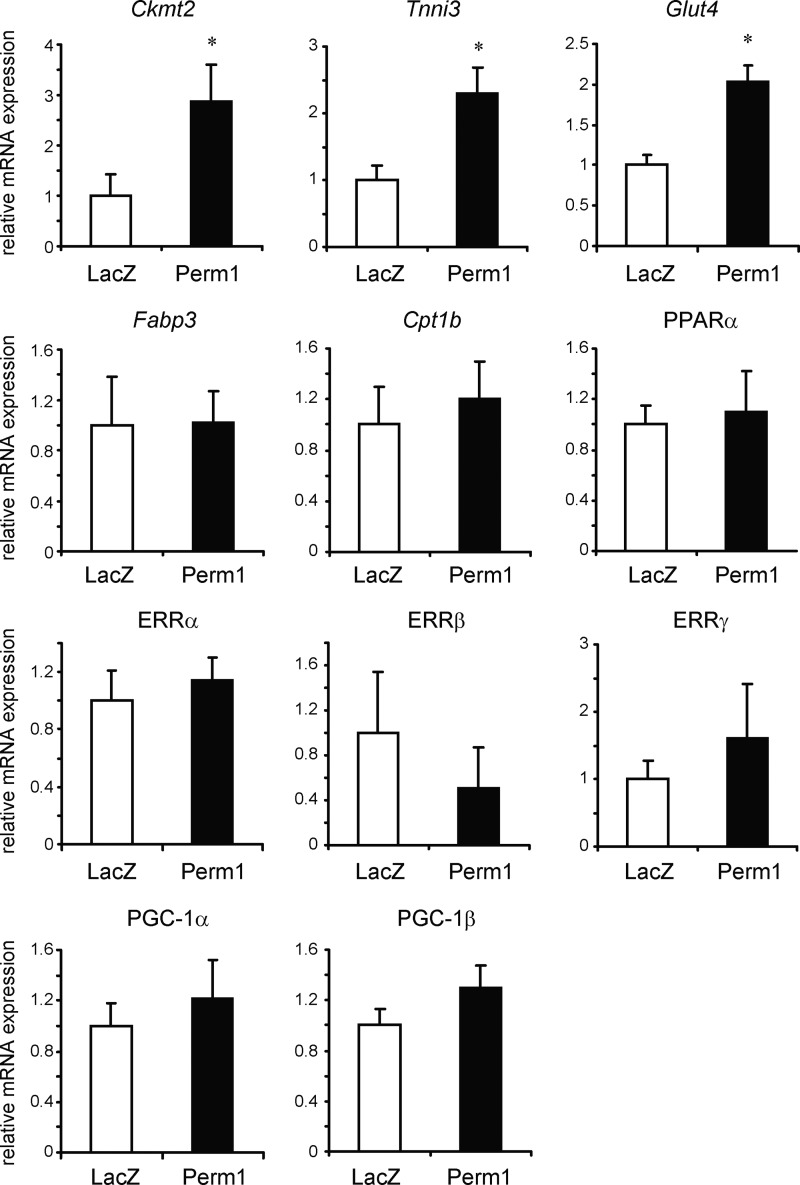
To test whether Perm1 affects the expression of PGC-1α and ERR gene targets, we first used a gain of function approach. C2C12 myotubes were infected with adenoviruses expressing LacZ (control) or Perm1, and the levels of known PGC-1α/β and ERRα/γ targets were determined by RT-qPCR. As shown in Fig. 3, Perm1 enhanced the expression of a subset of PGC-1 and ERR targets, such as Ckmt2, Tnni3, and Glut4; it did not affect the levels of other PGC-1/ERR targets, such as Fabp3, Cpt1b, and Ppara (Fig. 3). It also did not significantly alter the expression levels of PGC-1 and ERR family members (Fig. 3), suggesting that Perm1 does not act by altering general PGC-1 or ERR levels and activity.
FIGURE 3.
Overexpressed Perm1 up-regulates selective PGC-1α/ERR target genes in myotubes. C2C12 myotubes were infected on day 4 of differentiation with adenoviruses expressing LacZ (control) or Perm1. RNA was harvested on day 6. RNA levels for the indicated genes were determined by RT-qPCR, normalized to 36B4 levels, and expressed relative to the levels of each gene in control (LacZ) cells. Data are the mean of four experimental replicates from one of at least two representative experiments. Error bars represent S.D. *, p < 0.05.
To determine the extent to which endogenous Perm1 contributes to the expression of PGC-1/ERR targets, we next suppressed Perm1 levels in C2C12 myotubes using shRNAs specific to Perm1 (shPerm1) and measured the expression of target genes at the basal state and after induction by PGC-1α or ERRγ (ERRγ was chosen because it can efficiently activate ERR targets in a PGC-1-independent manner3). Perm1 mRNA and protein levels were efficiently knocked down by either of two distinct shPerm1 sequences in the absence and presence of PGC-1α or ERRγ (Fig. 4, A and B). Suppression of Perm1 expression did not affect the protein levels of exogenously expressed PGC-1α and ERRγ (Fig. 4C). Measurements of PGC-1/ERR target gene expression levels identified four classes of genes. In the first class were genes that relied on Perm1 for both their basal and their PGC-1α- and ERRγ-induced expression levels (e.g. Ckmt2, Tnni3, Glut4, Myl2, Fabp3, and ERRβ) (Fig. 4D). Perm1 did not seem essential for the response of these genes to PGC-1α or ERRγ (i.e. the genes were efficiently induced by PGC-1α and/or ERRγ in cells with suppressed Perm1 expression). However, this could be due to the incomplete Perm1 suppression; note that Perm1 protein levels were still significantly higher in PGC-1α/shPerm1- and ERRγ/shPerm1-expressing cells than in control cells (Fig. 4B). In the second class were genes that needed Perm1 for full induction by PGC-1α but not for basal expression (e.g. Cpt1b in Fig. 4D; more genes of this class are in Fig. 5). In the third class were PGC-1α-induced genes whose basal and induced levels were not affected by Perm1 knockdown, such as Sod2 (Fig. 4D). Finally, in the fourth class were genes that were expressed at higher levels in cells with suppressed Perm1 in basal and induced states, suggesting that they are negatively regulated by Perm1 (Pdk4; Fig. 4D). These results show that Perm1 modulates the expression of selective PGC-1α and ERR targets in a gene- and state (i.e. basal versus induced)-dependent manner.
FIGURE 5.
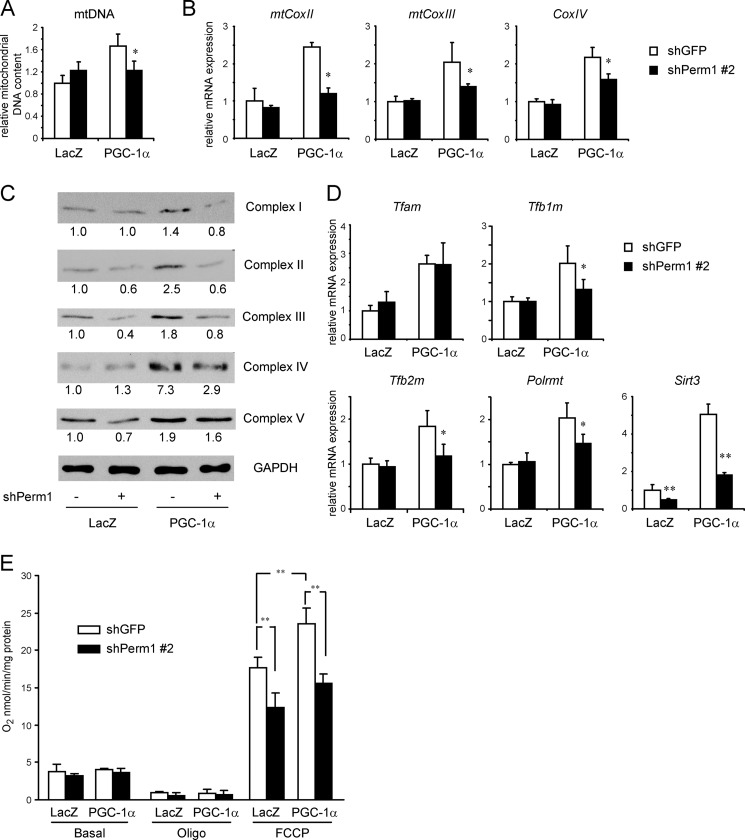
Perm1 is required for PGC-1α-induced mitochondrial biogenesis and maximal oxidative capacity. C2C12 myotubes were infected on day 4 of differentiation with adenoviruses expressing control (shGFP) or shPerm1 and on day 6 with adenoviruses expressing LacZ or PGC-1α. DNA, RNA, and protein were harvested on day 7. A, the relative mitochondrial DNA content was determined as the ratio of mitochondrial (CoxII) DNA to genomic (Nrip1) DNA copy numbers and expressed relative to the ratio seen in control (shGFP/LacZ) myotubes. B, mRNA levels for the indicated mitochondrial and nuclear DNA-encoded OxPhos genes were determined by RT-qPCR, normalized to 36B4 levels, and expressed relative to levels of each gene in control (shGFP/LacZ) myotubes. C, the levels of OxPhos complexes were measured by Western blot analysis using total protein lysates and the Total OxPhos Complex antibody mixture. The intensity of the bands was quantified using ImageJ software. The values are expressed relative to the signal intensity in control (shGFP/LacZ) myotubes. D, mRNA levels of genes with roles in mitochondrial biogenesis were determined and expressed as in B. In A, B, and D, data are the mean of three to four experimental replicates from one of at least two representative experiments. Error bars represent S.D. *, p < 0.05; **, p < 0.01. E, oxygen consumption rates of the myotubes were analyzed on day 7 in the absence (Basal) or presence of 2.5 μg/ml oligomycin (Oligo) and a 1.6 μm concentration of the uncoupling agent FCCP. Rates are normalized to the protein content of the cells and are the mean of three experimental replicates from one of two representative experiments. Error bars represent S.D. **, p < 0.01.
Perm1 Is Required for PGC-1α-induced Mitochondrial Biogenesis and Maximal Oxidative Capacity
PGC-1α enhances mitochondrial biogenesis and oxidative function in an ERRα-dependent manner (40, 49). The ability of Perm1 to regulate multiple PGC-1/ERR targets in C2C12 myotubes suggested that Perm1 could also regulate mitochondrial biogenesis and oxidative function. To test this hypothesis, we assessed the effects of Perm1 knockdown on mitochondrial DNA copy number, expression of genes encoding mitochondrial proteins important for oxidative phosphorylation and mitochondrial biogenesis, and respiratory capacity. As shown in Fig. 5A, knockdown of endogenous Perm1 did not affect basal mitochondrial DNA content but diminished the PGC-1α-induced increase in mitochondrial DNA. Consistent with decreased mitochondrial biogenesis in response to PGC-1α, Perm1 knockdown also prevented or diminished the ability of PGC-1α to enhance the RNA levels of mitochondrial DNA-encoded genes CoxII and CoxIII as well as nuclear genes encoding for OxPhos components, such as CoxIV (Fig. 5B). Similar results were observed with a different shPerm1 sequence (data not shown). To confirm the extent to which these changes were indeed affecting the abundance of mitochondrial OxPhos complexes, we assessed the levels of mitochondrial complexes I–V using antibodies against OxPhos subunits that are labile when not properly assembled in their complexes. Perm1 knockdown decreased the content of complexes II, III, and V in the basal state and abolished or attenuated the PGC-1α induction of complexes I–IV (Fig. 5C).
To gain insights into the pathway by which Perm1 affects mitochondrial biogenesis, we next assessed the expression levels of genes important for mtDNA replication and expression. Perm1 did not affect the expression of the replication/transcription factor Tfam but was required for the efficient induction of other biogenesis factors, such as Tfb1m, Tfb2m, and Polrmt, by PGC-1α (Fig. 5D). Perm1 was also necessary for both the basal and PGC-1α-induced levels of Sirt3, which codes for a mitochondrial deacetylase required for PGC-1α-induced mitochondrial biogenesis (55) (Fig. 5D).
To assess the functional consequences of the Perm1-dependent changes in gene and protein expression, we next determined the contribution of Perm1 to oxygen consumption rates in C2C12 myotubes. Knockdown of Perm1 had no effect on oxygen consumption rates in the basal state or when cells were treated with oligomycin but decreased maximal oxygen consumption rates measured in the presence of the uncoupler FCCP (Fig. 5E). The requirement of Perm1 for maximal mitochondrial oxidative capacity was seen in both the absence and presence of PGC-1α, although it may be more prominent in PGC-1α-expressing cells. In summary, Perm1 contributes to both basal and PGC-1α-induced levels of OxPhos complexes (Fig. 5, B and C), PGC-1α-induced mitochondrial biogenesis (Fig. 5, A and D), and maximal oxidative capacity (Fig. 5E).
DISCUSSION
The contractile activity of muscle requires a constant supply of ATP. The generation and efficient transfer of ATP to the cytoplasmic sites of ATP hydrolysis rely on multiple metabolic pathways, including glycogenolysis, glycolysis, oxidative metabolism, and the creatine shuttle (56). The signaling and transcriptional networks that control the expression of crucial components of these metabolic pathways are thus important determinants for the ability of muscle to maintain prolonged work and to adapt to different nutritional and physiologic states. To date, a number of transcription factors that control muscle metabolic pathways have been described, including coregulators, such as PGC-1α/β, NCoR1, and Rip140, and DNA binding factors, such as PPARs, FOXOs, MEF2, cAMP response element-binding protein, and ERRs (6, 31, 57–61). PGC-1α/β and ERRs in particular play a key role in mitochondrial biogenesis and oxidative metabolism. We show here that Perm1 is a novel downstream effector of PGC-1 and ERRs in myotubes. Perm1 is induced by PGC-1α/β and ERRs and required for maximal oxidative capacity. Suppression of basal Perm1 expression in C2C12 myotubes was sufficient to lead to decreases in levels of mitochondrial respiratory chain complexes and in maximal respiratory capacity. Furthermore, suppression of PGC-1α-induced Perm1 expression led to a loss in the PGC-1α-induced mitochondrial biogenesis at DNA, RNA, protein, and functional levels. Although some of the PGC-1α/ERR/Perm1-regulated genes and pathways (e.g. Ckmt2 and complex III) seem to rely on Perm1 even at the low endogenous levels of PGC-1 activity in C2C12 myotubes (62), others seem to use Perm1 predominantly for the efficient response to PGC-1α (e.g. Cpt1b, CoxIV, Tfb1m, Tfb2m, and Polrmt). Interestingly, there are also PGC-1/ERR targets that are not affected by Perm1 (e.g. Sod2 and Tfam) or are negatively regulated by Perm1 (e.g. Pdk4), suggesting that Perm1 does not act in a uniform manner at all PGC-1/ERR genes and pathways.
Besides playing a role in mitochondrial biogenesis and oxidative metabolism, basal Perm1 expression in C2C12 cells is required for the efficient expression of the glucose transporter Glut4, suggesting that Perm1 may affect glucose uptake, and of the mitochondrial creatine kinase Ckmt2. Ckmt2 uses mitochondrial ATP to phosphorylate creatine, thereby enabling the transfer of high energy phosphate from mitochondria to cytoplasmic ATP-hydrolyzing sites while maintaining high ADP levels at the mitochondria and stimulating oxidative metabolism (63). Interestingly, mice deficient in creatine kinase show impaired muscular contractile performance (64), and decreased Ckmt2 expression is associated with cardiomyopathy (65). Finally, Perm1 contributed to the expression of lipid uptake and utilization genes (e.g. Fabp3 and Cpt1b) and contractile genes (e.g. Tnni3 and Myl2). The roles of Perm1 in glucose and lipid metabolism as well as the contractile properties of muscle will be addressed in vivo in future studies.
The dramatic induction of Perm1 by PGC-1α in C2C12 myotubes depends on endogenous ERRα, which we show binds to ERREs in the first intron of the Perm1 gene and is necessary for the recruitment of PGC-1α at this site. The levels of PGC-1α and ERRα in human skeletal muscle are increased in response to exercise, which enhances oxidative capacity, and decreased in motor neuron disorders, such as ALS, which is characterized by atrophy and mitochondrial dysfunction (66, 67). PERM1 followed a similar pattern of induction by exercise and suppression in ALS patients, suggesting that the regulatory pathways seen in C2C12 cells reflect ones that are active in human exercise physiology and in the pathophysiology of neuromuscular disorders. Perm1 was also induced by PGC-1β, ERRβ, and ERRγ. It is possible that regulatory input via these factors may integrate distinct molecular pathways. Interestingly, ERRγ levels are also increased in skeletal muscle in response to exercise (38) and in skeletal myotubes by chronic low frequency electrical stimulation (36). Moreover, ERRγ levels are higher in muscle biopsies of physically active compared with sedentary humans (36). In contrast to the insights we have on the factors that may enable induction of Perm1 in response to physical activity, we know nothing about what drives its muscle-specific expression. Perm1 levels are high specifically in cardiac and skeletal muscle; the moderate Perm1 expression in brown adipose tissue may reflect the shared lineage and expression programs of brown adipose tissue and muscle (68). In support of our findings, a BioGPS-powered search for genes with close expression patterns shows that Perm1 expression correlates best with other known muscle-specific genes, such as the ones encoding γ-sarcoglycan, obscurin, β-taxilin, and Ckmt2 (44).
The mechanism by which Perm1 controls the expression of genes is currently unclear. Perm1 localizes both in the cytosol and nucleus, which would be compatible with Perm1 acting in the nucleus and modulating gene expression. However, Perm1 does not behave like a classical coactivator; we have not been able to detect a transcriptional activation function in Perm1 (i.e. Perm1 fused to the Gal4 DNA binding domain does not activate a Gal4-responsive reporter) or a physical interaction of Perm1 with ERRs or PGC-1α.3 Moreover, Perm1 acts differentially at different ERR targets in C2C12 cells (e.g. activates Ckmt2, has no effect on Sod2, and represses Pdk4). Perm1 is also unlikely to be a classical coactivator of other transcription factors required for mitochondrial biogenesis and expression of OxPhos components, such as nuclear respiratory factor 1 and GA-binding protein, because it does not activate Tfam, a well characterized nuclear respiratory factor 1- and GA-binding protein-responsive gene target (69). It is possible that Perm1 acts by regulating signaling pathways that control the activity of multiple transcription factors, possibly in a gene context-dependent manner. Future efforts to identify binding partners of Perm1 may provide mechanistic insights to its molecular function.
PGC-1s and ERRs are expressed in a wide range of tissues with high metabolic activity where they regulate a wide range of cellular pathways (e.g. lipid and glucose metabolism, mitochondrial biogenesis, mitochondrial function, oxidative stress responses, contractile proteins, ion transporters, and angiogenesis) (5–8, 31). Many of these pathways are regulated by PGC-1s and ERRs in a tissue-specific manner, suggesting the presence of other proteins/regulators, such as Perm1, that are expressed in a tissue-selective manner and that instruct (or enable) PGC-1/ERRs to induce specific sets of genes. There are other examples of such regulators: hepatic Lipin 1 acts as an amplifier and coregulator of PGC-1α in the activation of lipid metabolism genes (43). HIF2α is induced by PGC-1α/ERRα and acts as a downstream effector in the regulation of fiber type-specific proteins (70). Striated muscle activator of Rho signaling (Abra), which is also induced by PGC-1α and ERRα in skeletal muscle cells, acts as a downstream effector in the regulation of fatty acid oxidation genes (41). Such regulators may play central roles in shaping and specifying muscle adaptive metabolic responses to signals that induce PGC-1α, possibly integrating other physiologic and environmental cues.
In summary, identifying muscle-specific regulators in the PGC-1/ERR network of factors, such as Perm1, and elucidating their function will likely provide insights into how specific muscle PGC-1/ERR outputs are established. Such regulators may also provide new targets that could be used to modulate specific subsets of PGC-1/ERR pathways in the muscle (e.g. high capacity for ATP production) and address the bioenergetic defects seen in myopathies.
Acknowledgments
We thank Kralli laboratory members, especially Dr. Marin L. Gantner, for technical help and discussions; Drs. Enrique Saez and Shizuko Tachibana for helpful discussions and critical reading of the manuscript; and Dr. Sanna Ekholm Reed for technical help for the subcellular fractionation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK095686 from the NIDDK (to A. K.). This work was also supported by American Heart Association Postdoctoral Fellowship 12POST8610009 (to Y. C.).

This article contains supplemental Fig. 1 and Table 1.
The nucleotide sequences reported in this paper have been submitted to the GenBankTM/EBI Data Bank with accession numbers KF150175 and KF150176.
Y. Cho and A. Kralli, unpublished observations.
- PGC-1
- peroxisome proliferator-activated receptor γ coactivator 1
- ERR
- estrogen-related receptor
- Perm1
- PGC-1- and ERR-induced regulator in muscle 1
- OxPhos
- oxidative phosphorylation
- PPAR
- peroxisome proliferator-activated receptor
- ALS
- amyotrophic lateral sclerosis
- m.o.i.
- multiplicity of infection
- qPCR
- quantitative PCR
- ERRE
- ERR response element.
REFERENCES
- 1. Wallace D. C. (2000) Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am. Heart J. 139, S70–S85 [DOI] [PubMed] [Google Scholar]
- 2. DiMauro S. (2006) Mitochondrial myopathies. Curr. Opin. Rheumatol. 18, 636–641 [DOI] [PubMed] [Google Scholar]
- 3. van Adel B. A., Tarnopolsky M. A. (2009) Metabolic myopathies: update 2009. J. Clin. Neuromuscul. Dis. 10, 97–121 [DOI] [PubMed] [Google Scholar]
- 4. Ingwall J. S. (2009) On the control of metabolic remodeling in mitochondria of the failing heart. Circ. Heart Fail. 2, 275–277 [DOI] [PubMed] [Google Scholar]
- 5. Rowe G. C., Jiang A., Arany Z. (2010) PGC-1 coactivators in cardiac development and disease. Circ. Res. 107, 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Handschin C., Spiegelman B. M. (2006) Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 7. Arany Z. (2008) PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr. Opin. Genet. Dev. 18, 426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leone T. C., Kelly D. P. (2011) Transcriptional control of cardiac fuel metabolism and mitochondrial function. Cold Spring Harb. Symp. Quant. Biol. 76, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 10. Lehman J. J., Barger P. M., Kovacs A., Saffitz J. E., Medeiros D. M., Kelly D. P. (2000) Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 106, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 12. Meirhaeghe A., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J. K., Yeo G., Brand M. D., Cortright R. N., O'Rahilly S., Montague C., Vidal-Puig A. J. (2003) Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-γ co-activator 1α) gene in vitro and in vivo. Biochem. J. 373, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. St-Pierre J., Lin J., Krauss S., Tarr P. T., Yang R., Newgard C. B., Spiegelman B. M. (2003) Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 278, 26597–26603 [DOI] [PubMed] [Google Scholar]
- 14. Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B. M. (2007) The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 5, 35–46 [DOI] [PubMed] [Google Scholar]
- 15. Leone T. C., Lehman J. J., Finck B. N., Schaeffer P. J., Wende A. R., Boudina S., Courtois M., Wozniak D. F., Sambandam N., Bernal-Mizrachi C., Chen Z., Holloszy J. O., Medeiros D. M., Schmidt R. E., Saffitz J. E., Abel E. D., Semenkovich C. F., Kelly D. P. (2005) PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arany Z., He H., Lin J., Hoyer K., Handschin C., Toka O., Ahmad F., Matsui T., Chin S., Wu P. H., Rybkin I. I., Shelton J. M., Manieri M., Cinti S., Schoen F. J., Bassel-Duby R., Rosenzweig A., Ingwall J. S., Spiegelman B. M. (2005) Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 1, 259–271 [DOI] [PubMed] [Google Scholar]
- 17. Handschin C., Chin S., Li P., Liu F., Maratos-Flier E., Lebrasseur N. K., Yan Z., Spiegelman B. M. (2007) Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1α muscle-specific knock-out animals. J. Biol. Chem. 282, 30014–30021 [DOI] [PubMed] [Google Scholar]
- 18. Lelliott C. J., Medina-Gomez G., Petrovic N., Kis A., Feldmann H. M., Bjursell M., Parker N., Curtis K., Campbell M., Hu P., Zhang D., Litwin S. E., Zaha V. G., Fountain K. T., Boudina S., Jimenez-Linan M., Blount M., Lopez M., Meirhaeghe A., Bohlooly-Y M., Storlien L., Strömstedt M., Snaith M., Oresic M., Abel E. D., Cannon B., Vidal-Puig A. (2006) Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 4, e369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai L., Leone T. C., Zechner C., Schaeffer P. J., Kelly S. M., Flanagan D. P., Medeiros D. M., Kovacs A., Kelly D. P. (2008) Transcriptional coactivators PGC-1α and PGC-lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev. 22, 1948–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zechner C., Lai L., Zechner J. F., Geng T., Yan Z., Rumsey J. W., Collia D., Chen Z., Wozniak D. F., Leone T. C., Kelly D. P. (2010) Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 12, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 22. Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., Mandarino L. J. (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U.S.A. 100, 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandri M., Lin J., Handschin C., Yang W., Arany Z. P., Lecker S. H., Goldberg A. L., Spiegelman B. M. (2006) PGC-1α protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. U.S.A. 103, 16260–16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Russell A. P., Wada S., Vergani L., Hock M. B., Lamon S., Léger B., Ushida T., Cartoni R., Wadley G. D., Hespel P., Kralli A., Soraru G., Angelini C., Akimoto T. (2012) Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 49C, 107–117 [DOI] [PubMed] [Google Scholar]
- 25. Wenz T., Diaz F., Spiegelman B. M., Moraes C. T. (2008) Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 8, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Schilling J., Lai L., Sambandam N., Dey C. E., Leone T. C., Kelly D. P. (2011) Toll-like receptor-mediated inflammatory signaling reprograms cardiac energy metabolism by repressing peroxisome proliferator-activated receptor γ coactivator-1 signaling. Circ. Heart Fail 4, 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wenz T., Rossi S. G., Rotundo R. L., Spiegelman B. M., Moraes C. T. (2009) Increased muscle PGC-1α expression protects from sarcopenia and metabolic disease during aging. Proc. Natl. Acad. Sci. U.S.A. 106, 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Russell L. K., Mansfield C. M., Lehman J. J., Kovacs A., Courtois M., Saffitz J. E., Medeiros D. M., Valencik M. L., McDonald J. A., Kelly D. P. (2004) Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 94, 525–533 [DOI] [PubMed] [Google Scholar]
- 29. Choi C. S., Befroy D. E., Codella R., Kim S., Reznick R. M., Hwang Y. J., Liu Z. X., Lee H. Y., Distefano A., Samuel V. T., Zhang D., Cline G. W., Handschin C., Lin J., Petersen K. F., Spiegelman B. M., Shulman G. I. (2008) Paradoxical effects of increased expression of PGC-1α on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc. Natl. Acad. Sci. U.S.A. 105, 19926–19931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villena J. A., Kralli A. (2008) ERRα: a metabolic function for the oldest orphan. Trends Endocrinol. Metab. 19, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giguère V. (2008) Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 29, 677–696 [DOI] [PubMed] [Google Scholar]
- 32. Dufour C. R., Wilson B. J., Huss J. M., Kelly D. P., Alaynick W. A., Downes M., Evans R. M., Blanchette M., Giguère V. (2007) Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab. 5, 345–356 [DOI] [PubMed] [Google Scholar]
- 33. Huss J. M., Imahashi K., Dufour C. R., Weinheimer C. J., Courtois M., Kovacs A., Giguère V., Murphy E., Kelly D. P. (2007) The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 6, 25–37 [DOI] [PubMed] [Google Scholar]
- 34. Villena J. A., Hock M. B., Chang W. Y., Barcas J. E., Giguère V., Kralli A. (2007) Orphan nuclear receptor estrogen-related receptor α is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alaynick W. A., Kondo R. P., Xie W., He W., Dufour C. R., Downes M., Jonker J. W., Giles W., Naviaux R. K., Giguère V., Evans R. M. (2007) ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 6, 13–24 [DOI] [PubMed] [Google Scholar]
- 36. Gan Z., Rumsey J., Hazen B. C., Lai L., Leone T. C., Vega R. B., Xie H., Conley K. E., Auwerx J., Smith S. R., Olson E. N., Kralli A., Kelly D. P. (2013) Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J. Clin. Investig. 123, 2564–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Narkar V. A., Fan W., Downes M., Yu R. T., Jonker J. W., Alaynick W. A., Banayo E., Karunasiri M. S., Lorca S., Evans R. M. (2011) Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 13, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rangwala S. M., Wang X., Calvo J. A., Lindsley L., Zhang Y., Deyneko G., Beaulieu V., Gao J., Turner G., Markovits J. (2010) Estrogen-related receptor γ is a key regulator of muscle mitochondrial activity and oxidative capacity. J. Biol. Chem. 285, 22619–22629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huss J. M., Torra I. P., Staels B., Giguère V., Kelly D. P. (2004) Estrogen-related receptor α directs peroxisome proliferator-activated receptor α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24, 9079–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mootha V. K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Willy P. J., Schulman I. G., Heyman R. A., Lander E. S., Spiegelman B. M. (2004) Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U.S.A. 101, 6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wallace M. A., Hock M. B., Hazen B. C., Kralli A., Snow R. J., Russell A. P. (2011) Striated muscle activator of Rho signalling (STARS) is a PGC-1α/oestrogen-related receptor-α target gene and is upregulated in human skeletal muscle after endurance exercise. J. Physiol. 589, 2027–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mitra M. S., Schilling J. D., Wang X., Jay P. Y., Huss J. M., Su X., Finck B. N. (2011) Cardiac lipin 1 expression is regulated by the peroxisome proliferator activated receptor γ coactivator 1α/estrogen related receptor axis. J. Mol. Cell. Cardiol. 51, 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. (2006) Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 4, 199–210 [DOI] [PubMed] [Google Scholar]
- 44. Wu C., Macleod I., Su A. I. (2013) BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41, D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keller M. P., Choi Y., Wang P., Davis D. B., Rabaglia M. E., Oler A. T., Stapleton D. S., Argmann C., Schueler K. L., Edwards S., Steinberg H. A., Chaibub Neto E., Kleinhanz R., Turner S., Hellerstein M. K., Schadt E. E., Yandell B. S., Kendziorski C., Attie A. D. (2008) A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 18, 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schreiber S. N., Knutti D., Brogli K., Uhlmann T., Kralli A. (2003) The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα). J. Biol. Chem. 278, 9013–9018 [DOI] [PubMed] [Google Scholar]
- 47. Tiraby C., Hazen B. C., Gantner M. L., Kralli A. (2011) Estrogen-related receptor γ promotes mesenchymal-to-epithelial transition and suppresses breast tumor growth. Cancer Res. 71, 2518–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cartoni R., Léger B., Hock M. B., Praz M., Crettenand A., Pich S., Ziltener J. L., Luthi F., Dériaz O., Zorzano A., Gobelet C., Kralli A., Russell A. P. (2005) Mitofusins 1/2 and ERRα expression are increased in human skeletal muscle after physical exercise. J. Physiol. 567, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schreiber S. N., Emter R., Hock M. B., Knutti D., Cardenas J., Podvinec M., Oakeley E. J., Kralli A. (2004) The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun F. C., Wei S., Li C. W., Chang Y. S., Chao C. C., Lai Y. K. (2006) Localization of GRP78 to mitochondria under the unfolded protein response. Biochem. J. 396, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 53. Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 54. Russell A. P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C. A., Bell D. R., Kralli A., Giacobino J. P., Dériaz O. (2003) Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-γ coactivator-1 and peroxisome proliferator-activated receptor-α in skeletal muscle. Diabetes 52, 2874–2881 [DOI] [PubMed] [Google Scholar]
- 55. Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. (2010) Sirtuin 3, a new target of PGC-1α, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One 5, e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. MacIntosh B., Gardiner P., McComas A. (2005) Skeletal Muscle: Form and Function, 2nd Ed., pp. 208–223, Human Kinetics, Champaign, IL [Google Scholar]
- 57. Yamamoto H., Williams E. G., Mouchiroud L., Cantó C., Fan W., Downes M., Héligon C., Barish G. D., Desvergne B., Evans R. M., Schoonjans K., Auwerx J. (2011) NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seth A., Steel J. H., Nichol D., Pocock V., Kumaran M. K., Fritah A., Mobberley M., Ryder T. A., Rowlerson A., Scott J., Poutanen M., White R., Parker M. (2007) The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 6, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bassel-Duby R., Olson E. N. (2006) Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75, 19–37 [DOI] [PubMed] [Google Scholar]
- 60. Black B. L., Olson E. N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 61. Altarejos J. Y., Montminy M. (2011) CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Michael L. F., Wu Z., Cheatham R. B., Puigserver P., Adelmant G., Lehman J. J., Kelly D. P., Spiegelman B. M. (2001) Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. U.S.A. 98, 3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Greenhaff P. L. (2001) The creatine-phosphocreatine system: there's more than one song in its repertoire. J. Physiol. 537, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gorselink M., Drost M. R., Coumans W. A., van Kranenburg G. P., Hesselink R. P., van der Vusse G. J. (2001) Impaired muscular contractile performance and adenine nucleotide handling in creatine kinase-deficient mice. Am. J. Physiol. Endocrinol. Metab. 281, E619–E625 [DOI] [PubMed] [Google Scholar]
- 65. Schlattner U., Tokarska-Schlattner M., Wallimann T. (2006) Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta 1762, 164–180 [DOI] [PubMed] [Google Scholar]
- 66. Crugnola V., Lamperti C., Lucchini V., Ronchi D., Peverelli L., Prelle A., Sciacco M., Bordoni A., Fassone E., Fortunato F., Corti S., Silani V., Bresolin N., Di Mauro S., Comi G. P., Moggio M. (2010) Mitochondrial respiratory chain dysfunction in muscle from patients with amyotrophic lateral sclerosis. Arch. Neurol. 67, 849–854 [DOI] [PubMed] [Google Scholar]
- 67. Krasnianski A., Deschauer M., Neudecker S., Gellerich F. N., Müller T., Schoser B. G., Krasnianski M., Zierz S. (2005) Mitochondrial changes in skeletal muscle in amyotrophic lateral sclerosis and other neurogenic atrophies. Brain 128, 1870–1876 [DOI] [PubMed] [Google Scholar]
- 68. Timmons J. A., Wennmalm K., Larsson O., Walden T. B., Lassmann T., Petrovic N., Hamilton D. L., Gimeno R. E., Wahlestedt C., Baar K., Nedergaard J., Cannon B. (2007) Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. U.S.A. 104, 4401–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Scarpulla R. C. (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 70. Rasbach K. A., Gupta R. K., Ruas J. L., Wu J., Naseri E., Estall J. L., Spiegelman B. M. (2010) PGC-1α regulates a HIF2α-dependent switch in skeletal muscle fiber types. Proc. Natl. Acad. Sci. U.S.A. 107, 21866–21871 [DOI] [PMC free article] [PubMed] [Google Scholar]