Background: Recruitment of plasminogen is important for efficient dissemination of Borrelia burgdorferi.
Results: BBA70 of B. burgdorferi binds plasminogen, and following activation, bound plasmin can cleave fibrinogen and inactivate the key complement components C3b and C5.
Conclusion: BBA70 is a potent plasminogen-binding protein.
Significance: Investigation suggests that binding of plasminogen may aid in pathogen dissemination and inhibit bacteriolytic effects of the host complement system.
Keywords: Complement, Host-Pathogen Interactions, Infectious Diseases, Innate Immunity, Plasminogen, Borrelia, Immune Evasion, Spirochetes
Abstract
The Lyme disease spirochete Borrelia burgdorferi lacks endogenous, surface-exposed proteases. In order to efficiently disseminate throughout the host and penetrate tissue barriers, borreliae rely on recruitment of host proteases, such as plasmin(ogen). Here we report the identification of a novel plasminogen-binding protein, BBA70. Binding of plasminogen is dose-dependent and is affected by ionic strength. The BBA70-plasminogen interaction is mediated by lysine residues, primarily located in a putative C-terminal α-helix of BBA70. These lysine residues appear to interact with the lysine-binding sites in plasminogen kringle domain 4 because a deletion mutant of plasminogen lacking that domain was unable to bind to BBA70. Bound to BBA70, plasminogen activated by urokinase-type plasminogen activator was able to degrade both a synthetic chromogenic substrate and the natural substrate fibrinogen. Furthermore, BBA70-bound plasmin was able to degrade the central complement proteins C3b and C5 and inhibited the bacteriolytic effects of complement. Consistent with these functional activities, BBA70 is located on the borrelial outer surface. Additionally, serological evidence demonstrated that BBA70 is produced during mammalian infection. Taken together, recruitment and activation of plasminogen could play a beneficial role in dissemination of B. burgdorferi in the human host and may possibly aid the spirochete in escaping the defense mechanisms of innate immunity.
Introduction
Lyme borreliosis is the most prevalent vector-borne anthropozoonosis in the United States and Europe and is caused by spirochetes belonging to the Borrelia burgdorferi sensu lato complex. B. burgdorferi is transmitted to a variety of hosts through the bite of an infected Ixodes tick. Once deposited during tick feeding, spirochetes begin to spread outward from the bite site, with one of the early symptoms of infection being the characteristic bull's eye-shaped rash, termed erythema migrans. As a multisystemic disease, Lyme borreliosis may afflict various organs and, if left untreated, can result in a number of severe clinical manifestations, including arthritis, neuroborreliosis, acrodermatitis chronica atrophicans, and carditis. Key to its ability to affect multiple organs is the spirochete's remarkable ability to penetrate solid tissues and disseminate throughout the host (1–4).
Efficient dissemination requires that the spirochetes degrade components of the host extracellular matrix (ECM)3 and basement membranes surrounding blood vessels. These consist of various fibrous proteins, such as collagens, elastin, laminin, fibronectin, and proteoglycans. Whereas surface-bound or secreted bacterial proteases are used by other bacterial species for dissemination through the host (5), the Lyme disease spirochete is not known to produce any extracytoplasmic proteases. Instead, the spirochetes disseminate by hijacking the host protease plasmin(ogen) (1).
Plasmin is an important component of the human fibrinolytic system. The inactive proenzyme plasminogen is a 92-kDa glycoprotein consisting of an N-terminal preactivation peptide; five lysine-binding, disulfide-bonded kringle domains; and a serine protease domain (6, 7). The active serine protease, plasmin, is generated through proteolytic cleavage of plasminogen by activators such as urokinase-type (uPA) or tissue-type plasminogen activators (8). Bacterial activators, such as staphylokinase from Staphylococcus aureus and streptokinase, secreted by group A, C, and G streptococci, can also activate plasminogen (9–11). Plasmin has a relatively broad substrate spectrum, and in addition to fibrin(ogen), plasmin can cleave components of the host ECM, such as laminin (12), fibronectin (13), vitronectin (14, 15), and heparan sulfate proteoglycans (16).
Upon transmission through the bite of an infected tick, spirochetes encounter the human complement system, which plays a crucial role in recognition and clearance of invading pathogens (17). A triggered enzyme cascade, complement can be activated by the classical pathway, lectin pathway, or alternative pathway. Classical pathway activation is initiated by specific antibodies, whereas the lectin pathway is activated through recognition of carbohydrates (e.g. mannan). By contrast, alternative pathway activation occurs spontaneously. Activation of either pathway results in cleavage of the central protein C3 and deposition of the highly reactive C3b molecule on the surface of invading microorganisms. This, in turn, leads to opsonization (18) and the formation of the lytic membrane attack complex and resultant complement-mediated killing of the intruders.
Plasmin negatively regulates the complement system on several levels. Early evidence suggested that active plasmin can cleave various complement components (19). Recently, it has been demonstrated that plasmin efficiently degrades the central complement component C3b as well as C5. In addition to the alternative pathway, active plasmin inhibits the classical and lectin pathways. The proteolytically inactive proenzyme plasminogen also enhances complement factor I-mediated inactivation of C3b in the presence of factor H (20).
Several invasive human pathogens bind plasminogen, including Streptococcus pneumoniae (21), S. aureus (22), Pseudomonas aeruginosa (23), Haemophilus influenzae (24), Helicobacter pylori (25), and the yeast Candida albicans (26, 27). Several B. burgdorferi plasminogen-binding proteins have been identified, such as the infection-associated surface proteins CspA (complement regulator-acquiring surface protein 1; CRASP-1) and CspZ (CRASP-2) (28), ErpP (CRASP-3), ErpC (CRASP-4), ErpA (CRASP-5) (29), the 70-kDa surface protein BPBP (30), OspA (31), and OspC, which a recent study, employing live cell binding assays, has shown to be a plasminogen receptor on the surface of B. burgdorferi (32, 33). Additionally, B. burgdorferi enolase has recently been demonstrated to moonlight on the bacterial outer surface and serve as a plasminogen-binding protein (34–36).
Plasminogen-coated spirochetes, when exposed to or treated with activators, generate plasmin, which proteolytically degrades fibronectin, vitronectin, and laminin (37) and can penetrate endothelial cell monolayers (38). Although plasmin is not very efficient at degrading collagens (37), borreliae up-regulate expression and induce secretion of host matrix metalloproteinase MMP-1 (collagenase 1) and MMP-9 (gelatinase B) and stimulate activation of pro-MMP-9 (39). Acquisition of plasminogen by the spirochetes is required for transfer from ticks, and plasminogen-deficient mice show decreased spirochetemia (40). Taken together, these data indicate that acquisition of plasminogen by borreliae plays a significant role in virulence. Here we describe B. burgdorferi BBA70, a novel borrelial plasminogen-binding protein with a strong binding capacity, belonging to the paralog family 54 (PFam54) protein family.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
B. burgdorferi strain B31 was grown at 33 °C until mid-exponential phase (5 × 107 cells/ml) in Barbour-Stoenner-Kelly medium (Bio & SELL) supplemented with 6% heat-inactivated rabbit serum (Sigma-Aldrich). Density of spirochetes was assessed employing dark field microscopy and a Kova counting chamber (Hycor Biomedical). Escherichia coli JM109 cells (Promega) used for cloning experiments and protein expression were grown at 37 °C in yeast tryptone broth, supplemented with the appropriate antibiotics. For complement inactivation assays, E. coli DH5α cells grown at 37 °C in yeast tryptone broth were employed.
Proteins and Antisera
Plasminogen from Hematologic Technologies Inc. was used for all experiments. Activation to plasmin was accomplished using uPA from Chemicon. Fibrinogen was purchased from Sigma-Aldrich, as was the chromogenic substrate S-2251 (d-Val-Leu-Lys-p-nitroanilide dihydrochloride). Purified C3b, factor H, and factor I were all purchased from Complement Technology. Polyclonal antisera for plasminogen and fibrinogen from Acris Antibodies were used. Polyclonal C3 antiserum was purchased from Calbiochem. The polyclonal goat antiserum raised against C5 was obtained from Quidel. Monoclonal anti-hexahistidine antiserum from GE Healthcare was used. Horseradish peroxidase (HRP)-conjugated immunoglobulins were purchased from Dako. mAb LA22.1 (41) was used to detect the periplasmic FlaB protein, and for detection of the surface-exposed OspC protein, mAb 93-193/0246 was employed (42).
Generation of Recombinant, Polyhistidine-tagged Proteins
The BBA70-encoding gene (ORF ZSA70 of strain ZS7, which is 100% identical with BBA70 of strain B31) was amplified by PCR from plasmid pGEX-ZSA70 (43), using primers pGEX(+) and pGEX 2T SalI (Table 1). The PCR product was subsequently cloned into pQE-30 Xa (Qiagen). The resulting plasmid, pQE-A70 ZS7, was sequenced to exclude the possible introduction of mutations during the cloning process. In order to generate C-terminally truncated BBA70 constructs, plasmid pQE-A70 ZS7 was used as a template for PCR with primer pQE-FP-30 and either BBA70-166(−), BBA70-131(−), or BBA70-46(−). The respective PCR products were cloned into pQE-30 Xa. Both strands of the resulting plasmids were sequenced to ensure no mutations had been introduced during PCR and cloning procedures. In one case, a spontaneous mutation had been introduced, resulting in the generation of a stop codon at position 68. This truncated construct was also included in further studies. For recombinant BBA66, primers pGEX(+) and pGEX 2T SalI (Table 1) were used to amplify the encoding gene from plasmid pGEX-ZSA66 (43) (ZSA66 is 100% identical with BBA66). The resulting PCR product was then cloned into vector pQE-30 Xa (Qiagen). To rule out the possible introduction of mutations during cloning, the resulting plasmid termed pQE-A66 ZS7 was sequenced. Similarly, for recombinant ZQA70 primers, ZQ1 A70 SphI(+) and pGEX 2T SalI were used to amplify the encoding gene from plasmid pGEX ZQ1 A70 and cloned into pQE-30 Xa (Qiagen). The resulting plasmid was termed pQE-A70 ZQ1 and sequenced to verify that no mutations had been introduced during the cloning process. Production of purified ErpP and CspA has been described previously (29, 44). Recombinant proteins were expressed in E. coli JM109 cells (Promega). Following induction with isopropyl-β-d-thiogalactopyranoside, cells were harvested and lysed using a MICCRA D-9 dispersion device (Art Prozess- & Labortechnik GmbH) in buffer containing 10 mm imidazole, 300 mm NaCl, 50 mm NaH2PO4, and 1 mg/ml lysozyme. Cell debris was cleared by centrifugation, and proteins were purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen). Purity of recombinant proteins was determined by SDS-PAGE followed by silver staining. A bicinchoninic acid protein assay (Pierce) was used to measure concentrations of purified proteins.
TABLE 1.
Oligonucleotides used in the course of this study
Generation of Monoclonal BBA70 Antiserum
Monoclonal antibody 70-11/10.17 directed against BBA70 was generated by immunization of BALB/c mice with recombinant ZSA70 protein according to a method described elsewhere (45). Animal research was approved in advance by the Laboratory Animal Committee of the University of Heidelberg (RP Karlsruhe 35–9185.82/A-25/07). The animals were kept in a filter cabinet and given food and water ad libitum, with all maintenance performed according to German animal welfare guidelines.
Ligand Affinity Blotting
Recombinant polyhistidine-tagged BBA70, BBA66, ErpP, or BSA (250 ng each) were subjected to reducing 10% Tris/Tricine SDS-PAGE. After separation, proteins were transferred to a nitrocellulose membrane using Western blotting. Membranes were blocked using 1.5% nonfat dry milk powder in TBS containing 0.1% Tween 20 (TBS-T). After washing three times with TBS-T, membranes were incubated with 20 μg/ml plasminogen at room temperature for 1 h. Following three wash steps with TBS containing 0.2% Tween 20, bound plasminogen was detected using a goat polyclonal plasminogen antiserum and HRP-conjugated anti-goat immunoglobulins.
Protein Binding Assays
For ELISA, 500 ng of recombinant polyhistidine-tagged ErpP, BBA66, or BBA70 or equimolar amounts of C-terminally truncated BBA70 constructs in 100 μl of sodium carbonate buffer were immobilized onto 96-well microtiter plates (MaxiSorp, Nunc) at 4 °C overnight with gentle agitation. After washing three times with 0.05% PBS-T (PBS-T), wells were blocked with Blocking Buffer III (AppliChem). Following three wash steps, for each well 500 ng of plasminogen was added in 100 μl of PBS. Where indicated, tranexamic acid or varying concentrations of NaCl or NaBr were added. After incubation for 1 h at room temperature, unbound protein was removed by washing three times with PBS-T. Bound proteins were identified, employing a polyclonal plasminogen antiserum followed by anti-goat polyclonal HRP-conjugated immunoglobulins. Following three more wash steps, detection was performed utilizing ortho-phenylenediamide dihydrochloride (Sigma- Aldrich) as a substrate. Reactions were stopped by the addition of 2.6 m H2SO4, and absorbance was measured at 490 nm in a microtiter plate reader (PowerWave HT, BioTek). For ELISA with plasminogen constructs, 50 μl of 5 μg/ml polyhistidine-tagged BBA70 was immobilized onto microtiter plates at 4 °C overnight. After washing with PBS-T, wells were blocked with 0.2% gelatin in PBS. Following three wash steps, 1 μg of plasminogen or equimolar amounts of the plasminogen constructs were added. Bound constructs or full-length plasminogen was detected using a polyclonal plasminogen antiserum and HRP-conjugated anti-goat immunoglobulins. Tetramethylbenzidine was used as a substrate for HRP, and absorbance was measured at 450 nm. To investigate dose dependence of plasminogen binding to BBA70, 0.175 μm BBA70 was immobilized at 4 °C overnight and, after blocking, was incubated with varying amounts of plasminogen ranging from 0.002 to 2.174 μm. Detection of bound plasminogen was performed as described above.
Surface Plasmon Resonance (SPR) Protein Binding Analysis
Binding of plasminogen to immobilized His-tagged BBA70 was studied quantitatively using a BIAcore X100 and Sensor Chip NTA (GE Healthcare). The surface was Ni2+-coated with a 1-min injection of 2 mm NiCl2 at a flow rate of 10 μl·min−1. About 2000 response units of ligand (BBA70) were immobilized on the sensor chip. All measurements were carried out in the running buffer (5 mm KH2PO4, 77.5 mm NaCl, and 15 mm Na2HPO4, pH 7.4) at 25 °C. Binding kinetic traces were recorded when different concentrations of analyte (plasminogen) were passed over the loaded chip surface. The measured kinetics were not determined by diffusion limitations because higher flow rates (e.g. 30 μl·min−1) and lower analyte immobilization levels had no obvious effect on the kinetic traces. Injection of analyte in running buffer was accomplished in 3 min (association or contact phase), followed by a 4-min injection of running buffer (dissociation phase). The sensor chip surface was regenerated with a 3-min injection of strip buffer (0.5% SDS, 50 mm Na2EDTA, pH 8.0) and rinsed prior to the next binding experiment. The kinetic traces were corrected by taking into account the background dissociation of immobilized ligand from the sensor chip surface and nonspecific binding of analyte to the sensor chip surface during a blank run (no ligand added) prior to each binding experiment. The binding experiments at each concentration of analyte were performed at least in triplicate.
The sensograms were analyzed by curve-fitting with BIAevaluation software (version 4.1; Biacore AB, Uppsala, Sweden), where models were fitted for the single concentrations of analyte and fitted globally for the concentration series of analyte. Various reaction models, including the classical Langmuir 1:1 binding between analyte (A) and ligand (B), and a two-state reaction model, based on a 1:1 binding of analyte to an immobilized ligand followed by a conformational change (A + B ↔ AB ↔ AB*) were considered. For the determination of kon, only the middle portion (10–120 s) of the association curve was used for fitting. For determination of koff, all data, except the first 10 s, encompassing the dissociation phase, were used for fitting. All of our kinetic data were fit most adequately by assuming a two-state reaction (conformational change) model for interaction between soluble analyte and immobilized ligand.
Plasminogen Activation Assay
The plasminogen activation assay was performed as described previously (28). 96-Well microtiter plates (Maxisorb, Nunc) were coated with 500 ng of recombinant polyhistidine-tagged BBA70, BBA66, ErpP, or BSA in 100 μl of sodium carbonate buffer at 4 °C overnight with gentle agitation. Following three wash steps with 0.05% PBS-T, wells were blocked with Blocking Buffer III (AppliChem). After three wash steps, 500 ng of plasminogen was added in 100 μl of 50 mm Tris/HCl, pH 7.5, for each well. Where indicated, wells were supplemented with 50 mm tranexamic acid as a negative control. Plates were incubated for 3 h at room temperature. After washing three times, 96 μl of reaction buffer was added, containing 50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 0.003% Triton X-100, and 0.3 mg/ml S-2251 (d-Val-Leu-Lys-p-nitroanilide dihydrochloride). Finally, 4 μl of 2.5 μg/ml uPA was used to activate plasminogen to plasmin. Microtiter plates were then incubated at room temperature, measuring absorbance at 405 nm in a microtiter plate reader (PowerWave HT, BioTek) every 30 min over a time period of 24 h.
Fibrinogen Degradation Assay
Recombinant polyhistidine-tagged BBA70, BBA66, ErpP, or BSA (500 ng each) were immobilized onto 96-well microtiter plates (Maxisorb, Nunc) at 4 °C overnight in 100 μl of sodium carbonate buffer. After three wash steps with 0.05% PBS-T, wells were blocked with Blocking Buffer III (AppliChem). Three wash steps were performed, and then 500 ng of plasminogen was added for each well in 100 μl of 50 mm Tris/HCl, pH 7.5. For negative controls, 50 mm tranexamic acid was added to plasminogen. Incubation for 1 h at room temperature was followed by washing. Finally, a reaction mix containing 20 μg/ml fibrinogen and 0.16 μg/ml uPA in 50 mm Tris/HCl, pH 7.5, was added. Reactions were incubated at room temperature, and aliquots were taken at the indicated times. Reactions were stopped by the addition of SDS-PAGE sample buffer. Samples were then separated by 10% Tris/Tricine SDS-PAGE under reducing conditions and transferred to a nitrocellulose membrane by Western blotting. Fibrinogen and its degradation products were visualized using a polyclonal fibrinogen antiserum and corresponding HRP-conjugated anti-goat immunoglobulins.
C3b/C5 Degradation Assay
Degradation of C3b and C5 was assayed as described previously (20). Briefly, microtiter plates (Maxisorb, Nunc) were coated with 1 μg (for C3b degradation) or 2 μg (for C5 degradation) of recombinant BBA70, ErpP, BBA66, or BSA in 100 μl of sodium carbonate buffer at 4 °C overnight with gentle agitation. After washing with PBS-T and blocking with Blocking Buffer III (AppliChem), 1 μg (for C3b degradation) or 2 μg (for C5 degradation) of plasminogen in 50 mm Tris/HCl, pH 7.5, was added, and plates were incubated at room temperature for 1 h. Where indicated, 50 mm tranexamic acid was added as a negative control. Wells were washed three times with PBS-T, and a reaction mixture containing 5 μg/ml either C3b or C5 as well as 0.16 μg/ml uPA in 50 mm Tris/HCl, pH 7.5, was added. Microtiter plates were incubated at 37 °C for 24 h. Aliquots were taken at the indicated times, and reactions were stopped by the addition of SDS-PAGE sample buffer. The samples were separated by 12.5% glycine SDS-PAGE under reducing conditions, transferred to a nitrocellulose membrane, and assayed for C3b or C5 degradation products using polyclonal goat antisera raised against C3 or C5, respectively, and corresponding anti-goat, HRP-conjugated immunoglobulins.
Complement Inactivation Assay
Recombinant BBA70, ErpP, or ZQA70 (10 μg each) was allowed to bind to Dynabeads (Invitrogen). Following several wash steps, borrelial proteins coated to magnetic beads were incubated with 10 μg of plasminogen. After further washing, 200 μl of 2.5% normal human serum (NHS) in GVB++ buffer (Complement Technology) was added, together with 0.16 μg/ml uPA to activate bound plasminogen to plasmin for 1 h at 37 °C. Controls included omission of plasminogen and uPA, respectively, and the addition of 50 mm tranexamic acid. After 1 h, pretreated serum samples were added to E. coli DH5α (1 × 107 cells) and incubated for 2 h at 37 °C. Serial dilutions were plated on Mueller-Hinton agar and incubated overnight at 37 °C. Survival was determined by counting colony-forming units (cfu) on the following day.
Mice Infections
Four Bl/6 mice (Jackson Laboratories) were infected, under laboratory conditions, by vector tick bite (Ixodes scapularis), as described previously (46). Prior to tick feeding, a sample of 10 ticks was tested to ensure they were harboring B. burgdorferi (47). Mice, which were culture- and serology-positive for B. burgdorferi, were euthanized 1 year after tick transmission. Serum was separated from collected blood. In addition, control serum was collected from an uninfected animal, and all sera were stored at −20 °C until needed.
Line Blot Immunoassays and Serological Examination
Purified, nondenaturated borrelial proteins BBA70 (2–500 ng), VlsE (0.8 ng), and OspC (87.5 ng) as well as BSA (500 ng) in PBS were transferred onto a nitrocellulose membrane by a microdispensing method using a Desaga AS30 apparatus (Sarstedt). Immunogenicity of BBA70 was tested using control and infected mouse serum as described previously (48). Briefly, 1:800 dilutions of mouse serum were incubated with protein-coated nitrocellulose strips at room temperature for 2 h. Antibodies directed against BBA70 were detected by secondary incubation with donkey anti-mouse IgG/HRP diluted 1:25,000 (Santa Cruz Biotechnology, Inc.). Reactivity was determined via chemiluminescence (Thermo Scientific, Waltham, MA) and autoradiography. Experiments were repeated once to confirm reproducibility. Cross-reactivity with control serum was tested at 1:200, 1:400, and 1:800.
Reverse Transcription-PCR
Spirochetes were grown to mid-exponential phase and sedimented by centrifugation. After three wash steps with PBS, total RNA was isolated with the TRIzol Max bacterial RNA isolation kit (Invitrogen). Removal of contaminating genomic DNA was accomplished using TURBO DNA-free (Ambion). Concentrations of RNA samples were determined photometrically. 1 μg of RNA was subsequently used as a template to generate complementary DNA (cDNA) utilizing the avian myeloblastosis virus first strand cDNA synthesis kit for RT-PCR (Roche Applied Science) with random hexamer primers in accordance with the manufacturer's instructions. The obtained cDNA was employed as a template for PCR analysis. Primers BBA70-F and BBA70-R (Table 1) were used to generate amplicons corresponding to part of the bba70 gene. As controls, amplicons specific for the flaB and ospA gene were also generated, using primes Fla6 and Fla7 as well as OspA1 and OspA2, respectively (Table 1).
Triton X-114 Phase Separation
Phase separation was performed as described previously (49, 50). Briefly, a 300-ml culture of B. burgdorferi B31 was grown to mid-exponential phase. Spirochetes were pelleted by centrifugation and washed twice with PBS. 5 × 109 cells were resuspended in 1 ml of a solution containing 2% Triton X-114 in 5 mm EDTA, 10 mm Tris/HCl, pH 7.5. Following incubation with gentle agitation at 4 °C overnight, samples were centrifuged at 50,000 × g for 1 h at 4 °C. The supernatant was then incubated in a 37 °C water bath for 30 min, followed by centrifugation at 50,000 × g for 1 h at 25 °C. The hydrophilic phase was then separated from the hydrophobic phase. The hydrophobic phase was reextracted twice by washing with 20% (v/v) 10 mm Tris, 5 mm EDTA, followed by centrifugation at 20,000 × g at 25 °C. Protein fractions were then precipitated by the addition of 10-fold (v/v) ice cold acetone. Following centrifugation at 12,500 × g at 4 °C, precipitated protein was resolubilized in PBS. Protein concentrations were assayed utilizing a bicinchoninic acid protein assay (Pierce). 10 μg of the fractions corresponding to the periplasmic cylinder and the detergent phase were subjected to reducing 10% Tris/Tricine SDS-PAGE. Following separation, proteins were blotted onto a nitrocellulose membrane. BBA70 was visualized using mAb 70-11/10.17 and HRP-conjugated anti-mouse immunoglobulins.
Prediction of Secondary Structure
NetSurfP version 1.1 from the Center for Biological Sequence Analysis/Technical University of Denmark was used for prediction of the BBA70 secondary structure based on sequence information from GenBankTM CAH05016.1.
Statistical Analyses
A one-way ANOVA test with Bonferroni's multiple comparison post test was employed for statistical analysis of ELISA results using GraphPad Prism version 4. Results were deemed statistically significant for the following p values: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
BBA70 Is a Plasminogen-binding Protein
The B. burgdorferi CspA surface protein (also known as CRASP-1 or BBA68) binds both complement factor H and plasminogen (28, 42). Lyme disease spirochetes encode multiple, distinct proteins paralogous to CspA, constituting PFam54 (51). Although CspA is the only PFam54 member of strain B31 capable of binding factor H (52), the abilities of other paralogs to bind plasmin(ogen) had yet to be determined. Of these proteins, BBA70 displayed a strong affinity for plasminogen in a pull-down screening assay (data not shown).
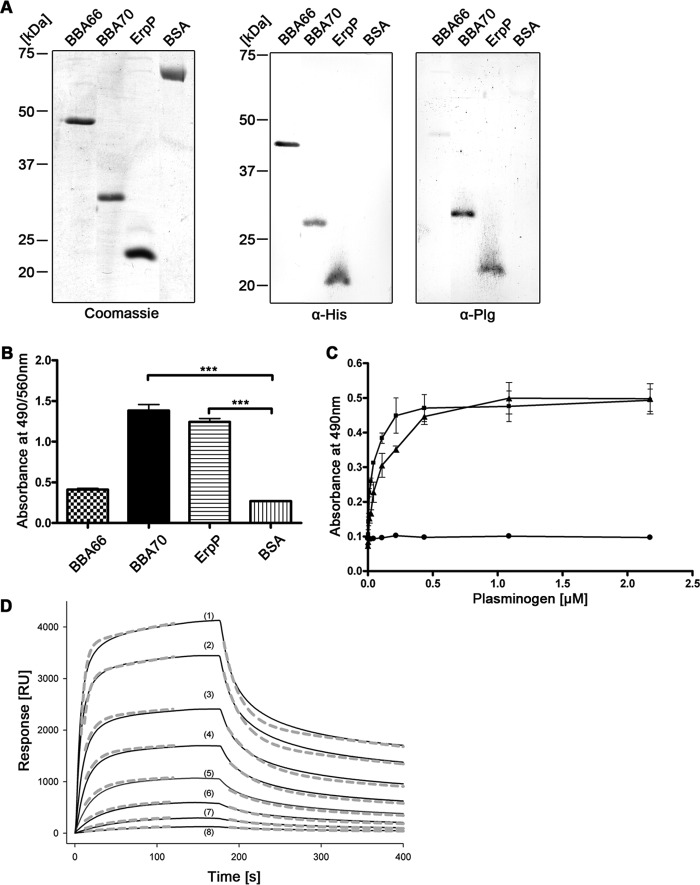
Affinity blots and ELISA were used for detailed analyses of interactions between BBA70 and plasminogen. Recombinant B. burgdorferi ErpP was used as a positive control protein (29). The borrelial protein BBA66 and BSA served as negative controls. Affinity blot analyses demonstrated binding of plasminogen to BBA70 and ErpP but not to BBA66 or BSA (Fig. 1A). ELISA also demonstrated that BBA70 and ErpP bound plasminogen in a dose-dependent manner (Fig. 1, B and C). Surface plasmon resonance analysis provided insight into the association and dissociation kinetics of the interaction between BBA70 and plasminogen. Typical sensograms obtained for interaction of BBA70 with different concentrations of plasminogen are shown in Fig. 1D. In all cases, the sensograms could not be fitted to a simple 1:1 binding model, and the best fits were attained using a two-state reaction model (Fig. 1D and Table 2), suggesting a conformational change after interaction of plasminogen with BBA70. Using this model, a dissociation equilibrium constant (Kd) of 55.1 nm for the BBA70-plasminogen interaction was calculated. Detailed binding parameters for the BBA70-plasminogen interaction are reported in Table 2.
FIGURE 1.
B. burgdorferi BBA70 binds human plasminogen. A, recombinant, hexahistidine-tagged borrelial proteins or BSA (1 μg each) were separated via SDS-PAGE for Coomassie staining in order to ensure sufficient purity of the recombinant proteins (left). For Western blotting, 250 ng of the respective borrelial proteins and BSA were subjected to SDS-PAGE and subsequently transferred to nitrocellulose. Detection of hexahistidine tag was performed as loading control (center), and ligand affinity blotting was employed to test for plasminogen binding. The membrane was incubated with 20 μg/ml purified human plasminogen, and bound plasminogen was detected using a polyclonal plasminogen antiserum (α-Plg) and HRP-conjugated secondary antibody (right). B, binding of plasminogen was verified for recombinant proteins using ELISA. Hexahistidine-tagged recombinant proteins or BSA (5 μg/ml each) was immobilized onto microtiter plates and incubated with 10 μg/ml purified human plasminogen. Bound plasminogen was detected using a polyclonal plasminogen antiserum and HRP-conjugated anti-goat immunoglobulins. Absorbance was measured at 490/560 nm. Experiments were each performed in triplicate, and graphs represent means from at least three independent experiments. Error bars, S.E. ***, p < 0.001 (one-way ANOVA with Bonferroni's multiple-comparison test). C, ELISA was performed to determine dose dependence. BBA70 (■), ErpP (▴), and BSA (●) (5 μg/ml each) were immobilized onto microtiter plates and incubated with increasing concentrations of purified human plasminogen. Detection was performed as described previously, and absorbance was measured at 490 nm. Data points represent means from three independent experiments, each performed in triplicate. Error bars, S.E. D, representative SPR binding and dissociation kinetics of human plasminogen to BBA70 immobilized on a Ni2+-nitrilotriacetic acid surface. Sensograms (solid lines) obtained when plasminogen solutions at the indicated concentrations were flowed across the sensor chip and the experimental data were fitted (dashed lines) with the two-state binding model, where A + B ↔ AB ↔ AB* (see “Experimental Procedures”). The resulting rate and affinity constants are given in Table 2 (mean ± S.D.). Plasminogen concentrations are 3410 nm (1), 2270 nm (2), 1136 nm (3), 620 nm (4), 324 nm (5), 133 nm (6), 67 nm (7), and 33 nm (8). RU, response units.
TABLE 2.
Rate and affinity constants for BBA70-plasminogen interaction
BBA70 was immobilized on a Ni2+-nitrilotriacetic acid surface, and varying concentrations of plasminogen were flowed over the sensor chip.
| kon 1 × 104 | koff 1 × 10−2 | kon 2 × 10−3 | koff 2 × 10−3 | Kd |
|---|---|---|---|---|
| m−1s−1 | s−1 | m−1s−1 | s−1 | nm |
| 9.2 ± 2.5 | 1.5 ± 0.1 | 3.9 ± 0.3 | 1.9 ± 0.5 | 55.1 ± 10.3 |
Roles of Lysine Residues and Ionic Strength in BBA70-Plasminogen Interaction
The lysine-binding kringle domains of plasminogen mediate binding to fibrin, components of the ECM, or lysine-containing host and bacterial binding proteins (6, 11, 53). To determine whether lysine residues in BBA70 are involved with binding of plasminogen, increasing concentrations of the lysine analog tranexamic acid were included in ELISA. Tranexamic acid, when added at 0.1 mm, decreased the BBA70-plasminogen interaction by more than 60%, whereas the addition of 1 mm tranexamic acid reduced the signal to background levels (Fig. 2A).
FIGURE 2.
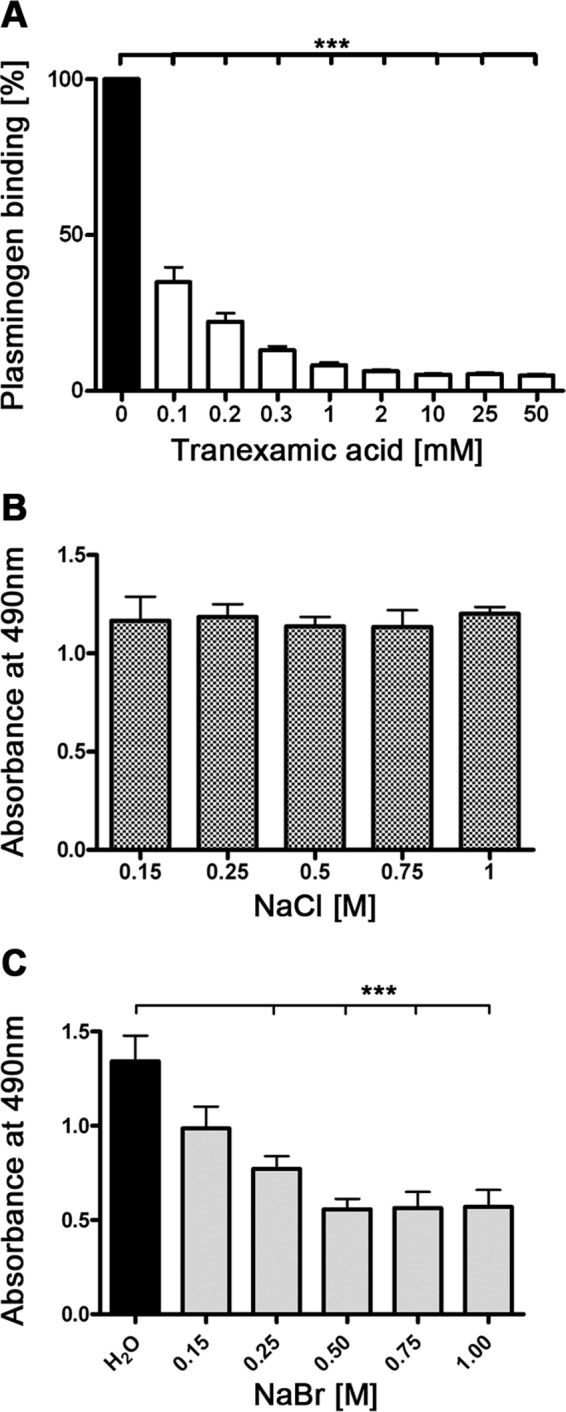
BBA70-plasminogen interaction is mediated by lysine residues and independent of ionic strength. A, recombinant, hexahistidine-tagged BBA70 (5 μg/ml) was immobilized onto microtiter plates and incubated with 10 μg/ml purified plasminogen in the presence of increasing concentrations of the lysine analog tranexamic acid. Plasminogen bound to BBA70 was detected with a polyclonal plasminogen antiserum and HRP-conjugated anti-goat immunoglobulins. Absorbance was measured at 490 nm, and binding of plasminogen by BBA70 in the absence of tranexamic acid was set to 100%. Values represent the means of three separate experiments conducted in triplicate, and error bars show S.E. ***, p < 0.001 (one-way ANOVA with post hoc Bonferroni's correction). B and C, microtiter plates were coated with 5 μg/ml recombinant BBA70 and subsequently incubated with human plasminogen and increasing concentrations of NaCl or NaBr. Bound plasminogen was detected using a specific antiserum. Absorbance was measured at 490 nm. Data represent means and S.E. of three separate experiments with three replicates per condition. Error bars, S.E. ***, p < 0.001 (one-way ANOVA with post hoc Bonferroni's correction).
Additionally, the effect of ionic strength on the BBA70-plasminogen interaction was investigated. Binding of plasminogen to BBA70 was measured in the presence of increasing concentrations of NaCl. Even at 6 times the physiological concentration of NaCl, the affinity of BBA70 for plasminogen remained unaffected (Fig. 2B). Because plasminogen is known to bind chloride anions, it is conceivable that changing the ionic strength with NaCl is insufficient. We thus also studied the effect of NaBr on plasminogen binding (Fig. 2C). Starting at a concentration of 0.25 m, a statistically significant reduction in plasminogen binding was detected. These data suggest that ionic strength does have an impact on the BBA70-plasminogen interaction.
The Putative C-terminal α-Helix of BBA70 Is Required for Binding of Plasminogen
Having demonstrated the involvement of lysine residues for the interaction of BBA70 with plasminogen, we sought to identify the plasminogen-binding regions within this borrelial protein. The predicted secondary structure for BBA70 included a number of putative α-helices (designated A–G), which contain a total of 36 lysine residues (Fig. 3A). To investigate the role of these structural elements of BBA70 in the interaction with plasminogen, we created a number of carboxyl-terminally truncated BBA70 constructs that lacked putative lysine-containing α-helices (Fig. 3A). The truncated constructs were then ELISA-tested for their ability to bind plasminogen. BBA70 or equimolar amounts of the carboxyl-terminally truncated constructs were immobilized on microtiter plates and incubated with plasminogen. Deletion of the putative C-terminal α-helix (G), containing 9 lysine residues (BBA70(1–166)), eliminated plasminogen binding (Fig. 3B), indicating that this predicted α-helix (G) of BBA70 is crucial for the interaction with plasminogen.
FIGURE 3.
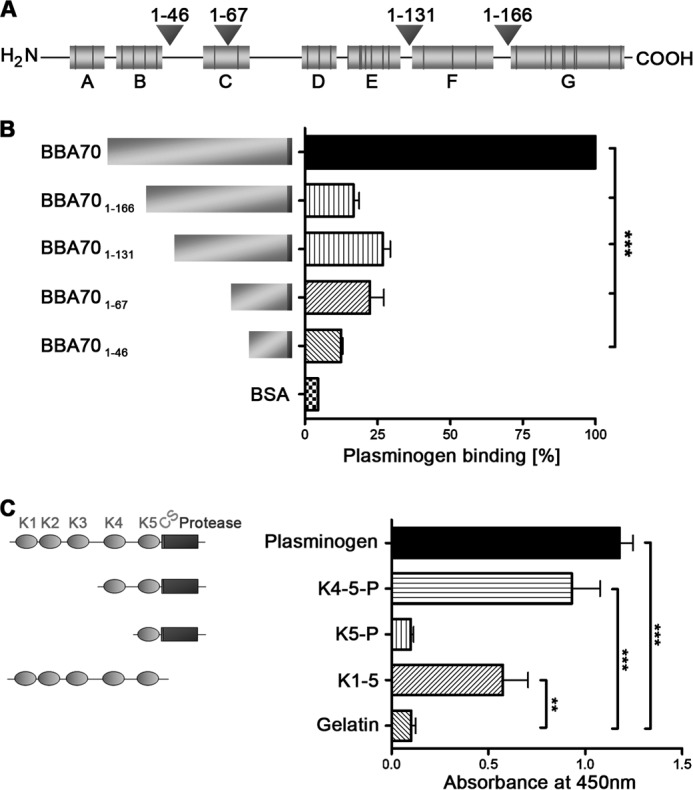
Further characterization of the BBA70-plasminogen interaction. A, schematic representation of the BBA70 molecule. Putative α-helices are shaded gray, with black vertical lines indicating lysine residues within the helices. Helices are designated A–G. Arrowheads indicate positions for the generation of C-terminally truncated BBA70 constructs and specify the lengths of the respective polypeptide chains. B, binding of purified plasminogen to C-terminally truncated BBA70 constructs was analyzed by ELISA. Full-length BBA70 (5 μg/ml) or equimolar amounts of C-terminally truncated constructs were immobilized onto microtiter plates and incubated with plasminogen (10 μg/ml). Bound plasminogen was detected using a polyclonal plasminogen antiserum and HRP-conjugated anti-goat immunoglobulins. Binding of plasminogen to full-length BBA70 was set to 100%. Lengths of BBA70 constructs are represented by the length of the area shaded gray, whereas the black vertical line indicates the N-terminal hexahistidine tag. C, microtiter plates were coated with 5 μg/ml recombinant, hexahistidine-tagged BBA70 and incubated with purified plasminogen (10 μg/ml) or equimolar amounts of various plasminogen fragments. K4–5-P, kringle domains 4 and 5 and the protease domain; K5-P, kringle 5 and the protease domain; K1–5, all five kringle domains but lacking the protease domain of plasminogen. Gelatin (10 μg/ml) was used as a nonspecific negative control. Plasminogen or the respective fragments bound to BBA70 were detected using a specific antiserum. Absorbance was measured at 450 nm. Three independent experiments were conducted in triplicate, and graphs represent means ± S.E. (error bars). ***, p < 0.001; **, p < 0.01 (one-way ANOVA with Bonferroni's multiple-comparison test).
BBA70-Plasminogen Interaction Depends on Kringle 4 Domain
Binding of plasminogen to host receptors and various bacterial proteins is mediated by plasminogen's five kringle domains (11, 54, 55). In order to narrow down the domain(s) of plasminogen required for the interaction with BBA70, plasminogen fragments comprising various lysine-binding kringle domains as well as the protease domain were assayed for their abilities to bind BBA70. Whereas a construct consisting of kringles 4 and 5 and the protease domain bound to BBA70, another protein containing only kringle 5 and the protease domain did not bind to BBA70 (Fig. 3C). Thus, kringle domain 4 plays an important role in BBA70-plasminogen interactions.
BBA70-bound Plasminogen Can Be Activated to Plasmin
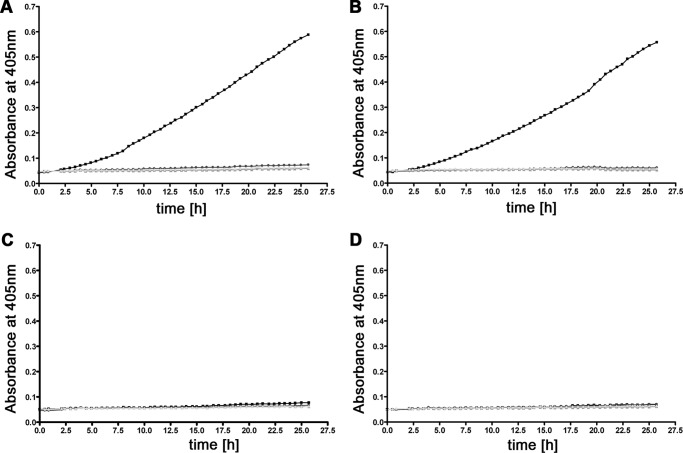
Plasminogen is converted to the serine protease plasmin by human activators, such as uPA or tissue-type plasminogen activator (56), and by bacterial activators like staphylokinase (57). In order for these activators to function, the appropriate domain of plasminogen needs to be accessible. Through use of the chromogenic substrate d-Val-Leu-Lys-p-nitroanilide dihydrochloride, we demonstrated that BBA70-bound plasminogen is accessible to human uPA and activated to proteolytically active plasmin. The positive control protein ErpP also bound activatable plasminogen (Fig. 4B). No cleavage of the chromogenic substrate was observed when using negative controls BBA66 or BSA (Figs. 4, C and D). Additional control reactions with the plasminogen inhibitor tranexamic acid, omitting plasminogen or plasminogen activator uPA, did not result in degradation of the chromogenic substrate.
FIGURE 4.
BBA70-bound plasminogen is converted to plasmin by uPA. Microtiter plate wells coated with 5 μg/ml recombinant BBA70 (A), ErpP (B), BBA66 (C), or BSA (D) were incubated with 10 μg/ml plasminogen. Upon washing, the plasminogen activator uPA was added (2.5 μg/ml) together with the chromogenic substrate S-2251 (d-Val-Leu-Lys-p-nitroanilide dihydrochloride) (■). As negative controls, either uPA (▴) or plasminogen (♦) was omitted, or the plasminogen inhibitor tranexamic acid (50 mm) was added to plasminogen (●). For clarity, graphs of negative controls are shaded gray. Microtiter plates were incubated for ∼26 h, and absorbance at 405 nm was measured at 30-min intervals. Three separate experiments were performed; data shown are from a representative experiment.
Plasmin Bound to BBA70 Degrades Fibrinogen
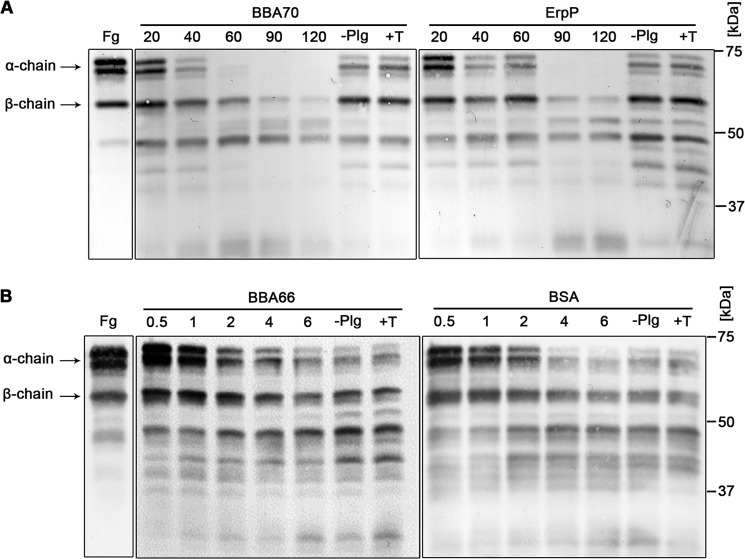
As a central component of the fibrinolytic system, active plasmin cleaves fibrin(ogen) (58). To determine if BBA70-bound active plasmin can degrade its natural substrate fibrinogen, we immobilized BBA70 on microtiter plates, incubated with plasminogen, and activated with uPA. Time course incubation with fibrinogen yielded almost complete substrate degradation within 60 min (Fig. 5A, left). In an identical experiment, using ErpP as a positive control, the fibrinogen α-chain was completely degraded after 90 min (Fig. 5A, right). By contrast, significant degradation was not observed for the negative controls BSA or BBA66 (Fig. 5B). Further controls were performed by the addition of the lysine analog tranexamic acid or by omission of plasminogen from the reaction mixture, with no significant degradation of fibrinogen occurring in either control.
FIGURE 5.
Plasmin bound to BBA70 can degrade the natural substrate fibrinogen. Recombinant, hexahistidine-tagged BBA70, ErpP, BBA66, and BSA were immobilized onto microtiter plates and incubated with 10 μg/ml plasminogen. After washing, the activator uPA (0.16 μg/ml) and fibrinogen (20 μg/ml) were added. As controls, either plasminogen was omitted from the reaction mixture or plasminogen was added together with the inhibitor tranexamic acid (50 mm). Microtiter plates were incubated at 37 °C, and aliquots were taken at various time intervals. Reactions were then separated by SDS-PAGE, and proteins were transferred to nitrocellulose. Fibrinogen or its degradation products were visualized using a polyclonal fibrinogen antiserum and appropriate HRP-conjugated secondary antibody. Fg, purified human fibrinogen with characteristic α-chain (63.5 kDa), β-chain (56 kDa), and γ-chain (48 kDa). −Plg, plasminogen was omitted; +T, the addition of 50 mm tranexamic acid. Two independent experiments were performed; Western blots show representative results. A, degradation of fibrinogen after 20, 40, 60, 90, and 120 min for BBA70 (left) and ErpP (right). Results of BBA66 (left) and BSA (right) after 0.5, 1, 2, 4, and 6 h shown in B.
Plasmin Bound to BBA70 Degrades Complement Components C3b and C5
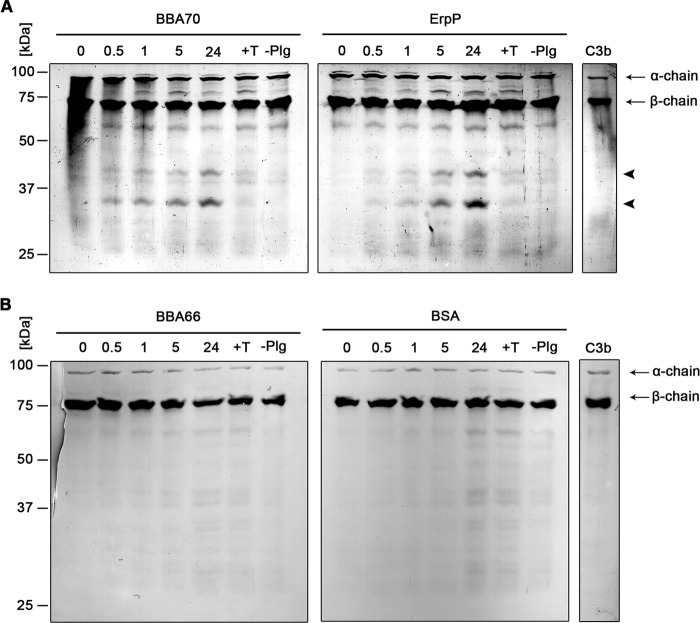
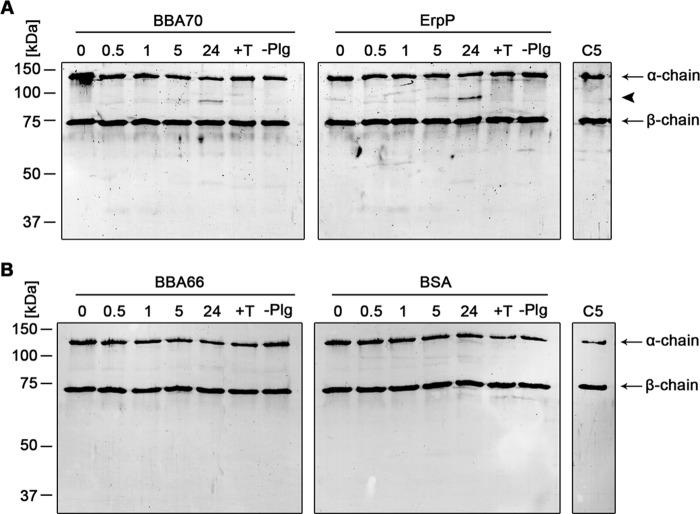
Active plasmin also degrades the key complement components C3b and C5 (20). Therefore, we examined the ability of BBA70-bound plasmin to degrade C3b and C5. Recombinant BBA70, ErpP, BBA66, or BSA was immobilized onto microtiter plates and incubated with plasminogen and then with uPA. Plasmin bound to BBA70 and ErpP cleaved C3b, as indicated by the appearance of degradation products with molecular masses of ∼30 and 40 kDa (Fig. 6A, arrowheads). These cleavage products were absent in experiments that omitted plasminogen or upon the inclusion of tranexamic acid. In addition, no prominent degradation of C3b was observed with the negative controls BBA66 and BSA (Fig. 6B). When BBA70- or ErpP-bound plasmin was incubated with C5, a cleavage product of ∼87 kDa appeared below the C5 α-chain (Fig. 7A, arrowhead). Degradation products were not observed in the negative controls with tranexamic acid, without plasminogen, or with BBA66 or BSA protein (Fig. 7B).
FIGURE 6.
BBA70-bound plasmin degrades complement component C3b. Microtiter plates were coated with recombinant, hexahistidine-tagged BBA70, ErpP, BBA66, and BSA (10 μg/ml). Following incubation with 10 μg/ml plasminogen, uPA (2.5 μg/ml) and purified C3b (5 μg/ml) were added. Microtiter plates were incubated at 37 °C for 24 h, and aliquots were taken at different time intervals. Following separation by SDS-PAGE, proteins were blotted onto nitrocellulose membranes, and C3b and its degradation products were detected, employing a polyclonal goat antiserum raised against C3b and a corresponding HRP-conjugated secondary antibody. Representative results of two independent experiments are shown. A, degradation of C3b by plasmin bound to BBA70 (left) and ErpP (right) after 0.5, 1, 5, and 24 h. Arrows, C3b α- and β-chain; arrowheads, C3b degradation products with a molecular mass of ∼30 and 40 kDa. B, results for BBA66 (left) and BSA (right). C3b, purified human C3b. Negative controls include the addition of 50 mm tranexamic acid (+T) as well as omission of plasminogen (−Plg).
FIGURE 7.
Degradation of C5 by plasmin bound to BBA70. Recombinant BBA70, ErpP, BBA66, and BSA (20 μg/ml) were immobilized onto microtiter plates and incubated with 20 μg/ml plasminogen. After washing, a reaction mixture containing 0.16 μg/ml activator uPA and 5 μg/ml C5 was added. Microtiter plates were incubated at 37 °C, and aliquots were taken at various time intervals. The mixtures were then separated via SDS-PAGE, and C5 as well as its degradation products were detected with a polyclonal goat anti-C5 antiserum and a corresponding anti-goat HRP-conjugated secondary antibody. Representative results of two separate experiments are shown. Arrows, α- and β-chain of C5; arrowhead, degradation product of C5 α-chain of ∼87 kDa. A, a visible degradation product appears after 24 h of incubation for plasmin bound to both BBA70 (left) and ErpP (right). B, no degradation products could be observed for BBA66 (left) or BSA (right). C5, purified human C5. Negative controls include the addition of 50 mm tranexamic acid (+T) as well as omission of plasminogen (−Plg).
BBA70-bound Plasmin Inhibits Bactericidal Activity of Complement
To further investigate the relevance of the observed degradation of purified C3b and C5, we employed an assay using NHS instead of purified complement proteins. Because borreliae produce a number of plasminogen-binding proteins, and a strain lacking all of these proteins is not available, we opted to use E. coli DH5α for these experiments. NHS was preincubated with BBA70-bound, activated plasmin. Following preincubation, serum-sensitive E. coli DH5α cells were incubated with the serum samples for 2 h at 37 °C, and cfu were determined. Plasmin bound to BBA70 and ErpP inhibited the bactericidal activity of complement, as shown by the survival rates of the sensitive E. coli DH5α cells of nearly 100% (Fig. 8, A and B) compared with the control incubated without NHS. By contrast, no statistically significant survival was observed for E. coli DH5α when NHS was preincubated with ZQA70 (Fig. 8C), which does not bind plasminogen (data not shown).
FIGURE 8.
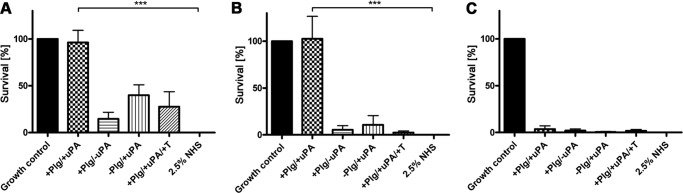
BBA70-bound plasmin inhibits bacteriolytic effects of complement. 10 μg recombinant BBA70, ErpP, or ZQA70 was bound to magnetic beads and incubated with plasminogen (Plg). After washing, 2.5% NHS was added together with a final concentration of 0.16 μg/ml uPA, and the serum was preincubated with the beads at 37 °C. After 1 h, the treated serum samples were incubated with 1 × 107 complement-sensitive E. coli DH5α cells. Following incubation at 37 °C for 2 h, serial dilutions were plated on agar plates, and cfu were determined the following day. DH5α cells incubated in GVB++ (growth control) without serum were presumed to have a survival rate of 100%. As a negative control, E. coli DH5α cells were incubated in the presence of 2.5% NHS, resulting in complete killing of the cells. A, for serum preincubated with BBA70-bound plasmin, E. coli show nearly 100% survival, whereas no statistically significant survival is seen for controls omitting either uPA or plasminogen or when adding 50 mm tranexamic acid to plasminogen (+T). B, when serum was preincubated with ErpP-bound plasmin, E. coli show 100% survival. C, no statistically significant survival was observed for E. coli when we used ZQA70, which does not bind plasminogen. Error bars, S.E.
Cellular Localization of BBA70
Many B. burgdorferi genes are not expressed by cultured bacteria, so we investigated whether the BBA70-encoding gene is expressed in culture. Total RNA was isolated from cultured spirochetes and subjected to RT-PCR. When using primers specific for the bba70 gene, an appropriately sized amplicon was produced (Fig. 9A). The flaB and ospA genes served as positive controls.
FIGURE 9.
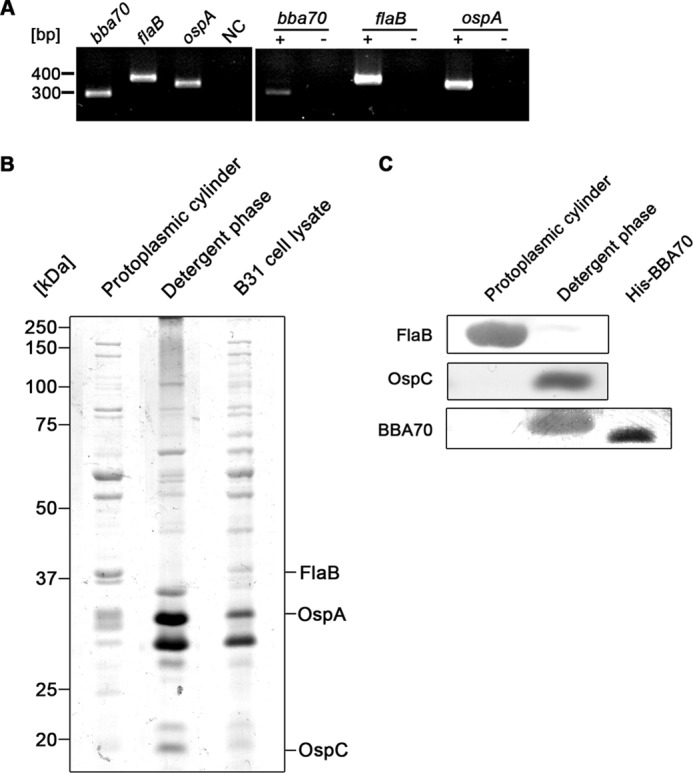
bba70 gene expression and membrane localization of BBA70 in vitro. A, total genomic RNA from a B. burgdorferi B31 culture in mid-exponential growth was isolated and reverse transcribed into cDNA. PCR was performed using cDNA as template to amplify products corresponding to the BBA70-encoding bba70 gene as well as the constitutively expressed flaB gene and the temperature-regulated ospA gene. The left panel shows PCR amplicons generated from B. burgdorferi B31 genomic DNA. Amplicons generated from cDNA are shown in the right panel. +, reverse transcriptase has been added to RT-PCR; −, omission of reverse transcriptase. B, Triton X-114 phase separation was performed with B. burgdorferi B31 cells grown to mid-exponential phase. Following phase separation, 10 μg of total protein from the fraction corresponding to the protoplasmic cylinder, 10 μg of protein from the detergent phase, and 10 μg of whole cell lysate of B. burgdorferi B31 were subjected to SDS-PAGE and stained with colloidal Coomassie. C, 10 μg of total protein from the fraction corresponding to the protoplasmic cylinder, the detergent phase, as well as 250 ng of recombinant, hexahistidine-tagged BBA70 (positive control) were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was then probed with mAb LA22.1 to detect periplasmic FlaB. mAb 93-193/0246 was used to detect the outer surface protein OspC, and mAb 70-11/10.17 was employed for the detection of BBA70.
Localization of the BBA70 protein in B. burgdorferi was addressed by Triton X-114 extraction (49, 50). Samples from the detergent phase containing the outer membrane protein-enriched fraction and the periplasmic cylinder as well as whole cell lysate from in vitro cultured spirochetes were separated by SDS-PAGE and stained with colloidal Coomassie (Fig. 9B). Proteins were also blotted onto a nitrocellulose membrane, and the presence of BBA70 in each fraction was detected using monoclonal antibody 70-11/10.17 (Fig. 9C). In addition, control blots were performed using monoclonal antibodies specific for the inner membrane-anchored FlaB or the outer membrane protein OspC. As expected, FlaB could only be detected in the fraction corresponding to the protoplasmic cylinder, whereas the outer surface protein OspC partitioned almost exclusively to the detergent phase. The BBA70 protein was detected only in the detergent fraction, indicating that this molecule is a component of the spirochetal outer membrane.
BBA70 Is Produced during Mammalian Infection
Line blot immunoassays were performed with sera from mice that had been infected with B. burgdorferi B31 via tick bite. Using a microdispersion device, nitrocellulose membranes were sprayed with various amounts of recombinant BBA70 protein (2–500 ng) to allow for optimization of both sensitivity and specificity. In addition, the known serological markers OspC and VlsE were included as positive control antigens, whereas BSA was used as a negative control. Sera from infected mice showed reactivity against the OspC and VlsE antigens but not BSA. Sera from the infected mice showed visible signals at 4 ng of recombinant BBA70, and 63 ng of recombinant BBA70 yielded strong signals, indicating that mice produced a robust antibody response directed against BBA70 (Fig. 10). In contrast, control serum obtained from an uninfected mouse did not exhibit activity against any of the borrelial antigens.
FIGURE 10.
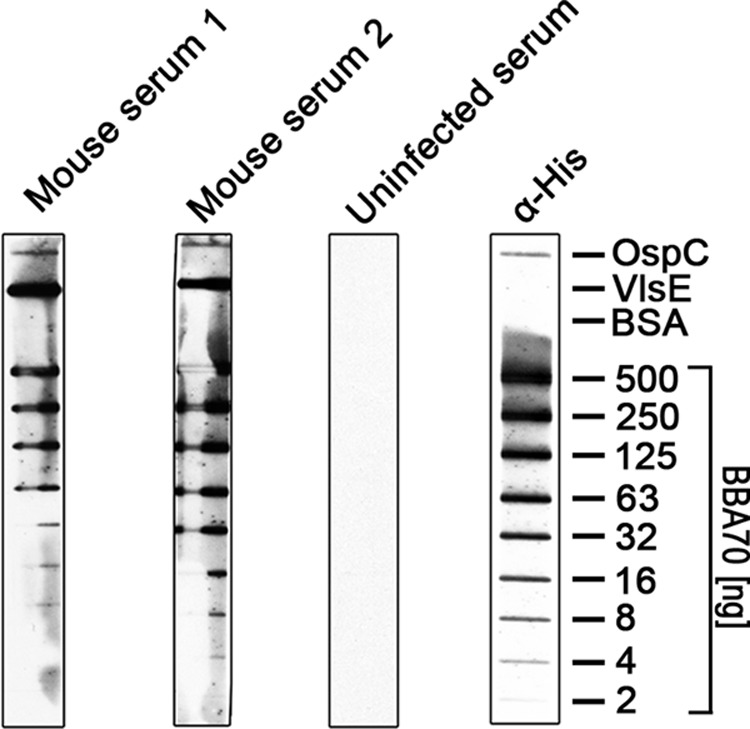
Immune sera from B. burgdorferi-infected mice show reactivity to BBA70. Varying amounts of recombinant, hexahistidine-tagged BBA70 as well as 87.5 ng of the control antigen OspC and 0.8 ng of VlsE were transferred onto a nitrocellulose membrane via microdispersion. 500 ng of BSA was used as a nonspecific negative control. Membranes were incubated with immune sera from two experimentally infected mice as well as serum from a control mouse. Reactivity of mouse immune sera to BBA70 and control antigens was detected using HRP-conjugated donkey anti-mouse immunoglobulins. Sera of infected mice show reactivity to both control antigens OspC and VlsE as well as to BBA70.
DISCUSSION
In the present study, we show that the PFam54 protein BBA70 is a plasminogen-binding protein. Binding of plasminogen is dose-dependent and mediated by lysine residues, primarily located in a putative C-terminal α-helix of the BBA70 protein. The interaction between plasminogen and BBA70 requires kringle domain 4 of plasminogen. We also demonstrated that BBA70-bound plasminogen is accessible to human uPA, can be activated to plasmin, and will cleave the natural substrate fibrinogen. BBA70 might also play a role in complement inactivation because BBA70-bound plasmin cleaved complement components C3b and C5. Furthermore, the bactericidal activity of complement was inhibited when NHS was preincubated with plasmin bound to BBA70. Consistent with these biochemical functions, the BBA70 protein is located in the borrelial outer membrane and is produced by B. burgdorferi during mammalian infection.
Binding of plasminogen represents an important virulence mechanism for a number of human pathogenic microbes. Several plasminogen-binding proteins have been characterized, including Tuf from P. aeruginosa (23), the PE protein of Haemophilus influenzae (59), Lsa20 from Leptospira interrogans (60), enolases from S. pneumoniae and Neisseria meningitidis (61, 62), and Gpm1p as well as Pra1 from the invasive yeast C. albicans (26, 27). For B. burgdorferi, a number of plasminogen-binding proteins have been reported, including OspA (31) and OspC (32), BPBP (30), and more recently B. burgdorferi enolase (34–36). Of note, the functionally related but genetically heterologous complement regulator-acquiring surface proteins CspA, CspZ, ErpP, ErpC, and ErpA all bind plasminogen in addition to binding various host-derived complement regulators (28, 29, 63–66). Other pathogenic microorganisms have proteins that also bind both plasminogen and complement regulators, such as factor H, factor H-like protein 1, factor H-related protein-1, or C4b-binding protein (23, 26, 27, 67–69). Notably, although BBA70 is orthologous to the borrelial factor H- and plasminogen-binding CspA protein, BBA70 is unable to bind any tested complement regulators (52).
Analysis of plasminogen binding to BBA70 indicates a high affinity interaction in the nanomolar range of Kd following a complex model with fast and slow components of kon and koff rate constants. Thus, kinetics data suggest that BBA70 binds to Glu-plasminogen in a two-stage process. Presumably, in the first stage, BBA70 interacts with the accessible kringle domain 1 of Glu-plasminogen (70) and promotes conformational changes resulting in unmasking of kringle domain 4 of Glu-plasminogen. In the second stage, the binding of BBA70 is stabilized via high affinity contacts with kringle domain 4 of plasminogen.
Changes in the conformational state of plasminogen as a result of interaction with other molecules, such as fibrin, have already been described previously (71, 72) and by analogy can be suggested for the BBA70 binding mechanism to plasminogen. The binding constant for BBA70 lies in a similar range of affinities reported for other plasminogen-binding proteins, such as B. burgdorferi enolase with a Kd of 125 nm (34), staphylokinase from S. aureus with a reported Kd of 9.3 nm (9), and DnaK and enolase from Bifidobacterium animalis with Kd values of 11 and 42 nm, respectively (73, 74).
Plasminogen interacts with a number of components of the fibrinolytic system via lysine binding sites, located within the kringle domains of the plasminogen molecule (7, 9). Numerous borrelial plasminogen-binding proteins bind plasminogen through lysine residues (11, 28, 29, 75), and the addition of the lysine analog tranexamic acid significantly reduced binding of plasminogen. Interestingly, although lysine residues play a role in the BBA70-plasminogen interaction, altering ionic strength with NaCl does not appear to affect plasminogen binding. By contrast, when NaBr is used to alter the ionic strength, a significant reduction of plasminogen binding by BBA70 is observed. Although it has been previously shown for other borrelial plasminogen-binding proteins (29) that changes in ionic strength using NaCl do not impact plasminogen binding, the literature also has numerous examples where the opposite is seen, and alterations in ionic strength using NaCl do affect plasminogen binding (20, 59). It is possible that changing ionic strength with NaCl in our experiments is not rigorous enough, due to the fact that plasminogen specifically binds chloride anions. NaBr thus seems more suited to alter ionic strength, and taken together, the results suggest that the BBA70-plasminogen interaction is dependent on ionic strength. Binding studies with recombinant plasminogen fragments determined that the kringle 4 domain has an essential role in the plasminogen-BBA70 interaction. Notably, this particular domain was shown to be important for the interaction with streptokinase (76). Prediction of the protein's secondary structure revealed that BBA70 consists of seven α-helices (designated A–G in Fig. 3A), and 17% of the total amino acids of BBA70 are lysine residues. Truncation of the putative C-terminal α-helix (G) resulted in abrogation of plasminogen binding, a finding that is consistent with the observation that C-terminal lysine residues also play a role in other bacterial plasminogen-binding proteins, such as α-enolase of S. pneumoniae (77).
BBA70-bound active plasmin degraded the physiological substrate fibrinogen. Recruitment of plasmin(ogen) to the borrelial surface is an important step in the pathogenesis of Lyme disease because thus far, no predicted endogenous, extracytoplasmic proteases have been reported for B. burgdorferi (78), and immobilization of plasmin(ogen) facilitates dissemination and penetration of endothelial cell monolayers (38, 40). In addition to degradation of ECM constituents, active plasmin degrades the central complement components C3b and C5 (20). BBA70-bound plasmin degraded C3b. This observation is similar to the activity of plasmin bound to the PE protein of H. influenzae (59). Similarly, plasmin-coated L. interrogans interferes with C3b and IgG, potentially decreasing opsonization of that pathogen (79). Furthermore, BBA70-bound plasmin cleaved the α-chain of C5. In addition to the degradation of purified complement components C3b and C5, BBA70-bound plasmin was able to inhibit the bacteriolytic activity of complement. Thus, binding of plasmin(ogen) might not be solely a means of invasion and dissemination but may also be a strategy for immune evasion by B. burgdorferi and other pathogenic microorganisms.
At least nine plasminogen-binding proteins have been described for B. burgdorferi, providing a large degree of functional redundancy. Because borreliae lack surface-exposed proteases of their own, they have to rely on coopting host proteases like plasminogen. Having multiple plasminogen-binding proteins provides a number of advantages. Given the complex enzootic life cycle of the Lyme disease spirochete, the various plasminogen-binding proteins may be produced at distinct stages of infection. For example, OspA is expressed primarily within the tick, whereas OspC expression is transient as the spirochetes are transmitted from the tick to the vertebrate host (80–85). In addition, various plasminogen-binding proteins may be expressed at different time periods during the infection. CspA is expressed during the initial stages of mammalian infection. By contrast, CspZ, ErpP, ErpC, and ErpA are expressed during established mammalian infection (86). A previous study investigating expression of the BBA70-encoding gene during persistent infection of mice reported no detectable bba70 transcripts in ear tissues, although specific antibody responses to BBA70 were demonstrated at late stages of murine infection (87). Similarly, the present study demonstrated that sera from infected mice contain robust levels of BBA70-targeting antibodies. Another advantage of having multiple plasminogen-binding proteins in a single mircroorganism could be explained by differences in affinity. While in the blood stream, at plasminogen concentrations of 200 μg/ml, low affinity binding proteins may be sufficient for recruitment of plasminogen to the spirochetal surface, whereas high affinity binding proteins may be required in compartments where the concentration of plasminogen is very low. In conclusion, our data suggest a dual role for binding of plasmin(ogen) by borreliae (Fig. 11), enabling efficient dissemination of the pathogens and possibly supporting evasion of the host innate immune response via cleavage of complement components.
FIGURE 11.
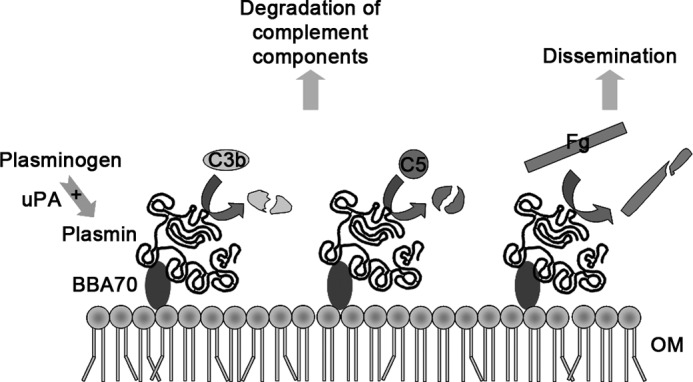
Possible roles of plasminogen binding by BBA70. Hypothetical model of the mechanisms by which human plasminogen bound to BBA70 might contribute to pathogen dissemination by degradation of components of the ECM and contribute to complement inactivation. BBA70 associated with the outer membrane (OM) of B. burgdorferi B31 is able to interact with plasminogen. BBA70-bound plasminogen is accessible to plasminogen activators, such as uPA, and converted to the active serine protease plasmin. Plasmin bound to BBA70 can cleave fibrinogen (Fg) and may thus contribute to efficient dissemination of borreliae within the host. In addition, BBA70-bound plasmin was able to cleave complement components C3b and C5 and inhibit bacteriolytic activity of complement. Based on our investigation, we suggest the BBA70-plasmin(ogen) interaction might also contribute to the spirochetes' evasion of innate immunity.
Acknowledgments
We are grateful to Jessica Günnewig and Axel Teegler for skillful and expert technical assistance. We also thank Ivan Dikic for providing access to the Biacore instrument at Goethe University, Frankfurt. D. P. acknowledges Thomas Meier and Klaas M. Pos for generous support of research.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant Kr3383/1-2 (to P. K.).
- ECM
- extracellular matrix
- uPA
- urokinase-type plasminogen activator
- NHS
- normal human serum
- Fg
- fibrinogen
- ANOVA
- analysis of variance
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Radolf J. D., Caimano M. J., Stevenson B., Hu L. T. (2012) Of ticks, mice and men. Understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steere A. C., Coburn J., Glickstein L. (2004) The emergence of Lyme disease. J. Clin. Invest. 113, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salazar J. C., Pope C. D., Sellati T. J., Feder H. M., Jr., Kiely T. G., Dardick K. R., Buckman R. L., Moore M. W., Caimano M. J., Pope J. G., Krause P. J., Radolf J. D. (2003) Coevolution of markers of innate and adaptive immunity in skin and peripheral blood of patients with erythema migrans. J. Immunol. 171, 2660–2670 [DOI] [PubMed] [Google Scholar]
- 4. Steere A. C. (2001) Lyme disease. N. Engl. J. Med. 345, 115–125 [DOI] [PubMed] [Google Scholar]
- 5. Goguen J. D., Hoe N. P., Subrahmanyam Y. V. (1995) Proteases and bacterial virulence. A view from the trenches. Infect. Agents Dis. 4, 47–54 [PubMed] [Google Scholar]
- 6. Ponting C. P., Marshall J. M., Cederholm-Williams S. A. (1992) Plasminogen. A structural review. Blood Coagul. Fibrinolysis 3, 605–614 [PubMed] [Google Scholar]
- 7. Wiman B., Lijnen H. R., Collen D. (1979) On the specific interaction between the lysine-binding sites in plasmin and complementary sites in α2-antiplasmin and in fibrinogen. Biochim. Biophys. Acta 579, 142–154 [DOI] [PubMed] [Google Scholar]
- 8. Danø K., Andreasen P. A., Grøndahl-Hansen J., Kristensen P., Nielsen L. S., Skriver L. (1985) Plasminogen activators, tissue degradation, and cancer. Adv. Cancer Res. 44, 139–266 [DOI] [PubMed] [Google Scholar]
- 9. Lijnen H. R., De Cock F., Van Hoef B., Schlott B., Collen D. (1994) Characterization of the interaction between plasminogen and staphylokinase. Eur. J. Biochem. 224, 143–149 [DOI] [PubMed] [Google Scholar]
- 10. Young K. C., Shi G. Y., Wu D. H., Chang L. C., Chang B. I., Ou C. P., Wu H. L. (1998) Plasminogen activation by streptokinase via a unique mechanism. J. Biol. Chem. 273, 3110–3116 [DOI] [PubMed] [Google Scholar]
- 11. Lähteenmaki K., Kuusela P., Korhonen T. K. (2001) Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25, 531–552 [DOI] [PubMed] [Google Scholar]
- 12. Salonen E. M., Zitting A., Vaheri A. (1984) Laminin interacts with plasminogen and its tissue-type activator. FEBS Lett. 172, 29–32 [DOI] [PubMed] [Google Scholar]
- 13. Salonen E. M., Saksela O., Vartio T., Vaheri A., Nielsen L. S., Zeuthen J. (1985) Plasminogen and tissue-type plasminogen activator bind to immobilized fibronectin. J. Biol. Chem. 260, 12302–12307 [PubMed] [Google Scholar]
- 14. Preissner K. T. (1990) Specific binding of plasminogen to vitronectin. Evidence for a modulatory role of vitronectin on fibrin(ogen)-induced plasmin formation by tissue plasminogen activator. Biochem. Biophys. Res. Commun. 168, 966–971 [DOI] [PubMed] [Google Scholar]
- 15. Kost C., Benner K., Stockmann A., Linder D., Preissner K. T. (1996) Limited plasmin proteolysis of vitronectin. Characterization of the adhesion protein as morpho-regulatory and angiostatin-binding factor. Eur. J. Biochem. 236, 682–688 [DOI] [PubMed] [Google Scholar]
- 16. Whitelock J. M., Murdoch A. D., Iozzo R. V., Underwood P. A. (1996) The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. J. Biol. Chem. 271, 10079–10086 [DOI] [PubMed] [Google Scholar]
- 17. Walport M. J. (2001) Complement. Second of two parts. N. Engl. J. Med. 344, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 18. Thurman J. M., Holers V. M. (2006) The central role of the alternative complement pathway in human disease. J. Immunol. 176, 1305–1310 [DOI] [PubMed] [Google Scholar]
- 19. Pillemer L., Ratnoff O. D., Blum L., Lepow I. H. (1953) The inactivation of complement and its components by plasmin. J. Exp. Med. 97, 573–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barthel D., Schindler S., Zipfel P. F. (2012) Plasminogen is a complement inhibitor. J. Biol. Chem. 287, 18831–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ullberg M., Kronvall G., Wiman B. (1989) New receptor for human plasminogen on gram positive cocci. APMIS 97, 996–1002 [DOI] [PubMed] [Google Scholar]
- 22. Kuusela P., Saksela O. (1990) Binding and activation of plasminogen at the surface of Staphylococcus aureus. Increase in affinity after conversion to the Lys form of the ligand. Eur. J. Biochem. 193, 759–765 [DOI] [PubMed] [Google Scholar]
- 23. Kunert A., Losse J., Gruszin C., Hühn M., Kaendler K., Mikkat S., Volke D., Hoffmann R., Jokiranta T. S., Seeberger H., Moellmann U., Hellwage J., Zipfel P. F. (2007) Immune evasion of the human pathogen Pseudomonas aeruginosa. Elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 179, 2979–2988 [DOI] [PubMed] [Google Scholar]
- 24. Ullberg M., Kronvall G., Karlsson I., Wiman B. (1990) Receptors for human plasminogen on gram-negative bacteria. Infect. Immun. 58, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ringnér M., Valkonen K. H., Wadström T. (1994) Binding of vitronectin and plasminogen to Helicobacter pylori. FEMS Immunol. Med. Microbiol. 9, 29–34 [DOI] [PubMed] [Google Scholar]
- 26. Luo S., Poltermann S., Kunert A., Rupp S., Zipfel P. F. (2009) Immune evasion of the human pathogenic yeast Candida albicans. Pra1 is a Factor H, FHL-1 and plasminogen binding surface protein. Mol. Immunol. 47, 541–550 [DOI] [PubMed] [Google Scholar]
- 27. Poltermann S., Kunert A., von der Heide M., Eck R., Hartmann A., Zipfel P. F. (2007) Gpm1p is a factor H-, FHL-1-, and plasminogen-binding surface protein of Candida albicans. J. Biol. Chem. 282, 37537–37544 [DOI] [PubMed] [Google Scholar]
- 28. Hallström T., Haupt K., Kraiczy P., Hortschansky P., Wallich R., Skerka C., Zipfel P. F. (2010) Complement regulator-acquiring surface protein 1 of Borrelia burgdorferi binds to human bone morphogenic protein 2, several extracellular matrix proteins, and plasminogen. J. Infect. Dis. 202, 490–498 [DOI] [PubMed] [Google Scholar]
- 29. Brissette C. A., Haupt K., Barthel D., Cooley A. E., Bowman A., Skerka C., Wallich R., Zipfel P. F., Kraiczy P., Stevenson B. (2009) Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77, 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu L. T., Pratt S. D., Perides G., Katz L., Rogers R. A., Klempner M. S. (1997) Isolation, cloning, and expression of a 70-kilodalton plasminogen binding protein of Borrelia burgdorferi. Infect. Immun. 65, 4989–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuchs H., Wallich R., Simon M. M., Kramer M. D. (1994) The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc. Natl. Acad. Sci. U.S.A. 91, 12594–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lagal V., Portnoï D., Faure G., Postic D., Baranton G. (2006) Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC-plasminogen affinity. Microbes Infect. 8, 645–652 [DOI] [PubMed] [Google Scholar]
- 33. Önder Ö., Humphrey P. T., McOmber B., Korobova F., Francella N., Greenbaum D. C., Brisson D. (2012) OspC is potent plasminogen receptor on surface of Borrelia burgdorferi. J. Biol. Chem. 287, 16860–16868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Floden A. M., Watt J. A., Brissette C. A. (2011) Borrelia burgdorferi enolase is a surface-exposed plasminogen binding protein. PLoS ONE 6, e27502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nogueira S. V., Smith A. A., Qin J. H., Pal U. (2012) A surface enolase participates in Borrelia burgdorferi-plasminogen interaction and contributes to pathogen survival within feeding ticks. Infect. Immun. 80, 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toledo A., Coleman J. L., Kuhlow C. J., Crowley J. T., Benach J. L. (2012) The enolase of Borrelia burgdorferi is a plasminogen receptor released in outer membrane vesicles. Infect. Immun. 80, 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coleman J. L., Roemer E. J., Benach J. L. (1999) Plasmin-coated Borrelia burgdorferi degrades soluble and insoluble components of the mammalian extracellular matrix. Infect. Immun. 67, 3929–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coleman J. L., Sellati T. J., Testa J. E., Kew R. R., Furie M. B., Benach J. L. (1995) Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 63, 2478–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gebbia J. A., Coleman J. L., Benach J. L. (2001) Borrelia spirochetes upregulate release and activation of matrix metalloproteinase gelatinase B (MMP-9) and collagenase 1 (MMP-1) in human cells. Infect. Immun. 69, 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coleman J. L., Gebbia J. A., Piesman J., Degen J. L., Bugge T. H., Benach J. L. (1997) Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 41. Hartmann K., Corvey C., Skerka C., Kirschfink M., Karas M., Brade V., Miller J. C., Stevenson B., Wallich R., Zipfel P. F., Kraiczy P. (2006) Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol. Microbiol. 61, 1220–1236 [DOI] [PubMed] [Google Scholar]
- 42. Kraiczy P., Skerka C., Brade V., Zipfel P. F. (2001) Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69, 7800–7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wallich R., Pattathu J., Kitiratschky V., Brenner C., Zipfel P. F., Brade V., Simon M. M., Kraiczy P. (2005) Identification and functional characterization of complement regulator-acquiring surface protein 1 of the Lyme disease spirochetes Borrelia afzelii and Borrelia garinii. Infect. Immun. 73, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cordes F. S., Roversi P., Kraiczy P., Simon M. M., Brade V., Jahraus O., Wallis R., Skerka C., Zipfel P. F., Wallich R., Lea S. M. (2005) A novel fold for the factor H-binding protein BbCRASP-1 of Borrelia burgdorferi. Nat. Struct. Mol. Biol. 12, 276–277 [DOI] [PubMed] [Google Scholar]
- 45. Kramer M. D., Schaible U. E., Wallich R., Moter S. E., Petzoldt D., Simon M. M. (1990) Characterization of Borrelia burgdorferi associated antigens by monoclonal antibodies. Immunobiology 181, 357–366 [DOI] [PubMed] [Google Scholar]
- 46. Bykowski T., Woodman M. E., Cooley A. E., Brissette C. A., Brade V., Wallich R., Kraiczy P., Stevenson B. (2007) Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect. Immun. 75, 4227–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jutras B. L., Liu Z., Brissette C. A. (2010) Simultaneous isolation of Ixodidae and bacterial (Borrelia spp.) genomic DNA. Curr. Protoc. Microbiol., Chapter 1, Unit 1E2 [DOI] [PubMed] [Google Scholar]
- 48. Brissette C. A., Rossmann E., Bowman A., Cooley A. E., Riley S. P., Hunfeld K. P., Bechtel M., Kraiczy P., Stevenson B. (2010) The borrelial fibronectin-binding protein RevA is an early antigen of human Lyme disease. Clin. Vaccine Immunol. 17, 274–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Herzberger P., Siegel C., Skerka C., Fingerle V., Schulte-Spechtel U., Wilske B., Brade V., Zipfel P. F., Wallich R., Kraiczy P. (2009) Identification and characterization of the factor H and FHL-1 binding complement regulator-acquiring surface protein 1 of the Lyme disease spirochete Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 299, 141–154 [DOI] [PubMed] [Google Scholar]
- 50. Radolf J. D., Chamberlain N. R., Clausell A., Norgard M. V. (1988) Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent Triton X-114. Infect. Immun. 56, 490–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Casjens S., Palmer N., van Vugt R., Huang W. M., Stevenson B., Rosa P., Lathigra R., Sutton G., Peterson J., Dodson R. J., Haft D., Hickey E., Gwinn M., White O., Fraser C. M. (2000) A bacterial genome in flux. The twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35, 490–516 [DOI] [PubMed] [Google Scholar]
- 52. Kraiczy P., Rossmann E., Brade V., Simon M. M., Skerka C., Zipfel P. F., Wallich R. (2006) Binding of human complement regulators FHL-1 and factor H to CRASP-1 orthologs of Borrelia burgdorferi. Wien. Klin. Wochenschr. 118, 669–676 [DOI] [PubMed] [Google Scholar]
- 53. Anglés-Cano E. (1994) Overview on fibrinolysis. Plasminogen activation pathways on fibrin and cell surfaces. Chem. Phys. Lipids 67, 353–362 [DOI] [PubMed] [Google Scholar]
- 54. Plow E. F., Herren T., Redlitz A., Miles L. A., Hoover-Plow J. L. (1995) The cell biology of the plasminogen system. FASEB J. 9, 939–945 [DOI] [PubMed] [Google Scholar]
- 55. Lottenberg R., Minning-Wenz D., Boyle M. D. (1994) Capturing host plasmin(ogen). A common mechanism for invasive pathogens? Trends Microbiol. 2, 20–24 [DOI] [PubMed] [Google Scholar]
- 56. Castellino F. J., McCance S. G. (1997) The kringle domains of human plasminogen. Ciba Found. Symp. 212, 46–60; discussion 60–45 [DOI] [PubMed] [Google Scholar]
- 57. Lähteenmäki K., Edelman S., Korhonen T. K. (2005) Bacterial metastasis. The host plasminogen system in bacterial invasion. Trends Microbiol. 13, 79–85 [DOI] [PubMed] [Google Scholar]
- 58. Lijnen H. R., Collen D. (1995) Mechanisms of physiological fibrinolysis. Bailliere's Clin. Haematol. 8, 277–290 [DOI] [PubMed] [Google Scholar]
- 59. Barthel D., Singh B., Riesbeck K., Zipfel P. F. (2012) Haemophilus influenzae uses the surface protein E to acquire human plasminogen and to evade innate immunity. J. Immunol. 188, 379–385 [DOI] [PubMed] [Google Scholar]
- 60. Mendes R. S., Von Atzingen M., de Morais Z. M., Gonçales A. P., Serrano S. M., Asega A. F., Romero E. C., Vasconcellos S. A., Nascimento A. L. (2011) The novel leptospiral surface adhesin Lsa20 binds laminin and human plasminogen and is probably expressed during infection. Infect. Immun. 79, 4657–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Knaust A., Weber M. V., Hammerschmidt S., Bergmann S., Frosch M., Kurzai O. (2007) Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J. Bacteriol. 189, 3246–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pancholi V., Fischetti V. A. (1998) α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273, 14503–14515 [DOI] [PubMed] [Google Scholar]
- 63. Hammerschmidt C., Hallström T., Skerka C., Wallich R., Stevenson B., Zipfel P. F., Kraiczy P. (2012) Contribution of the infection-associated complement regulator-acquiring surface protein 4 (ErpC) to complement resistance of Borrelia burgdorferi. Clin. Dev. Immunol. 2012, 349657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kraiczy P., Skerka C., Kirschfink M., Brade V., Zipfel P. F. (2001) Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and Factor H. Eur. J. Immunol. 31, 1674–1684 [DOI] [PubMed] [Google Scholar]
- 65. Kraiczy P., Hartmann K., Hellwage J., Skerka C., Kirschfink M., Brade V., Zipfel P. F., Wallich R., Stevenson B. (2004) Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 293, 152–157 [DOI] [PubMed] [Google Scholar]
- 66. Stevenson B., El-Hage N., Hines M. A., Miller J. C., Babb K. (2002) Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins. A possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70, 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Domingos R. F., Vieira M. L., Romero E. C., Gonçales A. P., de Morais Z. M., Vasconcellos S. A., Nascimento A. L. (2012) Features of two proteins of Leptospira interrogans with potential role in host-pathogen interactions. BMC Microbiol. 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C., Zipfel P. F. (2007) The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J. Immunol. 178, 5848–5858 [DOI] [PubMed] [Google Scholar]
- 69. Verma A., Brissette C. A., Bowman A. A., Shah S. T., Zipfel P. F., Stevenson B. (2010) Leptospiral endostatin-like protein A is a bacterial cell surface receptor for human plasminogen. Infect. Immun. 78, 2053–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bányai L., Patthy L. (1984) Importance of intramolecular interactions in the control of the fibrin affinity and activation of human plasminogen. J. Biol. Chem. 259, 6466–6471 [PubMed] [Google Scholar]
- 71. Weisel J. W., Petersen I. C. (1996) Plasminogen conformations. Ukr. Biokhim. Zh. 68, 41. [PubMed] [Google Scholar]
- 72. Weisel J. W., Nagaswami C., Korsholm B., Petersen L. C., Suenson E. (1994) Interactions of plasminogen with polymerizing fibrin and its derivatives, monitored with a photoaffinity cross-linker and electron microscopy. J. Mol. Biol. 235, 1117–1135 [DOI] [PubMed] [Google Scholar]
- 73. Candela M., Biagi E., Centanni M., Turroni S., Vici M., Musiani F., Vitali B., Bergmann S., Hammerschmidt S., Brigidi P. (2009) Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155, 3294–3303 [DOI] [PubMed] [Google Scholar]
- 74. Candela M., Centanni M., Fiori J., Biagi E., Turroni S., Orrico C., Bergmann S., Hammerschmidt S., Brigidi P. (2010) DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156, 1609–1618 [DOI] [PubMed] [Google Scholar]
- 75. Boyle M. D., Lottenberg R. (1997) Plasminogen activation by invasive human pathogens. Thromb. Haemost. 77, 1–10 [PubMed] [Google Scholar]
- 76. Joshi K. K., Nanda J. S., Kumar P., Sahni G. (2012) Substrate kringle-mediated catalysis by the streptokinase-plasmin activator complex. Critical contribution of kringle-4 revealed by the mutagenesis approaches. Biochim. Biophys. Acta 1824, 326–333 [DOI] [PubMed] [Google Scholar]
- 77. Bergmann S., Rohde M., Chhatwal G. S., Hammerschmidt S. (2001) α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40, 1273–1287 [DOI] [PubMed] [Google Scholar]
- 78. Klempner M. S., Noring R., Epstein M. P., McCloud B., Hu R., Limentani S. A., Rogers R. A. (1995) Binding of human plasminogen and urokinase-type plasminogen activator to the Lyme disease spirochete, Borrelia burgdorferi. J. Infect. Dis. 171, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 79. Vieira M. L., de Morais Z. M., Vasconcellos S. A., Romero E. C., Nascimento A. L. (2011) In vitro evidence for immune evasion activity by human plasmin associated to pathogenic Leptospira interrogans. Microb. Pathog. 51, 360–365 [DOI] [PubMed] [Google Scholar]
- 80. Crother T. R., Champion C. I., Whitelegge J. P., Aguilera R., Wu X. Y., Blanco D. R., Miller J. N., Lovett M. A. (2004) Temporal analysis of the antigenic composition of Borrelia burgdorferi during infection in rabbit skin. Infect. Immun. 72, 5063–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hodzic E., Feng S., Freet K. J., Barthold S. W. (2003) Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 71, 5042–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hodzic E., Feng S., Freet K. J., Borjesson D. L., Barthold S. W. (2002) Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 70, 3382–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liang F. T., Nelson F. K., Fikrig E. (2002) Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Montgomery R. R., Malawista S. E., Feen K. J., Bockenstedt L. K. (1996) Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo. Exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tilly K., Krum J. G., Bestor A., Jewett M. W., Grimm D., Bueschel D., Byram R., Dorward D., Vanraden M. J., Stewart P., Rosa P. (2006) Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74, 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bykowski T., Woodman M. E., Cooley A. E., Brissette C. A., Wallich R., Brade V., Kraiczy P., Stevenson B. (2008) Borrelia burgdorferi complement regulator-acquiring surface proteins (BbCRASPs). Expression patterns during the mammal-tick infection cycle. Int. J. Med. Microbiol. 298, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hughes J. L., Nolder C. L., Nowalk A. J., Clifton D. R., Howison R. R., Schmit V. L., Gilmore R. D., Jr., Carroll J. A. (2008) Borrelia burgdorferi surface-localized proteins expressed during persistent murine infection are conserved among diverse Borrelia spp. Infect. Immun. 76, 2498–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]