Background: The molecular basis of increased protein stability by N-glycosylation is incompletely understood.
Results: Glycosylation position rather than number is more important for protein thermostability in the xylanase A from Bacillus subtilis.
Conclusion: Glycans contribute both to stabilizing protein-glycan and less favorable glycan-glycan interactions. An extensive protein-glycan interface favors protein stability.
Significance: Formation of a protein-glycan interface provides a conceptual framework to understand glycoprotein stabilization.
Keywords: Glycoside Hydrolases, Glycosylation, Mass Spectrometry (MS), Protein Design, Protein Engineering, Protein Thermostability, Molecular Dynamics Simulation
Abstract
Protein glycosylation is a common post-translational modification, the effect of which on protein conformational and stability is incompletely understood. Here we have investigated the effects of glycosylation on the thermostability of Bacillus subtilis xylanase A (XynA) expressed in Pichia pastoris. Intact mass analysis of the heterologous wild-type XynA revealed two, three, or four Hex8–16GlcNAc2 modifications involving asparagine residues at positions 20, 25, 141, and 181. Molecular dynamics (MD) simulations of the XynA modified with various combinations of branched Hex9GlcNAc2 at these positions indicated a significant contribution from protein-glycan interactions to the overall energy of the glycoproteins. The effect of glycan content and glycosylation position on protein stability was evaluated by combinatorial mutagenesis of all six potential N-glycosylation sites. The majority of glycosylated enzymes expressed in P. pastoris presented increased thermostability in comparison with their unglycosylated counterparts expressed in Escherichia coli. Steric effects of multiple glycosylation events were apparent, and glycosylation position rather than the number of glycosylation events determined increases in thermostability. The MD simulations also indicated that clustered glycan chains tended to favor less stabilizing glycan-glycan interactions, whereas more dispersed glycosylation patterns favored stabilizing protein-glycan interactions.
Introduction
Glycoproteins are found in all cellular compartments with particular abundance both in cellular membranes and in extracellular secretions (1), and it has been estimated that over half the proteins in nature are glycosylated (2). Protein glycosylation is a complex process mediated by the sequential activity of at least 13 glycosyltransferases (3) and effectively generates structural diversity in proteins, thereby modulating their physicochemical and functional properties (3, 4). Glycosylation involves the covalent modification of either the amino side-chain group of Asn (N-glycosylation) or the hydroxyl group of Ser or Thr (O-glycosylation). Although N-glycosylation occurs at Asn residues in the Asn-Xaa-Ser/Thr recognition motif (where residue Xaa cannot be a proline), no consensus sequence has been identified in the case of O-glycosylation. The most common N-linked glycans consist primarily of N-acetylglucosamine and mannose or complex glycans containing GlcNAc, mannose, Gal, fucose, and sialic acid (3).
The biosynthesis, molecular composition, and functions of glycoproteins are well documented, and glycans can act as recognition markers (5–7); mediators of biomolecular interactions (8, 9); immune response modulators (10, 11); regulators of protein turnover and protein folding (12, 13); and markers for protein secretion (14), intracellular trafficking, and cell communication (13, 15). Due to their hydrophilicity and bulk, glycan modification can increase glycoprotein solubility, improve resistance against proteolysis, and reduce non-specific protein interactions (16). Furthermore, an auxiliary role for protein glycosylation in protein folding has been demonstrated (17, 18). Despite this wealth of information, in many cases, the answer to the central question of how glycosylation modulates protein function remains unclear.
Glycosylation plays a key role in the regulation of enzyme activity (19), where the covalent addition of glycans to the protein surface may modulate kinetic parameters and thermal stability (14). These effects demonstrate that glycosylation affects the protein energy landscape, and it has been suggested that protein stabilization by glycosylation may be due to an enthalpic effect resulting from interactions between the protein and the attached glycans (17, 20, 21). In silico studies using coarse-grained strategies corroborate the enthalpic stabilization of the Src-SH3 domain by different degrees of glycosylation (22). In contrast, other studies suggest that the stabilization might be due either to a “chaperone-like” activity of glycans (14, 17) or to hydrophobic collapse resulting from impaired solvation of the protein (23) and is largely entropic in origin (14, 17).
Knowledge of the molecular interactions between glycans, solvent, and polypeptide is the key to understanding the structural basis of glycoprotein function. Several experimental methods have been employed to investigate protein glycosylation, including x-ray crystallography and NMR spectroscopy. However, conformational flexibility of protein-linked glycans frequently hampers crystallization efforts (24). Moreover, when crystals are available, their quality is adversely affected by the high thermal motion of the glycans, compromising the reliability of molecular geometry determination beyond the rigid glycan core (25). NMR methods provide an ensemble of conformations; however, attempts to determine the complete conformational space of glycoproteins by this method can give rise to ambiguous interpretations (26).
An alternative approach uses molecular dynamics (MD)2 simulations to gain insights as to the structure, conformation, and dynamic aspects of glycoproteins. Molecular modeling methodologies, including MD simulations, are emerging as promising tools for structural and conformational investigations of glycoproteins and glycans with a high degree of accuracy not only in the spatial and time components but also in the prediction of geometry, flexibility, and intermolecular interactions (27, 28). Although MD simulation allows a description of the conformational properties of these molecules, only a limited number of glycoproteins have so far been studied using this method (23, 29, 30).
The methylotrophic yeast Pichia pastoris has been widely used as a cellular host for the expression of glycosylated recombinant proteins (31). As a eukaryote, P. pastoris offers an expression system for producing soluble, correctly folded proteins and conserves most of the post-translational glycosylation pathway. In contrast to glycoproteins from higher eukaryotes, P. pastoris glycans contain outer chains consisting primarily of mannose oligomers (32). Furthermore, glycoproteins expressed in P. pastoris generally have much shorter glycosyl chains than those expressed in S. cerevisiae, thus making P. pastoris an attractive host for the expression of recombinant glycoproteins (33).
The depolymerization of plant cell wall xylans by thermostable xylanases is of significant interest, with applications in the paper and cellulose industry and in the bioconversion of lignocellulosic material and agro-industrial wastes into fermentable sugars. Endo-β-1,4-xylanase A (XynA) is a mesostable GH11 glycosyl hydrolase secreted by Bacillus subtilis, and protein engineering studies have identified multiple factors that contribute to thermostabilization of the protein, including optimization of hydrogen bonds (34), hydrophobic interactions (35), optimization of the electrostatic surface of proteins (36), and the introduction of disulfide bonds (37). Here we present a novel approach to improve protein thermostability by rational design, in which the XynA expressed in P. pastoris has been used to investigate the effect of glycosylation on protein stability. We here describe significant differences in thermostability between enzymes with different glycan content and glycosylation position. These results have been correlated with MD simulations, which demonstrate that particular glycosylation patterns enhance glycan interactions with the protein surface and that this modulation of the protein-glycan interface can increase the thermostability of the protein.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Site-directed Mutagenesis
Genomic DNA of the PAP1158 strain from B. subtilis (BGSC code 168/168) was used as the template for PCR amplification of the gene encoding XynA using primers XylPN 5′-GGCAGCCTCGAGAAAAGAGAGGCTGAAGCTAGCACAGACTAC-3′ and XylPC 5′-GATCAGAATTCCGTTAGCTACCC-3′. The PCR product was digested with XhoI and EcoRI (recognition sites underlined in the oligonucleotide sequences) and cloned into plasmid pPIC9K (Invitrogen), modified to introduce a polyhistidine C-terminal tag (pPIC9K_CT) and create an in-frame fusion with the α-factor secretion signal. The vector was modified to introduce a C-terminal tag polyhistidine to the recombinant protein. The resulting pPIC9K plasmid containing the B. subtilis XynA is referred to as pPIC_BsXA.
Site-directed mutagenesis of the XynA was performed by PCR mutagenesis (38) to introduce the single mutations N8Q (5′-ACTGGCAACAGTGGACTGAT-3′), N20Q (5′-AAACGCTGTCCAGGGG TCT GGC-3′), N25Q (5′- CTGGCGGGCAGTACAGTGTT-3′), N29Q (5′-ACAGTGTTCAGTGGTCTAAT-3′), N141Q (5′-CGAAGAGACCAACCGGAAGCCAAGCTACA-3′), and N181Q (5′- GGAAGTTCTCAAGTAACAGTG-3′) (base changes underlined), and all constructs were fully sequenced to confirm the intended base changes. The XynA coding sequences containing single mutants were used as templates for additional cycles of site-directed mutagenesis to combine and accumulate mutations. The wild-type XynA and all mutant coding sequences were also subcloned into the expression vector pT7BsXA, which contains a constitutive promoter and an N-terminal secretion signal peptide (39). The vector was further modified to introduce a polyhistidine C-terminal tag, and the resulting vector is referred to as pT7BsXA_CT.
Protein Expression in P. pastoris GS115
All plasmids were linearized by digestion with StuI restriction enzyme digestion and integrated into P. pastoris GS115 by electroporation using a Gene PulserTM (Bio-Rad) according to the manufacturer's instructions (EasySelectTM Pichia Expression Kit, Invitrogen). P. pastoris transformant colonies were grown on MD solid medium for 3 days and were inoculated into 100 ml of buffered minimal glycerol medium (0.34% yeast nitrogen base, 1% ammonium sulfate, 4 × 10−5% biotin, 100 mm phosphate buffer, pH 6.0, 2% glycerol) and cultured under continuous shaking (200 rpm) at 30 °C. After 20 h, the culture reached an A600 of 4, and the cells were collected by centrifugation and resuspended in 100 ml of buffered methanol minimal medium (0.34% yeast nitrogen base, 1% ammonium sulfate, 4 × 10−5% biotin, 100 mm phosphate buffer, pH 6.0) containing 1% methanol as an inducer in a 1-liter Erlenmeyer flask. Over the course of 3–4 days, 1% (v/v) methanol was added daily, and the acidity of the culture was adjusted to pH 6 with 1 m K2HPO4.
Protein Expression in E. coli DH5α
Escherichia coli DH5α was transformed with plasmid pT7BsXA_CT carrying either the wild-type or mutant XynA, and single colonies were used to inoculate 100 ml of LB culture medium (1% tryptone, 0.5% yeast extract, 1% NaCl, and 100 mg/liter ampicillin). After an 18-h culture at 37 °C in an orbital shaker at 180 rpm, the culture was centrifuged at 5000 × g, and the recombinant protein was purified from the culture supernatant by nickel affinity chromatography.
Purification of Recombinant Proteins
Cells were removed from cultures by centrifugation at 5000 × g for 7 min at 4 °C and passed through a 0.2-mm filter to remove cellular debris. The filtered culture supernatant was concentrated, and the buffer was exchanged by an Amicon Stirred Cell, 10,000 molecular weight cut-off (Millipore Corp., Billerica, MA). The concentrated enzyme was applied to a HiTrapTM 3-ml Q HP column (Promega Biotecnologia do Brasil, São Paulo, Brazil), previously equilibrated with buffer containing 500 mm NaCl, 40 mm Hepes, pH 8.0. Proteins were eluted with the same buffer containing 300 mm imidazole. The flow rate was 1.5 ml/min, and the absorbance was monitored continuously at 280 nm. Eluted fractions containing enzyme were concentrated and desalted, and buffer was exchanged using an Amicon Ultra-15, 10,000 molecular weight cut-off (Millipore Corp.). Protein purity was routinely checked using SDS-polyacrylamide gel electrophoresis.
Far UV CD of Recombinant XynA
The far UV CD spectra of the XynA and mutants were measured using 100-μm path length quartz cuvettes at a concentration of 0.5 mg/ml in 20 mm KH2PO4 (pH 6.0) with a Jasco 810 spectropolarimeter (Jasco Corp., Tokyo, Japan) between wavelengths of 185 and 250 nm and a 4-nm bandwidth at a constant temperature of 298 K. The accumulated average of six protein spectra was corrected by subtraction of the spectra of the equivalent buffer in the absence of protein.
Enzyme Activity Assays
The effect of temperature on the catalytic activity of the XynA and mutants was evaluated by incubating 1 μg of protein with 1% oat spelt xylan in 100 mm sodium acetate, pH 6.0 for a period of 10 min over a temperature range of 25–65 °C. In all experiments, the reactions were halted by the addition of 3,5-dinitrosalicylic acid to a final concentration of 10 mm, and the mixture was heated to 100 °C for 5 min, followed by incubation for 5 min on ice. The production of xylose was assayed by measuring the A540 after the chromogenic conversion of the 3,5-dinitrosalicylic acid by the reducing sugar (40). One unit of enzyme was defined as the quantity of xylanase required to liberate 1 μmol of reducing sugar/min under the assay conditions described. Thermal inactivation was performed at 55 °C for controlled time intervals where the residual catalytic activity of a given protein after heating at different times was expressed relative to the activity of the same protein measured at 55 °C. The residual catalytic activity was determined in 20 mm sodium acetate, pH 6.0, in the presence of 1% oat spelt xylan substrate, and the production of xylose was assayed using the chromogenic 3,5-dinitrosalicylic acid assay.
Deglycosylation of Recombinant Proteins by Endoglycosidase H
Ten micrograms of recombinant proteins were boiled for 5 min in 50 mm sodium citrate buffer (pH 5.5) containing 1% SDS. After cooling, the sample was incubated with 250 units of endoglycosidase H (Endo H) (New England Biolabs, Ipswich, MA) and incubated at 37 °C for 3 h. Samples were subsequently analyzed by SDS-polyacrylamide gel electrophoresis (41). Endoglycosidase H cleaves the glycosidic bond in the diacetylchitobiose moiety of the glycan, generating a protein molecule with a single GlcNAc residue remaining on the asparagine side chain, and the presence of this modified Asn residue in peptides carrying glycosylation sites can be detected by a mass shift in mass spectrometry.
Mass Spectrometry Analysis
Slices of SDS-polyacrylamide gels containing Endo H-digested wild-type or mutant XynA were destained with 50% methanol and then reduced in 10 mm DTT for 1 h at 56 °C followed by alkylation in 55 mm chloroacetamide for 1 h at room temperature. After washing in 50 mm ammonium bicarbonate, the gel slices were shrunk in 100% acetonitrile. Protein hydrolysis was performed using trypsin in 50 mm ammonium bicarbonate for 8 h at 37 °C. The peptides were finally extracted in 90% acetonitrile, 0.5 m urea, lyophilized, and resolubilized in 5% acetonitrile (0.2% formic acid) prior to mass spectrometry analyses. LC-MS/MS analyses were performed on a nano-LC 2D pump (Eksigent) coupled to an LTQ-Orbitrap Elite hybrid mass spectrometer via a nanoelectrospray ion source (Thermo Fisher Scientific, Bremen, Germany). Peptides were loaded on a homemade 360-μm inner diameter × 4-mm C18 trap column prior to separation on a 150-μm inner diameter × 10-cm nano-LC column (Jupiter C18, 3 μm, 300 Å, Phenomex). Peptide elution was achieved using a linear gradient of 5–60% acetonitrile (0.2% formic acid) over 56 min with a flow rate of 600 nl/min. Each full MS spectrum was followed by 12 MS/MS spectra of the most abundant multiply charged ions. Tandem MS experiments were performed using collision-induced dissociation in the high energy collision cell prior to mass analysis on the Orbitrap analyzer. The data were processed using the 2.3 Mascot search engine (Matrix Science Inc., Boston, MA) with tolerance parameters set to 15 ppm and 0.02 Da for the precursor and the fragment ions, respectively. The selected variable modifications were carbamidomethyl (C), deamidation (NQ), oxidation (M), and HexNac (N). Tandem mass spectra were compared with a database composed of wild-type and mutated xylanases.
For intact mass profiling, the proteins were resolubilized in 5% acetonitrile (0.2% formic acid), and samples were separated on a C5 column (150 μm × 10 cm) using an Agilent 1100 nano-LC system interfaced to a 6520 Q-TOF mass spectrometer via a nanoelectrospray source (Agilent Technologies, Santa Clara, CA). A 56-min gradient from 10 to 60% acetonitrile including 0.2% formic acid was used to elute peptides from a reversed-phase column (150-μm inner diameter × 100 mm) with a flow rate set at 600 nl/min. Deconvolution of the protein isotope envelope was made with a maximum entropy algorithm as implemented in Masshunter B 04.00 (Agilent Technologies).
Molecular Dynamics Simulations
The initial atomic coordinates of the glycosylated xylanases were obtained from the GH11 xylanase from B. subtilis (Protein Data Bank code 1XXN) using the GlyProt tool (42). GlyProt works with the SWEET DB to build three-dimensional structures of glycoproteins, taking into account the stereochemical criteria for a given glycan composition. After obtaining the initial three-dimensional model of a given glycosylated variant, an initial energy minimization of the glycoprotein was performed using a steepest descent algorithm (43). Subsequently, the glycoprotein was solvated with SPC water molecules (44) at a concentration of ∼53.0 mol/liter in dodecahedron simulation boxes. The protonation state of the ionizable residues at physiological pH was determined by use of the H++ program (Poisson-Boltzmann-based pKa calculations) (45), and finally three Cl− ions were introduced in the most electrostatically favorable positions to neutralize the overall charge of the system. All glycoprotein systems were then equilibrated for ∼300 ps by position-restrained MD at 328 K to allow solvent relaxation and to avoid protein-water, protein-glycan, and glycan-water interaction artifacts. During the calculation phase, all systems were simulated in the NVT ensemble at pH 7.0 and 328 K, where the temperature was controlled by using the V-rescale thermostat. Covalent bonds involving hydrogen atoms and water molecules were constrained by the LINCS and SETTLE (46) algorithms, respectively. The leapfrog integration algorithm (47) was employed to solve the equations of motion with a time step of 2.0 fs. The initial velocities were sampled with the Maxwell-Boltzmann distribution. The long range interactions were treated using the particle mesh Ewald sum method (48) with a cut-off equal to 1.2 nm. The long range contributions of the electrostatic interactions were updated every 10 time steps. The MD simulation was monitored by calculation of the root mean square deviation at all time points using the backbone atoms of the protein and was halted when the root mean square deviation reached an equilibrium value, which in these simulations was around 0.20 nm. All MD runs and analyses were performed with the GROMACS version 4.5 software package (49) using the GROMOS-96 force field (50). The interaction potential energies (IPEs) were computed as the sum of all interaction energies (E) between all of the protein atoms (i) and the glycan atoms (j) according to the equation,
 |
where Np and Ng are the total number of protein and glycan atoms, respectively. The interaction energies (E) include all short and long range contributions.
RESULTS
MS Analysis of Wild-type Xylanse A
The XynA adopts a β-sheet jellyroll fold typical of the GH11 family. Using the analogy of a right hand, the active site cleft is formed by residues in the opposing “finger” and “thumb” domains, and the active site residues are located in the “palm” domain. The amino acid sequence of the xylanase A from B. subtilis (XynA; Fig. 1A) contains six predicted N-linked glycan sites, where all potential glycosylation sites are surface residues located on the finger domain of the protein (Fig. 1B). Mass spectrometry (MS) of chymotryptic peptides following Endo H digestion of the recombinant XynA expressed in P. pastoris assigned a single GlcNAc modification, referred to as HexNAc, to each of the four residues Asn20, Asn25, Asn141, and Asn181 (see supplemental Fig. S1). No mass modification was found on the Endo H-treated chymotryptic peptides containing Asn8 and Asn29, demonstrating the absence of glycosylation of these putative sites in the wild-type XynA. In these experiments, the sequence coverage was 100%, and the N-terminal peptide analysis revealed that the signal sequence peptidase present in P. pastoris cleaved the protein three residues upstream from the expected Kex2 signal peptide cleavage site (31). The N-terminal sequence of the mature XynA therefore contains three additional residues (EAE) derived from the yeast α-factor signal peptide (Fig. 1A).
FIGURE 1.
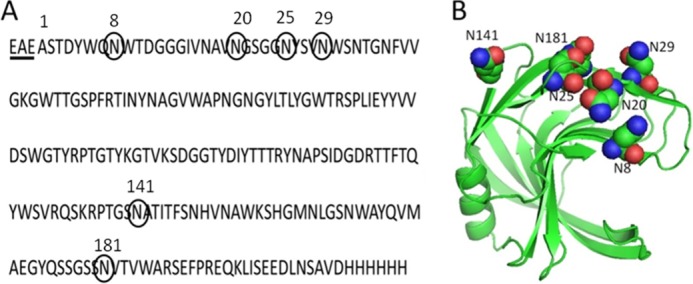
Amino acid sequence (A) and ribbon representation (B) of the crystal structure of xylanase A from B. subtilis showing the potential N-glycosylation sites. The sequence was modified to contain a C-terminal peptide with a polyhistidine. The underlined residues (EAE) are derived from the α-factor signal peptide. The numbers of amino acids correspond to the native sequence that does not include residues EAE.
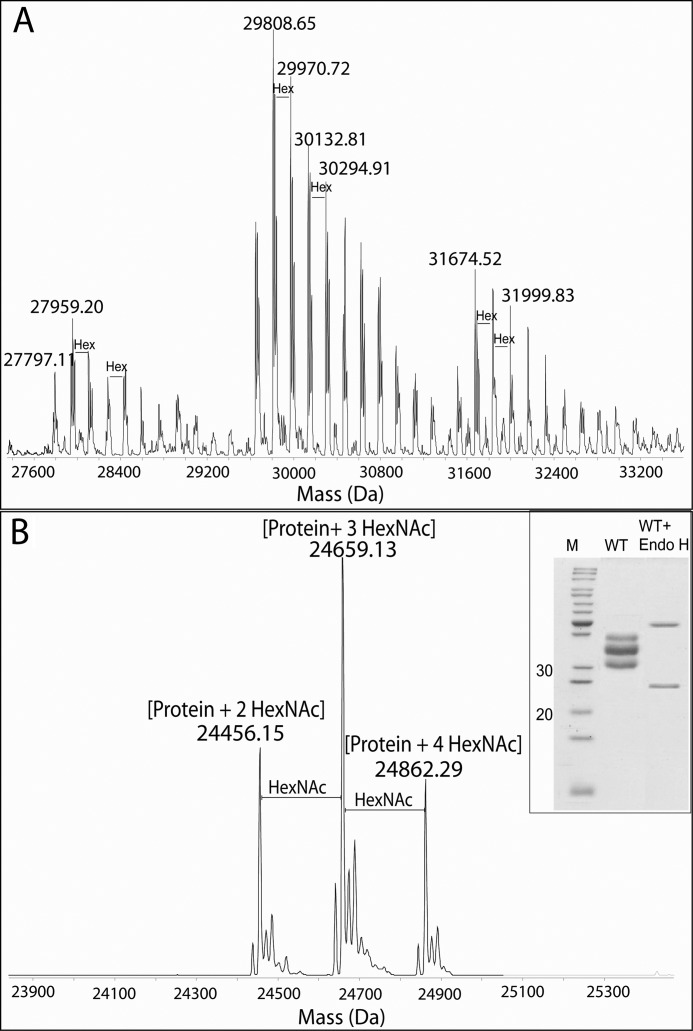
Glycopeptide mapping using higher-energy collisional dissociation on an Elite Orbitrap instrument revealed modification with N-linked high mannose-type glycans (Hex8–16 GlcNAc2) (supplemental Fig. S2). To further profile the glycan microheterogeneity, purified wild-type XynA expressed in P. pastoris was subjected to flow injection analysis by electrospray MS (Fig. 2A). The multiply charged ion envelopes were reconstructed as molecular mass profiles, revealing regular increments in the glycoform masses that are equivalent to hexose units (162 Da). Furthermore, the charged ion envelopes suggested three glycoform ensembles, each with a variable number of occupied glycosylation sites, which correspond to the three bands observed in SDS-PAGE (see Fig. 2B, inset). The percentages of the glycosylated species were estimated using the difference between the observed mass and the theoretical average mass. The relative proportion of glycans in the wild-type XynA with respect to the protein mass was calculated to be between 13% for the lighter detected mass and 27% for the heavier detected mass. After digestion of the XynA with Endo H, the measured protein masses were consistent with modification by two, three, or four HexNAc residues (Fig. 2B). These results demonstrate that a maximum of only four of the six potential N-glycosylation sites in the wild-type XynA sequence are glycosylated.
FIGURE 2.
LC-MS of B. subtilis xylanase A expressed in P. pastoris before (A) and after (B) Endo H digestion. Intact mass analyses indicate three main glycoform clusters with different extents of glycosylation. These data are supported by Endo H digestion, where the protein mass (24,050 Da) plus four, three, or two HexNAc monosaccharides was observed. The inset shows SDS-PAGE from purified xylanase before and after Endo H digestion.
Thermostability of Glycosylated and Unglycosylated Wild-type Xyn A
The wild-type XynA expressed in E. coli showed a measured temperature optimum of 55 °C, which is similar to that previously reported (37), whereas the XynA expressed in P. pastoris presents a temperature optimum of 45 °C (Fig. 3A). Increasing the temperature in a mixture of glycoproteins will result in the more rapid loss of the least stable component, resulting in a reduction of the total measured activity. This effect will be more evident at elevated temperatures; therefore, experiments measuring the catalytic activity at higher temperatures will tend to have lower measured activities, leading to a reduction of the apparent optimum temperature of catalysis of the mixture. This effect was confirmed for the glycosylated wild type that presented a decrease in optimum catalytic temperature (Topt) compared with the unglycosylated form expressed in E. coli, suggesting that the ensemble of glycosylated XynA contains glycoforms with a range of thermostabilities.
FIGURE 3.
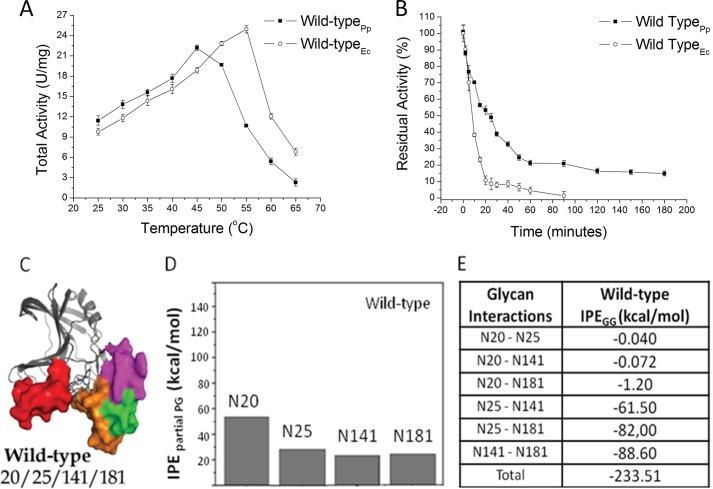
Biochemical and molecular dynamics simulations results for wild-type XynA. A, temperature dependence of catalytic activities of wild-type XynA expressed either in E. coli (Wild-typeEc) or P. pastoris (Wild-typePp). B, thermal inactivation profiles of the wild-type XynA at 55 °C. C, ribbon representations of MD simulations of the conformation of glycosylated XynA. The polypeptide portions of the molecules are represented by gray ribbons, whereas the glycan portions are represented as solid molecular surfaces in different colors according to their positions (glycosylation sites): red for the glycan at Asn20, green for the glycan at Asn25, blue for the glycan at Asn29, magenta for the glycan at Asn141, and orange for the glycan at Asn181. D, IPEpartial PG computed as time-average for the protein-glycan interactions for each glycosylated position during the MD simulation. E, IPE for glycan-glycan intermolecular interactions (IPEGG).
The thermostability curves of the glycosylated XynA expressed in P. pastoris and unglycosylated XynA expressed in E. coli (Fig. 3B and Table 1) showed that the time over which activity was reduced by 50% (t50%) for the wild-type glycosylated XynA was 2.5-fold longer as compared with the unglycosylated enzyme. Even after 180 min of incubation, the glycosylated XynA still retained 20% of the initial catalytic activity.
TABLE 1.
Summary of glycosylation sites occupancy and thermostability analyses for glycosylated and unglycosylated proteins
The occupancy was determined by LC-MS/MS analyses after Endo H/chymotrypsin digestion. Shown is the time of loss of the first 50% of activity (t50%) upon incubation at 55 °C of all proteins expressed in either P. pastoris (glycosylated) or E. coli (unglycosylated), confirming that altered stability was imparted by glycosylation rather than by mutation effects.
| Protein | Glycosylation site occupancy | Optimum temperature for glycosylated proteins | t50% at 55 °C for glycosylated proteins | Optimum temperature for unglycosylated proteins | t50% at 55 °C for unglycosylated proteins |
|---|---|---|---|---|---|
| °C | min | °C | min | ||
| XynA wild type | Asn-20/25/141/181 | 45 | 20 | 55 | 8 |
| N8Q | Asn-20/25/141/181 | 45 | 5 | 50 | 2 |
| N20Q | Asn-25/29/141/181 | 35 | 5 | 50 | 10 |
| N25Q | Asn-20/29/141/181 | 45 | 240 | 50 | 8 |
| N29Q | Asn-20/25/141/181 | 45 | 50 | 50 | 14 |
| N181Q | Asn-20/25/141 | 45 | 240 | 45 | 10 |
| N20/25Q | Asn-29/141/181 | 50 | 300 | 50 | 10 |
| N20/181Q | Asn-25/29/141 | 40 | 20 | 40 | 8 |
| N25/181Q | Asn-20/29/141 | 50 | 1380 | 50 | 8 |
| N20/25/181Q | Asn-29/141 | 50 | 120 | 50 | 8 |
| N20/25/141/181Q | Asn-29 | 45 | 10 | 45 | 2 |
| N20/25/29/141/181Q | 45 | 5 | 45 | 5 | |
| N8/20/25/29/141/181Q | 45 | 5 | 45 | 5 |
Molecular Dynamics Simulations of Wild-type Glycosylated XynA
A ribbon representation of the final three-dimensional structure from MD simulations of the wild type is presented in Fig. 3C, in which the glycans at different positions are shown by colored solid surfaces (see the legend to Fig. 3). The interactions between protein and glycans were estimated by calculation of the IPE, and the average total IPE for protein-glycan interactions over the final 20 ns of the MD simulations for the wild-type XynA showed a value of −125 ± 67 kcal·mol−1 (Fig. 4). The individual contribution of each glycan-protein interaction was denominated as its partial interaction potential energy (IPEpartial PG) and was computed as the time-average value of the protein-glycan interaction potential energy during the MD simulation. All glycosylated positions contributed to the stability of the protein-glycan interface with attractive interactions; however, the glycan at position 20 showed the most favorable energy (Fig. 3D). The glycan-glycan interaction potential energies (IPEGG; Fig. 3E) for the glycosylated wild-type XynA suggest that the combination of glycosylation at positions 141/181, 25/181, and 25/141 promotes glycan-glycan interactions. This observation led us to propose that combinations of glycosylation at given positions may favor the formation of glycan-glycan contacts involving sugar residues that might otherwise participate in the formation of protein-glycan contacts. The possibility that the thermostability of the enzyme might be modulated through the reorganization of glycan-glycan and glycan-protein interactions resulting from the alteration of the glycosylation pattern was evaluated by analysis of site-directed mutants of the XynA.
FIGURE 4.
IPETotal computed as time-average for protein-glycan interactions along the MD simulation. Error bars, S.D.
Molecular Dynamics Simulations, Thermostability, and Mass Spectrometry Analysis of the Glycosylated Mutant N181Q
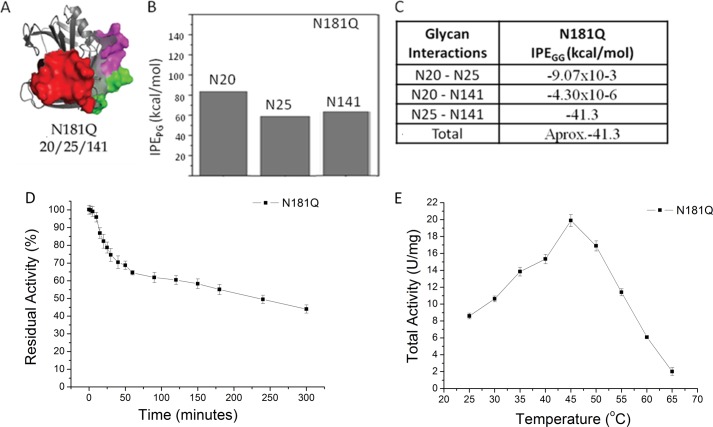
The differences in the contributions to the total IPE of the interaction potential energies for glycan-protein and glycan-glycan contacts prompted us to conduct a more detailed investigation with the aim of correlating the contributions of each glycan group to the global protein-glycan interface. We predicted that the N181Q mutant would eliminate glycosylation at position 181, and mass spectrometry analysis of this mutant after expression in P. pastoris confirmed the glycosylation at positions Asn20, Asn25, and Asn141 (Table 1). The N181Q mutant was evaluated by MD simulation, and the final three-dimensional structure with the three glycan chains represented as solid surfaces is shown in Fig. 5A. The average total IPE of −182 ± 15 kcal·mol−1 for protein-glycan interactions in this mutant was more negative in comparison with the wild-type enzyme (Fig. 4). The partial interaction potential energies (IPEpartial) showed that all three glycans contributed to the more negative energy of protein-glycan interactions (Fig. 5B) and concomitantly reduced the contribution of the glycan-glycan interaction potential energies (Fig. 5C) in comparison with wild-type glycosylated XynA.
FIGURE 5.
Molecular dynamics simulations and biochemical results for XynA mutant N181Q. A, ribbon representations of MD simulations of the conformation of glycosylated mutant. The polypeptide portions of the molecules are represented by gray ribbons, whereas the glycans portions are represented as solid molecular surfaces in different colors according to their positions (see the legend to Fig. 3). B, IPEpartial PG computed as time-average for the protein-glycan interactions for each glycosylated position during the MD simulation. C, IPEGG. D, temperature dependence of catalytic activities of XynA mutant N181Q expressed in P. pastoris. E, thermal inactivation profile at 55 °C of the XynA mutant N181Q expressed in P. pastoris. Error bars, S.D.
The thermostability and temperature/activity curves for the N181Q mutant are presented in Fig. 5, D and E. Although the optimum catalytic temperature (Topt) is unchanged when compared with the wild-type glycosylated protein, the N181Q mutant exhibits a significant increase in thermostability, with a 12-fold increase in the t50% (Table 1). These results supported a proposed mechanism of protein stabilization of XynA by glycosylation that is based on the creation and extension of a protein-glycan interface and prompted further investigation to evaluate the structural context of N-glycosylation and its importance for protein stabilization.
MS Analyses of Glycosylated Xylanase Mutants
In addition to the N181Q mutant, site-directed mutagenesis of the remaining single N-glycosylation sites resulted in the creation of the single mutants N8Q, N20Q, N25Q, N29Q, and N141Q. With the goal of progressively eliminating the N-glycosylation sites, single mutants were combined in the multiple mutants N20Q/N25Q, N20Q/N181Q, N25Q/N181Q, N20Q/N25Q/N181Q, N20Q/N25Q/Q141N/Q181Q, and N20Q/N25Q/N29Q/N141Q/N181Q. Despite exhaustive attempts, the N141Q single mutant could not be expressed and purified, and it was not studied further. The far UV CD spectra of the wild-type XynA and mutant proteins show essentially identical profiles (data not shown), and the minima at 216 nm and maxima at 194 nm are typical of native proteins rich in β-sheet secondary structure. The intact mass analyses of the single and combined mutants revealed heterogeneity in the glycosylation of all mutants (supplemental Figs. S3 and S4) The intact masses after Endo H digestion revealed a glycan modification in the single mutants N20Q and N25Q (supplemental Fig. S5) that was not observed in the wild-type XynA, and LC-MS/MS confirmed that position 29 is glycosylated in these proteins (Table 1); therefore, position 29 is glycosylated only in the absence of glycans at position 20 or 25. The intact masses of the N8Q and N29Q mutants were similar to that of the wild-type enzyme and show glycosylation at the same positions. In none of the mutants was the Asn8 position found to be glycosylated, even in the quintuple N20Q/N25Q/N29Q/N141Q/N181Q mutant (supplemental Figs. S4 and S6). The glycosylation site occupancy for all mutants is shown in Table 1. No O-glycosylation sites were detected, as confirmed by the absence of glycosylation in the mutant with all six potential sites mutated.
With the aim of characterizing the glycosylation pattern in each of the glycoform ensembles, the individual bands from SDS-PAGE of the wild-type XynA and mutants were excised, digested with Endo H, and analyzed by mass spectrometry. The LC-MS/MS of the Endo H/chymotryptic peptides of each glycoform band separated by SDS-PAGE revealed a preference for specific glycosylation sites. Positions 141 and 181 are glycosylated in almost all glycoforms, followed by a preference for positions 25, 20, and finally 29. In the wild-type XynA, positions 20, 25, 141, and 181 are glycosylated in the upper and the intermediate bands, as compared with positions 25, 141, and 181 in the lower band. Three combinations are possible in the intermediate and lower bands with three and two sites occupied, respectively. For the mutants N25Q and N20Q, position 29 is glycosylated only in the upper band, and a recurrent difference in the glycosylation pattern is the substitution at position 25 rather than position 20.
Thermostability of Glycosylated Mutant Enzymes
Almost all proteins expressed in P. pastoris presented a decrease in optimum catalytic temperature (Topt) compared with the unglycosylated form expressed in E. coli (Table 1). In general, increasing the number of mutations resulted in progressive decreases in the apparent Topt as compared with the same enzymes expressed in E. coli. Those mutants that show a rapid initial inactivation (for example N20Q and N20Q/N181Q) were those that also presented the lowest apparent Topt (35 and 40 °C, respectively).
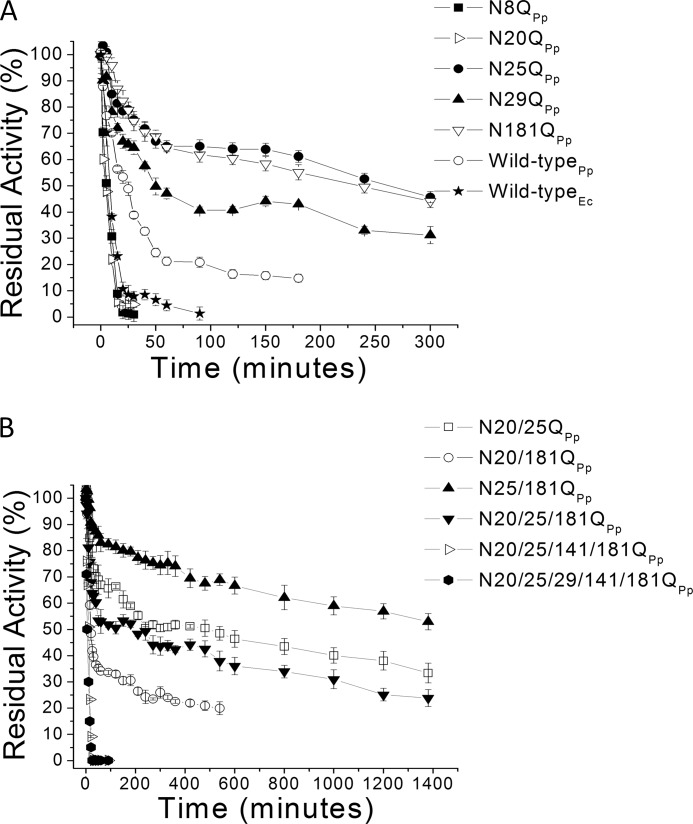
The apparent t50% of the XynA mutants was estimated from the thermostability curves (Fig. 6). An increased thermostability of the glycosylated protein in comparison with the unglycosylated protein was observed in most mutants (Table 1), and in the case of single mutants N25Q (glycans at positions 20, 29, 141, and 181) and N181Q (glycans at positions 20, 25, and 141), this stabilization effect results in a 30-fold increase in t50%. In contrast, the N20Q mutant (glycans at positions 25, 29, 141, and 181) is less stable with a 2-fold decrease in t50%. The double mutant N25Q/N181Q (glycans at positions 20, 29, and 141) is the most thermostable variant identified and retains 50% activity even after heating at 55 °C for 23 h. Glycosylation at position 181 in the presence of a glycan at position 25 resulted in decreased stability; however, mutations that eliminated glycans at either position 25 or 181 led to improved stability. Glycosylation at position 20 appears to play a key role in increasing thermostability when found in combination with glycans at positions 29, 25, and 141.
FIGURE 6.
Thermal inactivation profiles at 55 °C of the wild type and the mutants. A, wild-type xylanase A expressed either in E. coli (Wild-typeEc) or P. pastoris (Wild-typePp) and single mutants expressed in P. pastoris (see inset). B, results for double, triple, quadruple, and quintuple mutants. Error bars, S.D.
Although Table 1 clearly demonstrates the overall effects of glycosylation, the influence of individual amino acid substitutions on protein thermostability can also be observed by comparing the unglycosylated proteins expressed in E. coli. Mutants N8Q, N20Q/N25Q/N141Q/N181Q, N20Q/N25Q/N29Q/N141Q/N181Q, and N8Q/N20Q/N25Q/N29Q/N141Q/N181Q show reduced thermostability in relation to the wild-type protein. The reduced stability of these mutants in comparison with the wild-type XynA is also observed in the glycosylated proteins expressed in P. pastoris. The N29Q mutant expressed in E. coli presents an increased thermostability compared with the wild-type protein, with the t50% increasing from 8 to 14 min, and this stabilizing effect is also observed in the glycosylated protein expressed in P. pastoris, which shows an increased t50% from 20 to 50 min. Mass spectrometry analysis reveals glycan modification sites at positions 20/25/141/181, which is identical to the wild-type protein, indicating that the amino acid substitution contributes to the increased t50% of the glycosylated N29Q mutant. Finally, the elimination of all glycosylation sites (in the N20Q/N25Q/N29Q/N141Q/N181Q and N8Q/N20Q/N25Q/N29Q/N141Q/N181Q mutants) reduced the t50% from 8 to 5 min, and as expected, this value was the same for proteins expressed in both E. coli and P. pastoris.
Molecular Dynamics Simulations of Glycosylated Mutant Enzymes
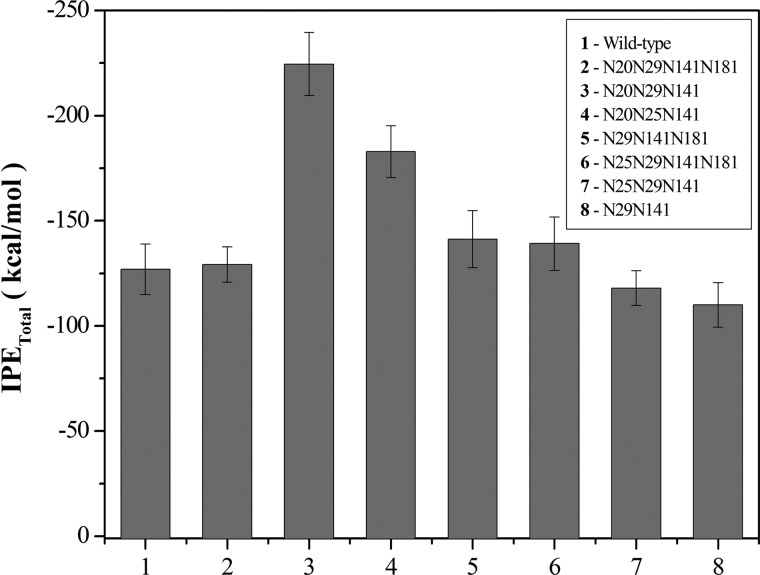
Ribbon representations of the final three-dimensional structures from MD simulations of the XynA mutants are presented in Fig. 7. Analysis of the contributions of the intermolecular interactions to the total IPE revealed that glycan-water interaction energies show no correlation to the experimentally observed stabilities and that the IPEs from protein-water interactions are similar in all glycosylated proteins, with an average value of −2180 ± 67 kcal·mol−1. The average total IPE of protein-glycan interactions for the mutants over the final 20 ns of the MD simulations are presented in Fig. 4. The most negative protein-glycan IPE values for protein-glycan interfaces are observed in two of the most thermostable mutants (N181Q and N25Q/N181Q). This indicates that of the three molecular interactions that contribute to the total IPE (protein-solvent, protein-glycan, and glycan-solvent), the protein-glycan interaction may play a predominant role in determining the increased thermostability of the glycoproteins.
FIGURE 7.
Snapshots of MD simulations of the conformations of glycosylated mutants of xylanase A. The structures are ribbon representations of the average stabilized structures obtained every 0.5 ps from MD calculations at 328 K over the final 15 ns of the trajectory. The polypeptide portions of the molecules are represented by gray ribbons, whereas the glycan portions are represented as solid molecular surfaces in different colors according to their positions (glycosylation sites): red for the glycan at Asn20, green for the glycan at Asn25, blue for the glycan at Asn29, magenta for the glycan at Asn141, and orange for the glycan at Asn181. These figures were generated using the PyMOL software (PyMOL Molecular Graphics System, version 1.5, Schrödinger, LLC).
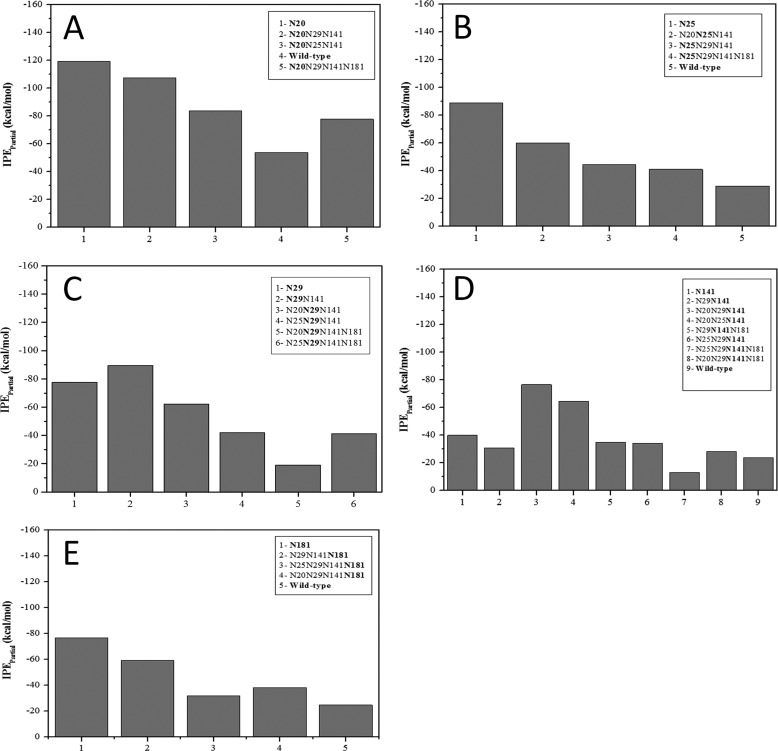
The IPEpartial values for the protein-glycan interactions of all mutants are presented in Fig. 8, in which each panel presents the IPE values of the glycan at a given position. For example, Fig. 8A shows the IPEpartial of the protein-glycan interaction at position 20 in all mutants in which glycosylation of Asn20 occurs. The contribution of the IPEpartial of each glycan to the overall stabilization can also be derived from Fig. 8. For example, Fig. 8A shows an IPEpartial of −82 kcal/mol for the glycan at position 20 in the N181Q mutant (glycans at positions 20/25/141), whereas Fig. 8B shows a IPEpartial of −60 kcal/mol for the glycan at position Asn25 in the same mutant. The IPEpartial values of glycans at positions 20, 25, 29, 141, and 181 (Fig. 8, A–E, respectively) indicate that glycosylation at position 20 promotes the greatest stabilization. This correlates with the experimentally observed increase in thermostability of those mutants glycosylated at this position. A correlation between the experimentally observed thermostability and the IPEpartial for glycosylation at position 181 was also observed. Finally, the lowest IPEpartial for the glycan at position 25 in the N181Q mutant (glycans at 20/25/141) appears to contribute to the experimentally determined increase in thermostability of this mutant.
FIGURE 8.
IPEpartial computed as time-average for the protein-glycan interactions for each glycosylated position during the MD simulation.
DISCUSSION
The present work has revealed that of the six consensus N-linked glycosylation sites in the wild-type XynA, only 2–4 asparagine residues are glycosylated by P. pastoris. Furthermore, LC-MS/MS analysis has revealed that multiple glycosylation combinations are present and that each combination displays a distinct and heterogeneous glycoform ensemble with alternative oligosaccharides occupying the differing positions. The presence of distinct glycoforms is consistent with the proposal that attached glycan groups alter the properties of the protein and indicates that functional differences are the consequences of both the number of occupied N-glycosylation sites and the extent of glycan decoration at each site (3). Elimination of glycosylation sites in the wild-type XynA resulted in both a significant decrease in the carbohydrate content of the heterologous mutants expressed in P. pastoris and in alterations in the thermostability of the enzyme. Although the specific catalytic activities were reduced in the glycosylated enzymes, the loss of activity is not proportional to the reduction in the carbohydrate content.
In the crystal structure of the unglycosylated XynA expressed on E. coli, the side chains of the asparagine residues in all potential glycosylation sites are exposed at the surface of the enzyme (see Fig. 1); therefore, the observed glycosylation preference for residues Asn141 = Asn181 > Asn20 = Asn25 > Asn29 of the wild-type and mutant XynA expressed in P. pastoris is not correlated with surface exposure of the target asparagine residue. A possible explanation of the preference is that an asparagine residue is more readily glycosylated in an Asn-Xaa-Thr sequon than in Asn-Xaa-Ser (31). Consistent with this proposal is the observation that glycosylation of Asn141 (in the triplet Asn-Ala-Thr) and Asn181 (in triplet Asn-Val-Thr) are preferred as compared with the Asn20 (in triplet Asn-Gly-Ser), Asn25 (in triplet Asn-Tyr-Ser), and Asn29 (in triplet Asn-Trp-Ser) residues. The low frequency for glycosylation of Asn29 in relation to Asn20 or Asn25 can be explained on the basis of the importance of the second amino acid residue in the recognition site, where bulky hydrophobic residues, such as tryptophan or phenylalanine, inhibit the initial step of glycan addition to the asparagine residue (3, 31). This explains the observed absence of glycosylation at Asn8 in the present study, where the Asn8 residue is found in a Asn-Trp-Ser triplet.
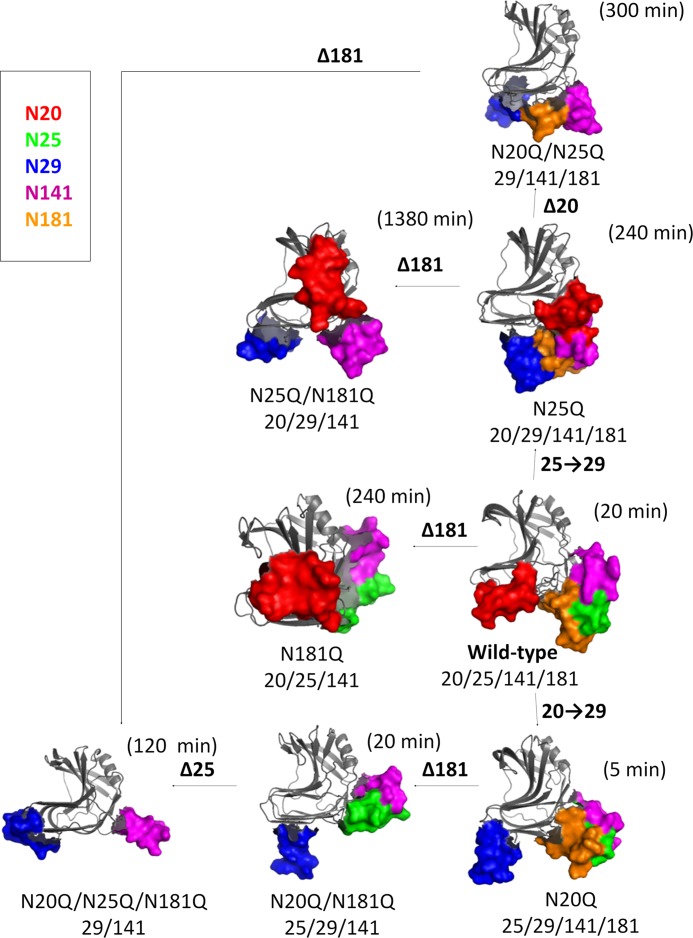
The t50% of the majority of mutant xylanases expressed in E. coli was between 8 and 10 min (with the exception of N8Q (2 min) and N29Q (14 min)), whereas the t50% of glycosylated enzymes varied from 5 min to 23 h (Table 1). To explain these effects, the combinations of positions of the N-linked glycan chains in the tertiary structure of the enzyme were examined. By merging data from thermostability experiments (Fig. 6) and glycosylation site occupation analysis, several conclusions can be drawn with respect to stabilizing glycosylation combinations. For example, glycans at positions 20 or 141 are observed in proteins with two sites occupied in the more stable mutants N25Q, N181Q, and N25Q/N181Q and can be considered a favorable combination. In contrast, the glycosylation combination of positions 25/181 or 25/141 appears to be destabilizing because these combinations are present in the mutants N20Q and N20Q/N181Q, both of which have a significantly reduced stability.
Positions 29 and 141 are glycosylated both in the most and the least thermostable mutants, and clearly the effect of glycosylation of these positions depends on the context of additional glycosylation site modifications. Results from MD simulations show that IPE values present significant variation, and glycosylation of position 29 has maximum stability in the presence of a glycan at position 141 (Fig. 8C), and both positions 29 and 141 are glycosylated in the N25Q/N181Q, N20Q/N25Q, and N20Q/N25Q/N181Q mutants. Glycosylation of positions 29/141 in the N20Q/N25Q/N181Q mutant might not seem to be a favorable combination, with a t50% of 120 min. However, the N20Q/N25Q/N181Q mutant presents a glycoform mixture that contains a high fraction of protein with a single glycosylation site occupied in Asn29 and Asn141 (supplemental Figs. S4D and S6D). The glycosylation at residue Asn141 alone presents a decreased stability (see Fig. 8D); therefore, this singly glycosylated protein is likely to be responsible for the fast phase of catalytic inactivation in the thermostability experiments. In addition, the thermal inactivation profile of the N20Q/N25Q/N141Q/N181Q mutant (glycan at position 29) shows a rapid decrease in activity, which is mainly due to the rapid denaturation of the unglycosylated mutant that is present in the glycoform mixture (supplemental Figs. S4E and S6E).
The combination of glycosylation at positions 20/141 favors thermostability and is observed in several proteins, including the wild-type XynA (glycans at positions 20/25/141/181). Although the combination of glycosylation of positions 20/141 is present in the N25Q mutant (glycans at positions 20/29/141/181), the modest stabilizing effect of the glycan at position 181 (see Fig. 8 (A and D) and Table 2) renders this mutant less stable as compared with the N25Q/N181Q mutant (glycans at positions 20/29/141). It is noteworthy that glycosylation at positions 20 and 141 in the N25Q/N181Q mutant results in a higher stability than the same combination in wild-type XynA (glycans at positions 20/25/141/181) and may be due to the destabilizing effect of glycans at positions 25/181.
TABLE 2.
IPE for glycan-glycan intermolecular interactions in the wild-type XynA and glycosylated variants N20N29N141N181 and N20N29N141
Energies are given in kcal/mol.
| Glycan interactions | Wild-type IPEGG | N20N29N141N181 IPEGG | N20N29N141 IPEGG |
|---|---|---|---|
| kcal/mol | kcal/mol | kcal/mol | |
| Asn20-Asn25 | −0.040 | ||
| Asn20-Asn29 | −2.09 | 0 | |
| Asn20-Asn141 | −0.072 | −29.60 | −0.0050 |
| Asn20-Asn181 | −1.20 | −5.57 | |
| Asn25-Asn141 | −61.50 | ||
| Asn25-Asn181 | −82,00 | ||
| Asn29-Asn141 | −0.070 | −0.0033 | |
| Asn29-Asn181 | −61.50 | ||
| Asn141-Asn181 | −88.60 | −71.10 | |
| Total | −233.51 | −169.93 | −0.0083 |
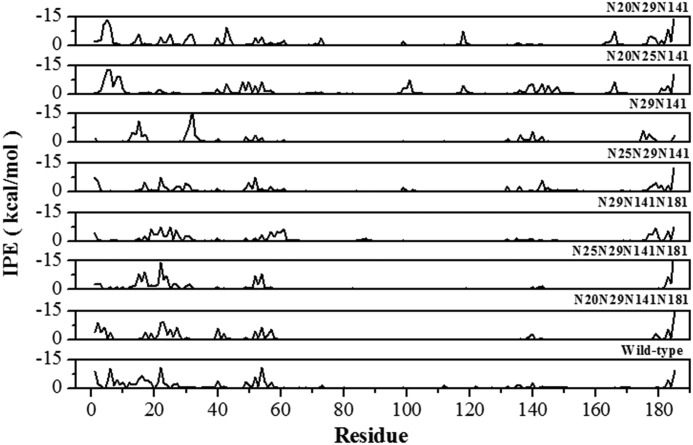
The effects of alternative glycosylation patterns on stability can be rationalized by further analysis of the MD simulations using the time-averaged IPE per amino acid residue for all protein-glycan interactions in the wild-type and thermostable mutants (Fig. 9), where IPE values <0 define attractive contacts between the glycan and protein residues. Although many protein-glycan contacts are conserved in all mutants, such as the region around residues 50–60 and the C terminus, the overall pattern of attractive protein-glycan interactions varies between mutants. Comparing the deletion of the Asn181 glycosylation site in the N25Q (glycans at positions 20/29/141/181) and N25Q/N181Q mutants (glycans at positions 20/29/141) reveals significant differences in the attractive energy profiles. Glycans at positions 20/29/141 present more attractive protein-glycan contacts and show increased thermostability as compared with the variant with glycans at positions 20/29/141/181, which demonstrates that position rather than number of glycans is important for thermostability. In the N25Q/N181Q mutant (glycans at positions 20/29/141), regions 4–6, 43–48, 71–73, 117–119, 163–166, 177–185, and the single residues 15, 25, 61, and 99 participate in attractive interactions. It is noteworthy that interactions in the N-terminal region (residues 1–60) with a glycan at position 181 make only a limited contribution to the overall attractive IPE and generally result in a significant reduction in protein-glycan contacts with residues 140–150. This is due to steric hindrance of the glycan at position 181, and the resulting reduction in IPE correlates with the overall destabilizing effect of glycosylation at this position.
FIGURE 9.
IPEs per residue calculated from the time-averaged protein residue-glycan interactions during the MD simulation for the indicated mutants.
Comparison of the glycan substitutions between positions 25 and 29 in the N181Q (glycans at 20/25/141) and N25Q/N181Q (glycans at positions 20/29/141) mutants shows that the first 10 residues of both proteins participate in attractive interactions that are conserved in all mutants with glycans at position 20. However, the N25Q/N181Q mutant (glycans at positions 20/29/141) gains attractive interactions with residues 10–40 in the N-terminal region and at position 120 but loses attractive protein-glycan interactions with residues 140–150. This suggests that glycosylation of position 29 complements the stability generated by glycosylation of position 20 by forming interactions with the first 40 residues in the N-terminal region. These results offer an explanation as to how a given mutation may have differing effects, depending on the context of the other glycosylated positions.
The experimental results indicate that glycosylation at position 20 significantly improves protein stability when combined with glycans at positions 25, 29, and 141, and glycosylation of position 20 is observed in thermostable mutants even in the presence of a destabilizing glycan at position 181. The IPE analyses from MD simulations (Fig. 8) clearly show that the glycan at position 20 covers the protein surface and maximizes protein-glycan interactions. The detailed effects of glycosylation of position 20 were revealed by comparing the effects of glycan substitution between positions 20 and 29 in the N181Q (glycans at positions 20/25/141) and N20/181Q (glycans at positions 25/29/141) mutants. The IPE per residue shows that the combination of glycans at positions 20/25/141 has greater attractive interaction energy than the combination at positions 25/29/141, particularly in the N-terminal region (residues 1–20), residues 140–150, and residues around position 120. Deletion of the Asn25 glycosylation site between the N20Q/N181Q (glycans at positions 25/29/141) and N20Q/N25Q/N181Q (glycans at positions 29/141) indicates that the increased thermostability of the N20Q/N25Q/N181Q mutant is a consequence of the stabilization of the protein-glycan interface involving regions 12–16 and 30–33 and residues 136, 140, and 174. Deletion of the glycosylation site at position 25 allows conformation change of the glycan at position 29, which finds a new minimum energy through the formation of interactions with residues in the N-terminal region. In contrast, protein-glycan contacts in the C terminus, residues 21–23, and two residues 52 and 143 were not present in the N20Q/N25Q/N181Q mutant (glycans at positions 29/141). Although different regions of the protein surface are covered by the glycans in the two mutants, an increase in the extent of the protein-glycan interface is a shared feature of these variants and supports the suggestion that increased protein-glycan interactions underlie the thermostabilization effect of glycosylation.
Not all monoglycosylated xylanase mutants have the lowest protein-glycan IPEpartial when compared with mutants with additional glycans (see, for example, glycans at positions 29 and 141 in Fig. 8, C and E), indicating that glycan-glycan interactions can also influence protein stability. For example, mutants with glycans at positions 20/29/141 (mutant N25Q/N181Q), 20/25/141 (N181Q), 29/141/181 (N20Q/N25Q), and 29/141 (N20Q/N25Q/N181Q) present relatively high thermostability. This suggests that substitutions and deletions of glycosylation sites produce different protein-glycan interaction patterns that can be additionally modulated by glycan-glycan interactions. The glycan-glycan interaction potential energies (IPEGG) of three different glycosylated variants, the wild type (glycans at positions 20/25/141/181), N25Q (20/29/141/181), and N25Q/N181Q (20/141/181) (Table 2), suggest that the combination of glycosylation at positions 141/181 increases glycan-glycan interaction and impairs the formation of the more stable protein-glycan interface. In particular, a glycan at position 181 may interact directly with glycans at all other positions; therefore, the combination of glycosylation at position 181 with any other glycosylated position can significantly modify the enzyme thermostability. Similarly, a glycan at position 25 is favorably located to interact with glycans at positions 141 and 181, and when position 25 is glycosylated in combination with positions 141 and 181, a glycan agglomerate covering the surface of the fingers domain in the XynA is formed (Table 2). Thus, although glycans at either position 25, 141, or 181 can be tolerated by adjustments of the conformation of the other glycans, combined glycosylation events at these positions favor glycan-glycan interactions and tend to reduce thermostability.
The study of factors influencing enzyme thermostability not only furthers the understanding of proteins in general but is also of considerable biotechnological interest. Here we have demonstrated that the thermostability of the bacterial wild-type XynA can be increased by glycosylation of four of the six potential glycosylation sites in the amino acid sequence. Site-directed mutagenesis of the glycosylation sites significantly alters both the glycosylation pattern and thermostability of the protein. There was no correlation between the degree of glycosylation either in thermostability or in protein-glycan interface, clearly demonstrating that the combination of glycosylation position rather than overall glycan content was the determinant of thermostability. Analyses of MD simulations indicated that glycosylated mutants with higher thermostability tend to minimize glycan-glycan interactions and optimize the protein-glycan contacts with the surface of protein. These results suggest that the structural basis for understanding the effects of glycosylation lies in the relative positions of occupied glycosylation sites. When the glycosylation sites are close together, crowding of the glycan chains leads to the formation of less favorable glycan-glycan interactions, whereas in contrast, more separated glycosylation sites enhance protein-glycan interactions, thereby optimizing the protein-glycan interface by permitting glycan conformations that cover the protein surface. If this proves to be a general principal shared by other proteins, we speculate that thermostable glycoproteins could be designed by introducing glycosylation sites with optimal spacing between them to enhance protein-glycan interactions.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grants 2010/18850-2, 2009/50838-5, 2008/57908-6, and 2010/52322-3; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Grants 574002/2008-1, 140124/2009-8, and 307795/2009-8; and Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES). The Institute for Research in Immunology and Cancer is supported in part by the Canadian Center of Excellence in Commercialization and Research (CECR), the Canadian Foundation for Innovation, and the Fonds de Recherche du Québec en Santé (FRQS).

This article contains supplemental Figs. S1–S6.
- MD
- molecular dynamics
- XynA
- endo-β-1,4-xylanase A
- Endo H
- endoglycosidase H
- IPE
- interaction potential energy
- IPEpartial PG
- time-average value of the protein-glycan IPE
- IPEGG
- glycan-glycan IPE.
REFERENCES
- 1. Rothman J. E., Lodish H. F. (1977) Synchronised transmembrane insertion and glycosylation of a nascent membrane protein. Nature 269, 775–780 [DOI] [PubMed] [Google Scholar]
- 2. Apweiler R., Hermjakob H., Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 3. Burda P., Aebi M. (1999) The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257 [DOI] [PubMed] [Google Scholar]
- 4. Taylor M. E., Drickamer K. (2011) Introduction to Glycobiology, Oxford University Press, United Kingdom [Google Scholar]
- 5. Ohtsubo K., Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 6. Jefferis R. (2009) Recombinant antibody therapeutics. The impact of glycosylation on mechanisms of action. Trends Pharmacol. Sci. 30, 356–362 [DOI] [PubMed] [Google Scholar]
- 7. Mulloy B., Corfield A. P. (2010) Structural glycobiology and human health. Biochem. Soc. Trans. 38, 1329–1332 [DOI] [PubMed] [Google Scholar]
- 8. Hecht M. L., Stallforth P., Silva D. V., Adibekian A., Seeberger P. H. (2009) Recent advances in carbohydrate-based vaccines. Curr. Opin. Chem. Biol. 13, 354–359 [DOI] [PubMed] [Google Scholar]
- 9. Jaeken J., Matthijs G. (2007) Congenital disorders of glycosylation. A rapidly expanding disease family. Annu. Rev. Genomics Hum. Genet. 8, 261–278 [DOI] [PubMed] [Google Scholar]
- 10. Marth J. D., Grewal P. K. (2008) Mammalian glycosylation in immunity. Nat. Rev. Immunol. 8, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawasaki N., Itoh S., Hashii N., Takakura D., Qin Y., Huang X., Yamaguchi T. (2009) The significance of glycosylation analysis in development of biopharmaceuticals. Biol. Pharm. Bull. 32, 796–800 [DOI] [PubMed] [Google Scholar]
- 12. Molinari M., Eriksson K. K., Calanca V., Galli C., Cresswell P., Michalak M., Helenius A. (2004) Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol. Cell 13, 125–135 [DOI] [PubMed] [Google Scholar]
- 13. Sitia R., Braakman I. (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 14. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 15. Helenius A., Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 16. Wang C., Eufemi M., Turano C., Giartosio A. (1996) Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry 35, 7299–7307 [DOI] [PubMed] [Google Scholar]
- 17. Shental-Bechor D., Levy Y. (2008) Effect of glycosylation on protein folding. A close look at thermodynamic stabilization. Proc. Natl. Acad. Sci. U.S.A. 105, 8256–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Straumann N., Wind A., Leuenberger T., Wallimann T. (2006) Effects of N-linked glycosylation on the creatine transporter. Biochem. J. 393, 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yi W., Clark P. M., Mason D. E., Keenan M. C., Hill C., Goddard W. A., 3rd, Peters E. C., Driggers E. M., Hsieh-Wilson L. C. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wyss D. F., Choi J. S., Li J., Knoppers M. H., Willis K. J., Arulanandam A. R., Smolyar A., Reinherz E. L., Wagner G. (1995) Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science 269, 1273–1278 [DOI] [PubMed] [Google Scholar]
- 21. Sinha S., Surolia A. (2007) Attributes of glycosylation in the establishment of the unfolding pathway of soybean agglutinin. Biophys. J. 92, 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shental-Bechor D., Levy Y. (2011) Communication. Folding of glycosylated proteins under confinement. J. Chem. Phys. 135, 141104. [DOI] [PubMed] [Google Scholar]
- 23. Cheng S., Edwards S. A., Jiang Y., Gräter F. (2010) Glycosylation enhances peptide hydrophobic collapse by impairing solvation. Chemphyschem. 11, 2367–2374 [DOI] [PubMed] [Google Scholar]
- 24. Petrescu A. J., Wormald M. R., Dwek R. A. (2006) Structural aspects of glycomes with a focus on N-glycosylation and glycoprotein folding. Curr. Opin. Struct. Biol. 16, 600–607 [DOI] [PubMed] [Google Scholar]
- 25. Petrescu A. J., Petrescu S. M., Dwek R. A., Wormald M. R. (1999) A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology 9, 343–352 [DOI] [PubMed] [Google Scholar]
- 26. Woods R. J. (1998) Computational carbohydrate chemistry. What theoretical methods can tell us. Glycoconj. J. 15, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuried E., Ramsland P. A. (2012) Structural Glycobiology, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 28. von der Lieth C., Luetteke T., Frank M. (2010) Bioinformatics for Glycobilogy and Glycomics: An Introduction, John Wiley & Sons, Inc., New York [Google Scholar]
- 29. Woods R. J., Edge C. J., Dwek R. A. (1994) Protein surface oligosaccharides and protein function. Nat. Struct. Biol. 1, 499–501 [DOI] [PubMed] [Google Scholar]
- 30. Lu D., Yang C., Liu Z. (2012) How hydrophobicity and the glycosylation site of glycans affect protein folding and stability. A molecular dynamics simulation. J. Phys. Chem. B 116, 390–400 [DOI] [PubMed] [Google Scholar]
- 31. Daly R., Hearn M. T. (2005) Expression of heterologous proteins in Pichia pastoris. A useful experimental tool in protein engineering and production. J. Mol. Recognit. 18, 119–138 [DOI] [PubMed] [Google Scholar]
- 32. Bretthauer R. K., Castellino F. J. (1999) Glycosylation of Pichia pastoris-derived proteins. Biotechnol. Appl. Biochem. 30, 193–200 [PubMed] [Google Scholar]
- 33. Kukuruzinska M. A., Bergh M. L., Jackson B. J. (1987) Protein glycosylation in yeast. Annu. Rev. Biochem. 56, 915–944 [DOI] [PubMed] [Google Scholar]
- 34. Vieira D. S., Degrève L. (2009) An insight into the thermostability of a pair of xylanases. The role of hydrogen bonds. Mol. Phys. 107, 59–69 [Google Scholar]
- 35. Georis J., de Lemos Esteves F., Lamotte-Brasseur J., Bougnet V., Devreese B., Giannotta F., Granier B., Frère J. M. (2000) An additional aromatic interaction improves the thermostability and thermophilicity of a mesophilic family 11 xylanase. Structural basis and molecular study. Protein Sci. 9, 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Torrez M., Schultehenrich M., Livesay D. R. (2003) Conferring thermostability to mesophilic proteins through optimized electrostatic surfaces. Biophys. J. 85, 2845–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruller R., Deliberto L., Ferreira T. L., Ward R. J. (2008) Thermostable variants of the recombinant xylanase A from Bacillus subtilis produced by directed evolution show reduced heat capacity changes. Proteins 70, 1280–1293 [DOI] [PubMed] [Google Scholar]
- 38. Nelson R. M., Long G. L. (1989) A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal. Biochem. 180, 147–151 [DOI] [PubMed] [Google Scholar]
- 39. Ruller R., Rosa J. C., Faça V. M., Greene L. J., Ward R. J. (2006) Efficient constitutive expression of Bacillus subtilis xylanase A in Escherichia coli DH5α under the control of the Bacillus BsXA promoter. Biotechnol. Appl. Biochem. 43, 9–15 [DOI] [PubMed] [Google Scholar]
- 40. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 [Google Scholar]
- 41. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 42. Bohne-Lang A., von der Lieth C. W. (2005) GlyProt. In silico glycosylation of proteins. Nucleic Acids Res. 33, W214–W219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arfken G. (1985) The Method of Steepest Descents in Mathematical Methods for Physicists. 3rd Ed., pp. 428–436, Academic Press, Orlando, FL [Google Scholar]
- 44. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Hermans J. (1981) Interaction models for water in relation to protein hydration. in Intermolecular Forces (Pullman B., Reidel D., eds) pp. 331–334, Dordrecht, The Netherlands [Google Scholar]
- 45. Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., Onufriev A. (2005) H++. A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyamo S., Kollman P. A. (2002) SETTLE. An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 [Google Scholar]
- 47. van Gunsteren W. F., Berendsen H. J. C. (1988) A leap-frog algorithm for stochastic dynamics. Mol. Simul. 1, 173–185 [Google Scholar]
- 48. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald. An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 49. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4. Algorithms for highly efficient, load-balanced and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 50. van Gunsteren W. F., Billeter S. R., Eising A. A., Hünenberger P. H., Krüger P., Mark A. E., Scott W. R. P., Tironi I. G. (1996) The GROMOS96 Manual and User Guide. in Biomolecular Simulation, Biomos, Groningen, The Netherlands [Google Scholar]