Background: Derlins are a family of membrane proteins involved in ER-associated degradation with unknown mechanism.
Results: Derlin2 is an integral component of the HRD1-SEL1L complex required for retro-translocation of SHH and NHK, independent of its glycosylation status.
Conclusion: Derlin2 plays an essential role in HRD1-mediated retro-translocation of SHH and NHK.
Significance: DERL2 but not DERL1 or DERL3 is required for retro-translocation of HRD1-mediated SHH and NHK.
Keywords: Endoplasmic Reticulum (ER), ER-associated Degradation, Protein Degradation, Protein Translocation, Protein Turnover, HRD1, Derlin, Retro-translocation, Sonic Hedgehog
Abstract
Endoplasmic reticulum-associated degradation (ERAD) is an important system that eliminates misfolded proteins from the ER. Three derlins have been implicated in this process, but their precise function remains unknown. In this study, we report that although both derlin1 and derlin2 are capable of binding the ERAD-specific ubiquitin ligase HRD1, they associate with the HRD1-containing complex with different affinities. Accordingly, these derlins have nonredundant functions in ERAD with derlin2 being an essential functional partner for HRD1-mediated ERAD of SHH and NHK. We show that derlin2, but not derlin1 or derlin3, is required for ERAD of both glycosylated and nonglycosylated SHH, as well as NHK. Derlin2 appears to act at a post-targeting step for HRD1-dependent retro-translocation. Without derlin2, the assembly of HRD1 into a functional retro-translocation homo-oligomer proceeds normally, and substrate targeting to the HRD1 complex also occurs. However, the ERAD substrate SHH-C is largely trapped inside the ER lumen. These observations raise the possibility that derlin2 may regulate the movement of substrates through the HRD1-containing retro-translocon. Our study is the first to report that derlin2 functions with HRD1 in ERAD of certain substrates independent of their glycosylation status. The mammalian ERAD system may require multiple derlins that each functions with a distinct E3 partner to eliminate a specific subset of substrates. This is different from the model in Saccharomyces cerevisiae, in which Hrd1p alone is sufficient for retro-translocation.
Introduction
Proteins that misfold in the endoplasmic reticulum (ER)2 are cleared through a ubiquitin-proteasome-dependent mechanism known as ER-associated degradation (ERAD) (1). Many misfolded proteins are recognized by chaperones and lectins in the ER and delivered to the ubiquitin ligase HRD1 complex in the ER membrane (1, 2). By a not yet understood mechanism, the substrates are retro-translocated from the ER lumen to the cytosol, ubiquitinated by the RING (really interesting gene) domain of HRD1, extracted by the AAA (ATPase associated with diverse cellular functions) ATPase p97, and targeted to the proteasome for degradation. When overexpressed, Hrd1p (HMG-CoA reductase degradation), the homolog of HRD1 in Saccharomyces cerevisiae, can carry out the retro-translocation function in the absence of any known interacting partners (3), suggesting that it plays an essential role in retro-translocation. However, the identity of the retro-translocon(s) and the precise mechanism of retro-translocation remain unclear.
Derlins are a family of membrane proteins involved in ERAD (4, 5). Although the interactions of derlins with other retro-translocation machineries have been characterized, the role of derlins in ERAD remains unclear. Studies in S. cerevisiae revealed that Der1p, one of the two derlin orthologs, is part of the Hrd1p membrane complex required for the degradation of ERAD substrates with luminal lesions, but not of substrates with membrane lesions (6–9). It links to the Hrd1p-Hrd3p complex through Usa1p (10, 11). As demonstrated by in vivo cross-linking studies, Der1p also interacts directly with an ERAD substrate, and the interaction is independent of Hrd1p, its associated components Hrd3p, and Yos9p, an ER lectin (3, 12). Thus, Der1p is proposed to function in substrate binding and delivery to a putative retro-translocon containing Hrd1p (3, 12). Dfm1p, the other derlin ortholog in S. cerevisiae, does not appear to function in ERAD (9, 13).
In mammalian cells, three derlins, derlin1 (DERL1), derlin2 (DERL2), and derlin3 (DERL3), have been identified. DERL1 is involved in virally induced degradation of major histocompatibility complex (MHC) class I through interacting with the transmembrane domain (TMD) of the human cytomegaloviral protein US11 (4, 5). DERL1, but not DERL2, also facilitates the retro-translocation of cholera toxin (14). On the other hand, DERL2 and DERL3 are more homologous to each other than to DERL1 (15, 16). Oda et al. reported that DERL2 and DERL3 are involved in the degradation of null Hong Kong (NHK), a glycosylated luminal ERAD substrate, but not its nonglycosylated variant (15). It is unclear whether the glycan dependence is applicable to other ERAD substrates. In addition, Oda et al. showed that DERL1 does not associate with ERAD substrate or other derlins (15). However, other studies reported that DERL1 and DERL2 can co-immunoprecipitate with each other and with the HRD1 complex (16, 17), although more DERL2 precipitates with HRD1 and/or SEL1L than DERL1 (16, 18). Given the controversy (15–18), the function of different derlins in ERAD remains to be clarified. The mechanism that controls the differential requirement of derlins for ERAD is also unknown. Furthermore, how derlins affect the process of retro-translocation is to be investigated.
Previously, we discovered that sonic hedgehog (SHH) undergoes an autocleavage reaction in the ER, resulting in an N-terminal fragment for signaling and a C-terminal fragment (19). The C-terminal fragment of SHH (SHH-C) contains an N-glycan and is degraded by an ERAD pathway requiring HRD1, SEL1L, and p97 (19). To understand the roles of derlins in ERAD, we have evaluated the contribution of the individual derlin for ERAD of SHH-C and its nonglycosylated variant N278A. Interestingly, we found that only DERL2 is essential for ERAD of SHH-C, independent of its glycosylation status. Further analysis revealed that retro-translocation and subsequent ubiquitination of SHH proteins do not occur when DERL2 is absent. The substrates are trapped inside the ER with a fraction of it in complex with the HRD1-SEL1L complex. The results reveal a critical role for DERL2 in HRD1-mediated retro-translocation of SHH-C.
EXPERIMENTAL PROCEDURES
Materials
The following materials were used in this study. Anti-HA was purchased from Roche Applied Science. Anti-p97, Myc, GAPDH, HRD1, calnexin,and calreticulin antibodies were from GeneTex Inc., Taiwan. Anti-SEL1L, XTP3-B, and ubiquitin antibodies were from Santa Cruz Biotechnology. Anti-OS-9 antibodies were from Abcam (Novus). Anti-FLAG and M2 beads were from Sigma. Anti-VIMP was from Abnova. Anti-BiP antibodies were from BD Biosciences. Protein G-Sepharose beads were from GE Healthcare. Anti-DERL1 antibody was described previously (4). The DERL2 antibody was generated using the peptides NH2-EERPGGFAWGEGQRLGG-COOH and NH2-KAIFDTPDPNYN-COOH. Anti-HERP antibody was a generous gift from Dr. K. Kokame (National Cerebral and Cardiovascular Center, Suita, Osaka, Japan). Stealth siRNA duplexes were custom synthesized by Invitrogen. TransIT-siQUEST was from Mirus. MG132 was from Enzo Life Science. DSS (disuccinimidyl suberate), DSP (dithiobis-succinimidyl propionate), and EGS (ethylene glycolbis-succinimidyl succinate) were purchased from Thermo Scientific.
DNA Constructs for Mammalian Cell Transfection
Full-length SHH with an HA tag in pIRES-EGFP (Takara Bio, Inc.) was described before (19). Expression plasmids coding for DERL2-FLAG and WASP-FLAG were generated by PCR from human cDNA libraries and cloned to pIRES-EGFP vector. Site-directed mutagenesis was carried out using the QuikChange kit (Stratagene) to generate N278A on SHH and mutations/truncations on DERL2-FLAG. Different expression plasmids for HRD1 plasmids were kindly provided by Billy Tsai (University of Michigan, Ann Arbor, MI) (14).
Cell Culture, Generation of Stable Cells, and Transfection of Plasmids and siRNAs
Human 293T cells were grown in DMEM supplemented with 10% fetal bovine serum, 0.1% penicillin, and streptomycin. To generate DERL2-FLAG stable cell lines, constructs encoding DERL2-FLAG in pIRES-EGFP vector were transfected to 293T cells and selected for cells expressing the proteins by flow cytometry. The expression of the DERL2-FLAG proteins was confirmed by Western blot analysis with both FLAG and DERL2 antibodies. Transfection of plasmids was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs were carried out as described (19). siRNA duplexes were transfected using a transfection reagent (TransIT-siQUEST) at a final concentration of 50 nm siRNA according to the manufacturer's instructions. siRNA transfection was performed twice, on days 1 and 3 of the experiments. On day 5, the cells were harvested, and the cell lysates were prepared. Proteins were analyzed by SDS-PAGE and immunoblotting. siRNA sequences used are shown in Ref. 19 and Table 1. Except in Fig. 1 as labeled, siRNA oligonucleotide DERL2-A was used in the rest of the RNA interference studies. Plasmid coding for NHK was a kind gift from Dr. N. Hosokawa, Kyoto University, Kyoto, Japan.
TABLE 1.
Sequences of siRNA duplexes used in this study
| Gene | Nucleotides | Primers (forward and reverse) |
|---|---|---|
| DERL1 | 3′ | CAACUGCGUUCUGGCUAACACUGUU |
| AACAGUGUUAGCCAGAACGCAGUUG | ||
| DERL2-A | 381 | AGGCCUUUACAAUAAUGCUCGUCUA |
| UAGACGAGCAUUAUUGUAAAGGCCU | ||
| DERL2-B | 435 | UUGAGAAGGCCGAAGAAGUUCAUGC |
| GCAUGAACUUCUUCGGCCUUCUCAA | ||
| DERL3-A | 447 | UCAACUUCUUCGGCCUGCUCACUUU |
| AAAGUGAGCAGGCCGAAGAAGUUGA | ||
| DERL3-B | 309 | CCGACUUCGUCUUCAUGUUUCUCUU |
| AAGAGAAACAUGAAGACGAAGUCGG | ||
| VIMP | 418 | CAGAAGAUUGAAAUGUGGGACAGCA |
| UGCUGUCCCACAUUUCAAUCUUCUG | ||
| HRD1–7 | 159 | CCUACUACCUCAAACACCAGUUCUA |
| UAGAACUGGUGUUUGAGGUAGUAGG | ||
| SEL1L-2 | 450 | GAAACCAGCUUUGACCGCCAUUGAA |
| UUCAAUGGCGGUCAAAGCUGGUUUC |
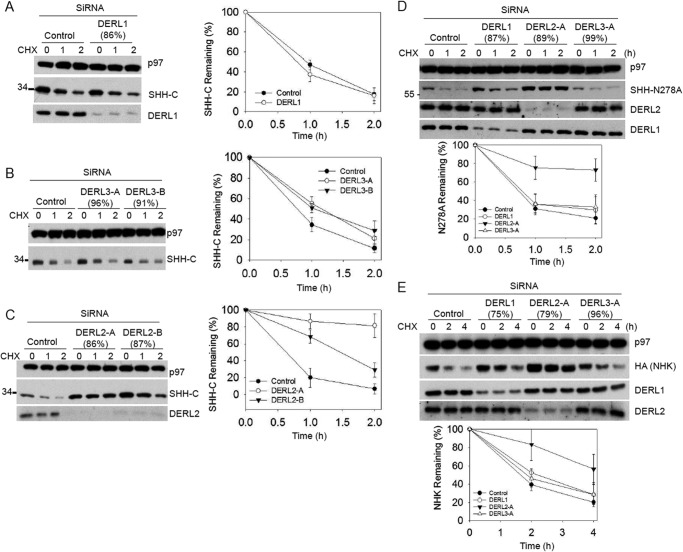
FIGURE 1.
DERL2 is required for the degradation of SHH-C, SHH N278A, and NHK. The extent of the depletion (in parentheses) was determined by quantitative RT-PCR (shown in parentheses on top of the figure) and immunoblotting if the antibodies were available. Immunoblotting with p97 antibodies was used as loading control. A, 293T cells stably expressing SHH-HA were depleted of DERL1, and the fate of glycosylated SHH-C proteins was followed after cycloheximide (CHX) addition. Controls were treated with a negative control siRNA duplex (Invitrogen). All samples were analyzed by SDS-PAGE and immunoblotting with HA antibodies. The right graph shows quantification of SHH-C in the experiment. Error bars, S.E. B is as in A, but with depletion of DERL3 using two different siRNA oligonucleotides. C is as in A, but with depletion of DERL2 using two different siRNA oligonucleotides. D is as in A, but with 293T cells stably expressing N278A-HA and depletion of DERL1, DERL2, and DERL3 by siRNA oligonucleotides. E is as in D, but with 293T cells stably expressing NHK.
Cycloheximide Chase Assay
A cycloheximide chase assay was carried out as described (19). Cells were cultured in 6-well plates and incubated with 100 μg/ml cycloheximide in culture medium at 37 °C. At the times indicated in the figures, cells were removed and treated on ice for 10 min in lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1 mm EDTA supplemented with protease inhibitors and 1% Triton X-100. The lysate was centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant was collected and denatured in SDS-PAGE sample buffer before separation on SDS-PAGE.
Quantitative RT-PCR Analysis
Total RNA was extracted with TRIzol reagent, and cDNA was synthesized with reverse transcriptase (ImProm-IITM; Promega). Quantitative PCR was performed on an ABI Prism 7900 cycler using SYBR Green PCR Master Mix (Applied Biosystems). The degree of siRNA knockdown was calculated relative to HPRT (hypoxanthine guanine phosphoribosyl transferase) mRNA levels. The primers used to quantify mRNA knockdown were shown in our previous publication (19) and Table 2.
TABLE 2.
Primer sequences for quantitative PCR used in this study
| Homo sapiens gene | Nucleotides | Primers (forward and reverse) |
|---|---|---|
| DERL2 | 358–491 | GTCTATGTGTGGAGCCGAAGGA |
| AACAAGGAAAATCCCATGAGCA | ||
| DERL3 | 314–459 | GCGTCCTTATGACCCTGCTGG |
| GAACGGTGCCTGGAAAGTGAGC | ||
| VIMP | 444–544 | CCTGAAACGGAAATCGGACA |
| CTCTGCGTCCAGGTCTCCAG |
Immunoblotting and Immunoprecipitation
For immunoprecipitation of any components of HRD1-SEL1L-DERL2-p97 complex, cells were lysed on ice for 20 min in HKM buffer (20 mm HEPES, pH 7.4, 100 mm KOAc, and 2 mm MgOAc) supplemented with protease inhibitors and 1% digitonin (or 1% deoxy-bigChap), unless indicated. The lysate was centrifuged for 20 min at 4 °C and 20,000 × g before immunoprecipitation. For detecting ubiquitinated SHH-HA, the cells were lysed in denaturing buffer with 0.8% SDS, 4 mm DTT, and 5 mg/ml N-ethylmaleimide with protease inhibitors. Afterward, the lysates were diluted 5-fold with denaturing buffer without SDS before immunoprecipitation. For interaction between derlins and ERAD substrates (see Fig. 2), the lysis and immunoprecipitation were carried out in the buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1% Nonidet P-40 with protease inhibitors as described (15).
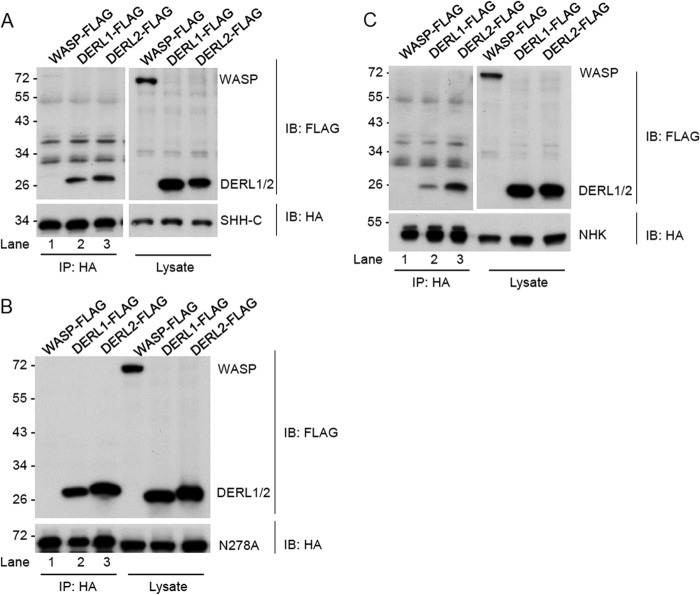
FIGURE 2.
Interaction of derlins with the ERAD substrates. A, 293T cells stably expressing SHH-HA precursor were transfected with FLAG-tagged DERL1, DERL2, or WASP plasmids as indicated. The cells were lysed in buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1% Nonidet P-40 with protease inhibitors and immunoprecipitated (IP) by HA antibodies. Precipitates were separated on SDS-PAGE and immunoblotted (IB) with FLAG antibodies to detect the interaction with SHH. B is as in A, but with cells stably expressing N278A proteins. C is as in A but with cells stably expressing NHK.
Chemical Cross-linking
Cross-linking with DSS, DSP, and EGS was carried out according to the manufacturers' instructions. Specifically, the cross-linkers were prepared and diluted to a final concentration of 1 mm for the treatment as suggested by the manufacturers. The cells on the culture dishes were washed by PBS first before the addition of the cross-linker. The cross-linker was directly added on top of the intact cells and incubated at room temperature for 1 h. 10–20 mm Tris-HCl, pH 7.5, was added to stop the reaction before the cells were harvested by centrifugation. The cell lysate were prepared and subjected to SDS-PAGE analysis followed by immunoblotting with HRD1 and DERL2 antibodies.
RESULTS
DERL2 Is Required for ERAD of HRD1 Substrates
To understand the function of derlins in ERAD, we first used siRNA to knock down individual derlins in cells stably expressing the glycosylated SHH. Whenever possible, immunoblotting was used to determine the knockdown efficiency. Otherwise, quantitative RT-PCR was used to monitor the mRNA depletion. Depletion of DERL1 did not affect the degradation of SHH-C (Fig. 1A), consistent with our previous study (19). Depletion of DERL3 using two different siRNA oligonucleotides also had no significant effect on SHH-C turnover (Fig. 1B). By contrast, two different siRNA oligonucleotides targeting DERL2 both significantly inhibited the degradation of glycosylated SHH-C (Fig. 1C).
To investigate the function of derlins in the ERAD of nonglycosylated substrate, we have generated a mutant SHH variant carrying a point mutation (N278A) at the predicted glycosylation site. Upon entry into the ER lumen, SHH undergoes a cholesterol-dependent cleavage (19), but the N278A mutation abolished this cleavage (Fig. 1D). Cycloheximide chase experiments showed that N278A mutants were similarly degraded by a HRD1-dependent mechanism (data not shown), suggesting that the N-glycan may not be important for ERAD of SHH. Depletion of individual derlin was carried out in cells stably expressing N278A proteins. As shown, depletion of DERL2 but not DERL1 or DERL3 inhibited the degradation of N278A (Fig. 1D).
To test whether the specific involvement of DERL2 in HRD1-dependent ERAD may be applicable to other ERAD substrates, we established a 293T cell line stably expressing a misfolded α1-antitrypsin variant, NHK, which was previously established as a HRD1 substrate (20). Once again, depletion of DERL2, but not DERL1 or DERL3, inhibited the degradation of NHK (Fig. 1E). Thus, DERL2, but not DERL1 or DERL3, is required for HRD1-dependent ERAD.
Interaction of Derlins with ERAD Substrates
We next wished to determine whether the function of DERL2 in ERAD involves specific substrate engagement. Because ERAD substrates are constantly transferred from one machinery protein to another during retro-translocation, the interaction of ERAD substrates with endogenous HRD1 components is difficult to detect in cells expressing low levels of substrates (15, 20, 21). Therefore, to detect the interaction of derlins with ERAD substrates, we transiently expressed FLAG-tagged derlins or as a control, an unrelated protein WASP (Wiskott-Aldrich syndrome protein) in 293T cells stably expressing the HA-tagged ERAD substrates. Cell extracts were subjected to immunoprecipitation with HA antibodies. As expected, all three substrates, SHH-C, N278A, and NHK, did not precipitate WASP proteins (Fig. 2A–C, lanes 1). By contrast, these substrates could readily precipitated DERL2 and DERL1 (Fig. 2A–C, lanes 2 and 3). Interestingly, DERL1 bound SHH-C and N278A with a slightly reduced affinity, but its interaction with NHK was significantly weaker than DERL2 (lane 2 versus 3). All together (Figs. 1 and 2), our results suggest that even though both DERL1 and DERL2 associate with SHH and NHK, only DERL2 plays a major role in ERAD of these substrates independent of their glycosylation status. In comparison, DERL1 is dispensable despite its ability to associate with these HRD1 substrates. DERL1 may tag along these ERAD substrates through the reported hetero-oligomerization with DERL2 (17).
DERL2 Is Part of the HRD1-SEL1L Retro-translocation Complex
Both SHH-C and N278A proteins require HRD1-SEL1L for their retro-translocation.3 (19). We therefore determined whether endogenous DERL2 is part of the HRD1-SEL1L complex in 293T cells using immunoprecipitation with HRD1 antibodies, or as a negative control, with IgG. Immunoblotting showed that HRD1 antibody, but not control IgG, pulled down HRD1 together with the known HRD1-associated components SEL1L and HERP as well as derlins. By contrast, the HRD1 antibody did not precipitate calnexin or the abundant ER chaperone BiP, demonstrating the specificity of the observed interactions. In accordance with the notion that DERL2 is a functional partner of HRD1, a larger fraction of DERL2 was detected in complex with HRD1 than DERL1 (Fig. 3A). The results are consistent with a previous report done with a different experimental setup (16, 17).
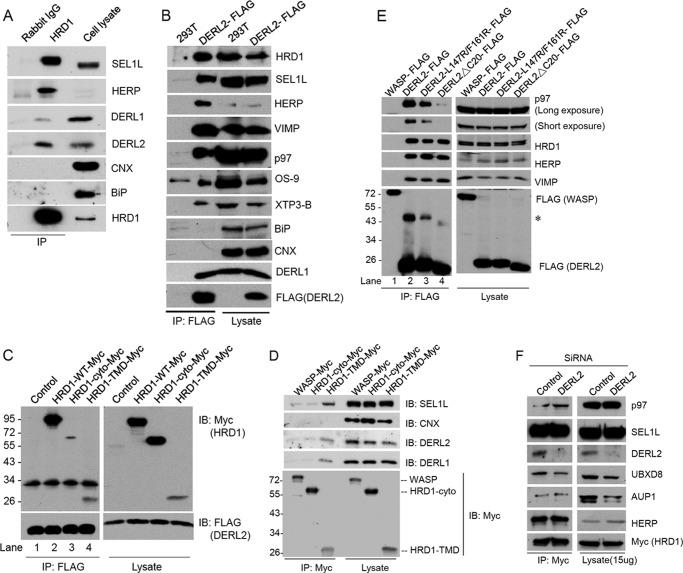
FIGURE 3.
DERL2 associates with HRD1-SEL1L complex requiring TMD of HRD1. A, 293T cells were lysed in HKM buffer with 1% deoxy-bigChap and protease inhibitors and immunoprecipitated (IP) with HRD1 antibodies. The precipitates were separated in SDS-PAGE and immunoblotted (IB) with the indicated antibodies. B is as in A but with 293T cells stably expressing DERL2-FLAG and immunoprecipitated with FLAG-M2 beads. Immunoblotting was carried out with the indicated antibodies. C, Myc-tagged HRD1 expression plasmids were transfected to 293T cells stably expressing DERL2-FLAG. The cells were lysed in HKM buffer with protease inhibitors and 1% digitonin and immunoprecipitated by FLAG-M2 beads. Precipitates were separated on SDS-PAGE and immunoblotted with FLAG or Myc antibodies. D, Myc-tagged HRD1 or WASP plasmids were transfected to 293T cells. The cells were lysed in HKM buffer with 1% Triton X-100 and protease inhibitors, immunoprecipitated by Myc antibodies, and immunoblotted with Myc, SEL1L, calnexin (CNX), DERL1, and DERL2 antibodies. E, 293T cells were transfected with various FLAG-tagged DERL2 plasmids as indicated. Cells were lysed, and immunoprecipitation was carried out with FLAG-M2 beads. Precipitates were separated on SDS-PAGE and immunoblotted with the indicated antibodies. The asterisk (*) marks the dimeric DERL2. F, 293T cells stably expressing Myc-HRD1 proteins were depleted of DERL2 by siRNA oligonucleotides. The cells were lysed and immunoprecipitated using Myc antibodies in HKM buffer containing 1% deoxy-bigChap. The immunoprecipitate and cell lysates were separated on SDS-PAGE and immunoblotted with the indicated antibodies.
To further test the interaction of DERL2 with the HRD1 complex, we established a 293T cell line stably expressing FLAG-tagged DERL2 at a level comparable with endogenous DERL2 (supplemental Fig. 1A). The expression of DERL2-FLAG did not affect the degradation of SHH-C, indicating that DERL2-FLAG is functional (supplemental Fig. 1B). The components that interact with DERL2 were analyzed by immunoprecipitation with FLAG antibody followed by immunoblotting. As anticipated, HRD1 and its associated ERAD components including SEL1L, HERP, VIMP, DERL1, and p97 were all readily co-precipitated with DERL2 (Fig. 3B). The soluble ER lectins OS-9 and XTP3-B were also co-precipitated with DERL2 (Fig. 3B), likely through their interactions with SEL1L (20, 22, 23). By contrast, the abundant ER chaperons such as calnexin and BiP were undetectable in the precipitates, indicating that the interactions detected for DERL2 were specific (Fig. 3B). The interaction of DERL2 with the HRD1-SEL1L complex could also be demonstrated by immunoprecipitation using SEL1L antibody (see below, Fig. 5C). These data indicate that DERL2 is an integral component of a HRD1-SEL1L-containing retro-translocation complex.
FIGURE 5.
DERL2 depletion does not affect the amount, stability, and composition of HRD1-SEL1L complex. A, 293T cells stably expressing SHH-HA were depleted of individual component of ERAD as indicated. The cells were lysed in buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100. Cell lysates were separated on SDS-PAGE and immunoblotted with the indicated antibodies. Immunoblotting with GAPDH antibodies was used as loading control. The extent of the depletion (in parentheses) was determined by quantitative RT-PCR (shown in parentheses on top of the figure). B is as in A but depleted by two different siRNA oligonucleotides. Immunoblotting with DERL2 antibodies was used as loading control. C, 293T cells were depleted of DERL2 by siRNA oligonucleotides. The cells were lysed and immunoprecipitated (IP) using SEL1L antibodies in HKM buffer containing 1% deoxy-bigChap. The immunoprecipitate and cell lysates were separated on SDS-PAGE and immunoblotted with the indicated antibodies.
To determine how DERL2 interacts with the HRD1 membrane complex, we analyzed the interaction of DERL2 with HRD1 and a few HRD1 truncation mutants carrying different portions of this E3 enzyme. HRD1-TMD contains only the TMDs of HRD1, whereas HRD1-cyto consists of the cytosolic portion of HRD1 including the catalytic RING finger (14). DERL2-FLAG-expressing cells were transfected with plasmids expressing Myc-tagged HRD1 variants. Immunoblotting showed that HRD1-TMD was expressed at a much lower level than full-length HRD1, but the relative level of HRD1-TMD co-precipitated with DERL2-FLAG was comparable with full-length HRD1 (Fig. 3C, lane 4 versus 2). By contrast, very little HRD1-cyto could be co-immunoprecipitated with DERL2 despite its high level of expression (Fig. 3C, lane 3). Reciprocal immunoprecipitation of HRD1-Myc further confirmed that HRD1 associates with DERL2 directly or indirectly through its TMD regions (Fig. 3D).
Interaction of DERL2 with p97
DERL1 contains a conserved SHP motif at the C-terminal region that mediates its interaction with p97, a large oligomeric ATPase complex proposed to extract polyubiquitinated ERAD substrates from the ER membrane (17, 24). A similar motif is present in DERL2. We constructed a mutant DERL2 lacking the SHP box by deleting the last 20 amino acids (DERL2-ΔC20) and tested whether this motif is required for p97 interaction by co-immunoprecipitation. As demonstrated previously, wild-type DERL2 but not an unrelated control protein WASP interacted with p97 and several previously identified HRD1-interacting partners such as HERP and VIMP (Fig. 3E, lane 2). By contrast, immunoprecipitation of DERL2-ΔC20 did not precipitate p97 (Fig. 3E, lane 4). However, this mutant still interacted with HRD1 and its associated protein HERP and VIMP similarly to wild type DERL2 (Fig. 3E, lane 4). Thus, the interaction of DERL2 with HRD1 is independent of p97 binding. Another DERL2 mutant that had two hydrophobic residues in the predicted TM5 domain replaced by charged Arg (L147R/F161R) still interacted with p97 and HRD1 (Fig. 3E, lane 3). Taken together, our data show that DERL2 interacts with p97 and HRD1 independent of each other.
To test whether DERL2 may serve as an adaptor in ERAD by linking the ubiquitination and substrate extraction machineries together, we investigated whether DERL2 depletion would abolish the interaction of p97 with HRD1-Myc. Co-immunoprecipitation experiments showed that the amount of p97 co-precipitated with HRD1-Myc was comparable in control and DERL2 knockdown cells, suggesting that DERL2 was not required for an association of HRD1 with p97 (Fig. 3F). However, our results did not exclude the possibility that DERL2 may modulate the interaction between p97 and HRD1 to coordinate their activities in ERAD.
DERL2 Is required for Retro-translocation of SHH-C
To understand how DERL2 facilitates ERAD, we wished to determine the step in ERAD that is specifically affected when DERL2 is depleted. We took advantage of the fact that SHH-C is a glycosylated protein that undergoes ubiquitination and deglycosylation once retro-translocated into the cytosol (19). Thus, deglycosylated SHH accumulated upon proteasome inhibition reflects the retro-translocation activity. To determine whether depletion of DERL2 affects retro-translocation of SHH, we first compared the percentage of deglycosylated SHH-C in proteasome-inhibited control and DERL2-depleted cells. HRD1 knockdown cells were also included in this study to serve as a positive control given the proposed role of HRD1 in retro-translocation. In control cells, ∼46% of SHH-C accumulated in the deglycosylated form after treatment with MG132 (Fig. 4, A and B). However, in HRD1-depleted cells, the deglycosylated SHH-C levels were reduced to ∼7% (Fig. 4, A and B). This is consistent with the proposed role of HRD1 in retro-translocation. Similar to HRD1 depletion, DERL2 knockdown also significantly reduced the amount of deglycosylated SHH-C (∼11%) (Fig. 4, A and B), indicating that DERL2 is required for HRD1-dependent retro-translocation of SHH-C. Consistent with this idea, less ubiquitinated SHH-C was detected in DERL2- and HRD1-depleted cells compared with control cells (Fig. 4C).
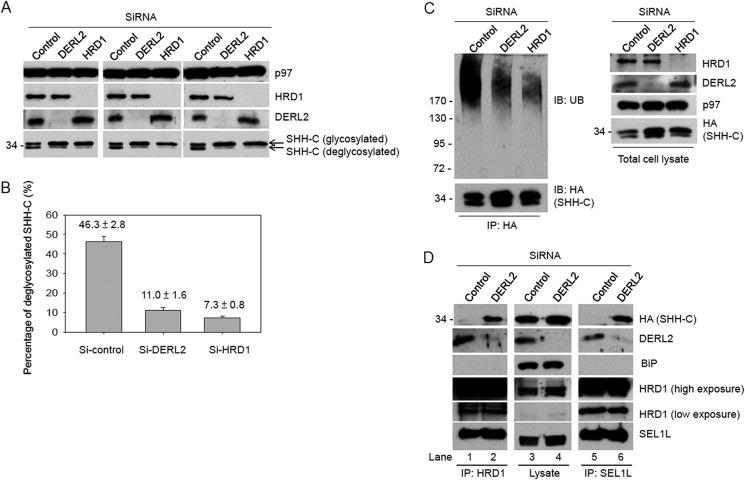
FIGURE 4.
Depletion of DERL2 traps the substrates inside ER. The extent of the depletion was carried out by immunoblotting with the indicated antibodies. A, cells stably expressing SHH-HA were depleted of DERL2 or HRD1 by siRNA. Cells were then treated with MG132 (150 mm) for 4 h before lysed in lysis buffer containing 1% Triton X-100 and protease inhibitors. Cell lysates were separated in SDS-PAGE and immunoblotted with HA antibodies. Immunoblotting with p97 was used as loading control. SHH-C, glycosylated or not, was as indicated. Three independent repeats are shown. B, quantifications of the percentage of deglycosylated SHH-C based on A were carried out and are shown. C is as in A, but lysed in denaturing buffer containing SDS (28). Immunoprecipitation (IP) was carried out with HA antibodies as described under “Experimental Procedures.” The precipitates were separated on Bis-Tris gradient gel and immunoblotted (IB) with ubiquitin (UB) antibodies. Total cell lysates were also separated on SDS-PAGE and immunoblotted with HRD1, DERL2, p97, and HA antibodies. Immunoblotting with p97 was used as loading control. D, cells stably expressing SHH-HA were depleted of DERL2 by siRNA oligonucleotides. The cells were lysed in HKM buffer containing 1% deoxy-bigChap. The cell lysates were immunoprecipitated with HRD1 or SEL1L antibodies. The precipitates and the total cell lysates were separated on SDS-PAGE and immunoblotted with the indicated antibodies.
DERL2 Acts at a Post-targeting Step to Promote ERAD
Lack of SHH-C retro-translocation in DERL2 knockdown cells might be due to defective substrate delivery to the HRD1-SEL1 complex. To test this possibility, we investigated whether DERL2 depletion affects the association of SHH-C with SEL1L or HRD1 in a cell line stably expressing SHH. SHH-C was barely detectable when cell extract from control cells was subject to immunoprecipitation with either HRD1 or SEL1L antibodies (Fig. 4D, lanes 1 and 5), consistent with the notion that ERAD substrates do not form stable interaction with the machinery proteins under normal condition. However, when DERL2 was depleted, a significant amount of SHH-C was detected in HRD1 or SEL1L precipitates (Fig. 4D, lanes 2 and 6), indicating that the substrate was delivered to the HRD1-SEL1L complex, but it fails to properly engage this retro-translocation machinery to be dislocated. Taken together (Fig. 4), we conclude that DERL2 acts at a post-targeting step to facilitate HRD1-dependent retro-translocation.
DERL2 Depletion Did Not Affect the Stability of ERAD Component and HRD1 Oligomerization
To understand how DERL2 depletion led to a retro-translocation defect, we investigated several possibilities. First, we tested whether DERL2 depletion changed the stability of individual retro-translocation components associated with HRD1. To this end, we depleted the indicated components including HRD1, SEL1L, VIMP, DERL1, DERL2, or p97 in cells stably expressing SHH precursors. Knockdown of these ERAD essential genes all resulted in the accumulation of SHH-C at the steady state due to inhibition of its degradation (Fig. 5A). In comparison, depletion of DERL1 or a nonrelated control siRNA had no effect on SHH degradation (Fig. 5A). Immunoblotting by antibodies against various ERAD components showed that depletion of DERL2 did not affect the steady-state levels of these proteins, nor their stability as demonstrated by chase experiments (Fig. 5A). By contrast, depletion of SEL1L with two different siRNA oligonucleotides significantly reduced the amount of HRD1 proteins (Fig. 5B). This result was different from a previous report (25), but consistent with the reported regulation of Hrd1p by Hrd3p (the SEL1L homolog) in S. cerevisiae (26, 27). Collectively, we concluded that DERL2 depletion did not affect ERAD by altering the stability of other HRD1-associated ERAD components.
We then determined whether DERL2 depletion affects the composition of the HRD1-SEL1L complex by immunoprecipitation of endogenous SEL1L from 293T cell extracts prepared from either control or DERL2 knockdown cells. In control cells, HRD1, DERL2, DERL1, and OS-9 were all detected in complex with SEL1L (Fig. 5C). BAG6, a soluble holdase working downstream of ERAD (28), was not stably associated with SEL1L pulldowns (Fig. 5C). The interactions of the SEL1L-HRD1 complex with other ERAD components were maintained in DERL2 knockdown cells, indicating that DERL2 is not required for the assembly of the HRD1-SEL1L complex.
In yeast, Hrd1p homo-oligomerization is essential for its activity during retro-translocation (3). In mammalian cells, exogenously expressed HRD1 can co-immunoprecipitate with endogenous HRD1 or overexpressed HRD1 bearing a different tag, suggesting that HRD1 oligomerization is evolutionarily conserved (14, 29). To determine whether DERL2 regulates HRD1 oligomerization, we first used chemical cross-linking to confirm that endogenous HRD1 forms homo-oligomers in mammalian cells. Upon treating cells with various cross-linkers (e.g. DSP, DSS, and EGS), a band corresponding to the size of HRD1 dimer was readily detected by immunoblotting using HRD1-specific antibodies (Fig. 6A, lanes 3, 5, and 7 versus 1). The identity of this cross-linking product was further verified by siRNA-mediated knockdown of HRD1, which abolished both HRD1 monomer and dimer (Fig. 6B). As expected, cross-linking by the disulfide-containing cross-linker DSP, but not DSS or EGS, could be reversed by treatment with the reducing agent DTT (Fig. 6A, lane 4 versus 6 and 8). siRNA-mediated depletion of DERL2 did not affect the cross-linking pattern of HRD1 (Fig. 6B), suggesting that DERL2 is not required for HRD1 homo-oligomerization. Collectively, these results rule out the possibility that DERL2 regulates the stability or the composition of the HRD1-SEL1L complex or the oligomerization of HRD1.
FIGURE 6.

Depletion of DERL2 does not affect oligomerization of HRD1. A, 293T cells were treated with chemical cross-linkers DSP, DSS, or EGS as indicated by the manuals provided by the manufacturers. Cell lysates were incubated with the sample buffers with or without DTT (50 mm) before being separated on SDS-PAGE. Immunoblotting (IB) with HRD1 antibodies was carried out. The asterisk (*) marks a nonspecific band. B, 293T cells were depleted of DERL2 or HRD1 by siRNA oligonucleotides. The cell lysates were prepared after EGS or DSS treatment to preserve the HRD1 dimers and separated on SDS-PAGE. Immunoblotting with HRD1 and DERL2 antibodies was carried out.
DISCUSSION
DERL1 and DERL2 are essential proteins in mammals. Mice lacking either of them die during embryonic development (30, 31). Given that the ERAD system is not essential in yeast, mammalian derlins likely regulate other essential pathways. How derlins act in ERAD and related cellular processes is unclear. In yeast, the derlin ortholog Der1p is required only for turnover of misfolded luminal substrate (6–9). In mammals, a handful of model ERAD substrates have been discovered, but their degradations are mostly unaffected by depletion of individual derlins, prompting the idea that derlins may play redundant roles in protein quality control at the ER. Lack of a robust derlin substrate and/or the potential functional redundancy has hampered our understanding of the functions of this important class of cellular factors.
In this study, we report novel ERAD substrates that require specifically DERL2, but not DERL1 or DERL3, for degradation. Our study is the first to report that a nonglycosylated ERAD substrate (SHH N278A) also requires DERL2 for degradation. The finding provides a new tool for future study of DERL2-dependent ERAD of nonglycosylated proteins. Our study shows that a small amount of DERL1 is present in the HRD1 complex together with ERAD substrates, consistent with a previous report (16). However, our data indicate that DERL1 is not functional in ERAD of HRD1 substrates in 293T cells (Fig. 1). DERL1 and DERL2 form a complex with each other, with ERAD substrates, and with HRD1. However, the association of DERL1 with HRD1 and the substrates is much weaker than that of DERL2 (Figs. 2 and 3A).
When viewed in the context of previous work (16, 17), our study demonstrates that these derlins are functionally different. Only DERL2 is required for ERAD of several HRD1 substrates investigated here, including SHH-C, SHH N278A, and NHK. DERL1 may function primarily through a HRD1-independent pathway. In accordance with this idea, DERL1 has been implicated in US11-mediated degradation of MHC class I protein (4, 5), which depends on an E3 ligase other than HRD1 (32). Our observation that DERL2, but not DERL1 or DERL3, has a more significant and specialized role in ERAD of NHK is different from a previous report, which established DERL3, in addition to DERL2, as a regulator of NHK degradation (15). In previous study, NHK was overexpressed by transient transfection, and the degradation kinetics was slow even for control cells (15). By contrast, in our study, we used a stable cell line that expresses NHK at a low level to avoid potential artifact of protein overexpression. The clearance of NHK was much more efficient, and the differential effect of derlin knockdown on the degradation of NHK was obvious and reproducibly observed (Fig. 1E). Moreover, our model is consistent with a previous report showing that DERL2 is the major derlin in complex with HRD1 (16). In further support of our conclusion, DERL2, but not DERL1, interacted preferentially with the HRD1-interacting proteins such as SEL1L and UBXD2 as well as with the HRD1 substrate NHK (Fig. 2C) (18).
Mammalian derlins may each act with a distinct E3 partner(s) to promote turnover of different substrates. In line with this view, a recent proteomic study reveals that DERL1 and DERL2 have very different interactomes (18). Interestingly, other than the HRD1 complex, DERL2 was also reported to interact with another ERAD-specific ubiquitin ligase gp78, which was confirmed by our study (supplemental Fig. 2 and Ref. 18). Whether DERL2 has a role for gp78-mediated ERAD awaits further investigation using gp78-specific substrates. The majority of endogenous derlins in cells appear to exist as homo-oligomers that have distinct E3 partners. On top of E3 diversity, different modes of E3 engagement as well as additional ERAD accessory proteins may also contribute to the division of labor for derlins.
We demonstrated that within the HRD1-SEL1L complex, DERL2 functions to facilitate the ERAD of SHH and NHK. DERL2 appears to be specifically involved at the step of retro-translocation because in its absence, the ERAD substrate SHH-C accumulates in the ER lumen as a glycosylated retro-translocation precursor that remains associated with the HRD1-SEL1L complex. Our results also suggest that upon docking of substrate at the HRD1-SEL1 complex, DERL2 may be required to move the substrates through the HRD1 retro-translocation complex. Regarding the role(s) of DERL2 in ERAD, there are several possibilities. DERL2 might be part of the retro-translocon essential for ERAD. Alternatively, DERL2 might prime ERAD substrates for subsequent retro-translocation, as suggested previously for DERL1 (17). It is also possible that DERL2 might serve as a gating factor that controls the entry of the substrates into the putative HRD1-containing retro-translocon. Future studies using purified proteins in an in vitro reconstituted ERAD assay would be required to define how DERL2 modulates the HRD1-SEL1L-p97 pathway to facilitate ERAD.
Our results suggest that DERL2 may interact with p97 through its C-terminal SHP box on the cytosolic side of the ER membrane. This function appears to be conserved among derlin family members in humans (17, 24). The association of DERL2 with the HRD1 complex may be mediated by the TMDs of these proteins because a HRD1 truncation mutant bearing just the TMDs is sufficient to interact with DERL2. Without DERL2, p97 still binds HRD1, suggesting that DERL2 is not necessary for the interaction (Fig. 3F). This is not entirely surprising because p97 has been shown to interact with several components of the HRD1-SEL1L complex including HRD1, VIMP, and UBXD8 (4, 17, 18, 33). On the other hand, without the SHP motif, the DERL2-ΔC20 mutant may interfere with binding to p97, even though it can still associate with HRD1.
Our results demonstrate that the retro-translocon(s) in mammalian cells is more complex than previously thought, requiring many factors, including DERL2, to work together with HRD1. This is in sharp contrast to the study from S. cerevisiae, in which overexpression of Hrd1p alone was sufficient for retro-translocation of an ERAD substrate from ER to cytosol without the presence of any other known ERAD components, such as Hrd3p, Usa1p, and Der1p (3). The findings in yeast argue against the model that the derlins form the pore of a retro-translocon. Recent structure modeling suggests that by itself, derlins are unlikely to be the elusive retro-translocon because they do not have a pore-like structure (24). On the other hand, several lines of evidence suggest that HRD1 is most likely the key component of this elusive retro-translocon at least for HRD1-mediated ERAD (3). Hrd1p was proposed to go through an oligomerization process during ERAD in S. cerevisiae (3). We detected endogenous HRD1 in oligomeric forms in mammalian cells (Fig. 6). It is not clear whether the change of oligomerization state of HRD1 occurs in mammalian cells. In vitro reconstitution of retro-translocation process will shed light on the composition and regulation of the retro-translocon.
Acknowledgments
We thank Dr. Pedro Carvalho for reading the manuscript and discussion; Dr. Ming-Jing Hwang, Dr. Ruey-Hwa Chen, Ching-Shu Suen, Dr. Mei-Ru Lin, Ya-Han Zhuang, and Chih-Ting Wang for discussions and help on this project.
This work was supported by NRPGM (National Research Program for Genomic Medicine), National Science Council, Taiwan and National Health Research Institutes, Taiwan, Republic of China.

This article contains supplemental Figs. 1 and 2.
H. Y. Tang, C. H. Huang, J. Christianson, Y. H. Zhuang, Y. R. Chu, and X. Chen, unpublished data.
- ER
- endoplasmic reticulum
- Bis-Tris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- DERL1
- derlin1
- DERL2
- derlin2
- DERL3
- derlin3
- DSP
- dithiobis-succinimidyl propionate
- DSS
- disuccinimidyl suberate
- EGS
- ethylene glycolbis-succinimidyl
- ERAD
- endoplasmic reticulum-associated degradation
- NHK
- null Hong Kong
- SHH
- sonic hedgehog
- SHH-C
- C-terminal fragment of SHH
- TMD
- transmembrane domain
- WASP
- Wiskott-Aldrich syndrome protein.
REFERENCES
- 1. Smith M. H., Ploegh H. L., Weissman J. S. (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoseki J., Ushioda R., Nagata K. (2010) Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 147, 19–25 [DOI] [PubMed] [Google Scholar]
- 3. Carvalho P., Stanley A. M., Rapoport T. A. (2010) Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 5. Lilley B. N., Ploegh H. L. (2004) A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 6. Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. (2003) Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278, 35903–35913 [DOI] [PubMed] [Google Scholar]
- 7. Carroll S. M., Hampton R. Y. (2010) Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J. Biol. Chem. 285, 5146–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vashist S., Ng D. T. (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato B. K., Hampton R. Y. (2006) Yeast derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast 23, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 10. Carvalho P., Goder V., Rapoport T. A. (2006) Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 11. Horn S. C., Hanna J., Hirsch C., Volkwein C., Schütz A., Heinemann U., Sommer T., Jarosch E. (2009) Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell 36, 782–793 [DOI] [PubMed] [Google Scholar]
- 12. Stanley A. M., Carvalho P., Rapoport T. (2011) Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocross-linking. FEBS Lett. 585, 1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hitt R., Wolf D. H. (2004) Der1p, a protein required for degradation of malfolded soluble proteins of the endoplasmic reticulum: topology and Der1-like proteins. FEMS Yeast Res. 4, 721–729 [DOI] [PubMed] [Google Scholar]
- 14. Bernardi K. M., Williams J. M., Kikkert M., van Voorden S., Wiertz E. J., Ye Y., Tsai B. (2010) The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol. Biol. Cell 21, 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006) Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lilley B. N., Ploegh H. L. (2005) Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W., Kopito R. R. (2012) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen X., Tukachinsky H., Huang C. H., Jao C., Chu Y. R., Tang H. Y., Mueller B., Schulman S., Rapoport T. A., Salic A. (2011) Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J. Cell Biol. 192, 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cormier J. H., Tamura T., Sunryd J. C., Hebert D. N. (2009) EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol. Cell 34, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. (2008) Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283, 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greenblatt E. J., Olzmann J. A., Kopito R. R. (2011) Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α1-antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 18, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iida Y., Fujimori T., Okawa K., Nagata K., Wada I., Hosokawa N. (2011) SEL1L protein critically determines the stability of the HRD1-SEL1L endoplasmic reticulum-associated degradation (ERAD) complex to optimize the degradation kinetics of ERAD substrates. J. Biol. Chem. 286, 16929–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. (2000) Endoplasmic reticulum degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plemper R. K., Bordallo J., Deak P. M., Taxis C., Hitt R., Wolf D. H. (1999) Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci. 112, 4123–4134 [DOI] [PubMed] [Google Scholar]
- 28. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kny M., Standera S., Hartmann-Petersen R., Kloetzel P. M., Seeger M. (2011) Herp regulates Hrd1-mediated ubiquitylation in a ubiquitin-like domain-dependent manner. J. Biol. Chem. 286, 5151–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eura Y., Yanamoto H., Arai Y., Okuda T., Miyata T., Kokame K. (2012) Derlin-1 deficiency is embryonic lethal, Derlin-3 deficiency appears normal, and Herp deficiency is intolerant to glucose load and ischemia in mice. PLoS One 7, e34298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dougan S. K., Hu C. C., Paquet M. E., Greenblatt M. B., Kim J., Lilley B. N., Watson N., Ploegh H. L. (2011) Derlin-2-deficient mice reveal an essential role for protein dislocation in chondrocytes. Mol. Cell. Biol. 31, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stagg H. R., Thomas M., van den Boomen D., Wiertz E. J., Drabkin H. A., Gemmill R. M., Lehner P. J. (2009) The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 186, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P. M., Seeger M. (2005) The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum-associated degradation pathway. J. Mol. Biol. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]