Background: Silkworm ApoLp protein binds S. aureus cell surfaces and inhibits hemolysin gene expression.
Results: ApoLp protein binds LTA. The ApoLp inhibitory effect was attenuated in an LTA synthetase knockdown mutant.
Conclusion: ApoLp protein suppressed S. aureus virulence via LTA binding.
Significance: A non-protein macromolecule on the bacterial cell surface functions as a receptor in the bacterial signal transduction pathway.
Keywords: Bacterial Protein Kinases, Bacterial Signal Transduction, Lipoprotein, Lipoprotein Receptor, Receptors, Silkworm, Staphylococcus aureus, Transcription Regulation
Abstract
We previously reported that a silkworm hemolymph protein, apolipophorin (ApoLp), binds to the cell surface of Staphylococcus aureus and inhibits expression of the saePQRS operon encoding a two-component system, SaeRS, and hemolysin genes. In this study, we investigated the inhibitory mechanism of ApoLp on S. aureus hemolysin gene expression. ApoLp bound to lipoteichoic acids (LTA), an S. aureus cell surface component. The addition of purified LTA to liquid medium abolished the inhibitory effect of ApoLp against S. aureus hemolysin production. In an S. aureus knockdown mutant of ltaS encoding LTA synthetase, the inhibitory effects of ApoLp on saeQ expression and hemolysin production were attenuated. Furthermore, the addition of anti-LTA monoclonal antibody to liquid medium decreased the expression of S. aureus saeQ and hemolysin genes. In S. aureus strains expressing SaeS mutant proteins with a shortened extracellular domain, ApoLp did not decrease saeQ expression. These findings suggest that ApoLp binds to LTA on the S. aureus cell surface and inhibits S. aureus hemolysin gene expression via a two-component regulatory system, SaeRS.
Introduction
Understanding the molecular interactions between host and bacteria is important toward elucidating the mechanisms of infectious diseases. Bacteria express various virulence genes according to the host microenvironment (1). To recognize environmental signals, bacteria possess a two-component system comprising a receptor with histidine kinase activity and a transcription factor activated by phosphorylation. Host animals, on the other hand, recognize invading bacteria by diverse receptors and induce immune responses to kill bacteria.
Staphylococcus aureus is a human pathogenic bacterium present in the nares of 30% of healthy individuals that causes many diseases including sepsis and meningitis. S. aureus secretes several hemolysins and damages host cells. S. aureus hemolysin gene deletion mutants exhibit attenuated virulence against mammals (2, 3), indicating that S. aureus hemolysins are essential for its virulence. S. aureus has 16 two-component systems, which are assumed to recognize the environment and express adequate virulence genes, including hemolysin genes (4). Within the two-component systems, agr, arlRS, and saeRS are required for the expression of hemolysin genes (5–11). The sensor protein, SaeS, of the two-component system SaeRS is a transmembrane protein with two transmembrane helixes and a short extracellular region of nine amino acids (12). Such sensor proteins are called intramembrane-sensing kinases, most of which are involved in resistance against cell wall stress and antibiotics (12). The saeRS locus contains two promoters; saeP-saeQ-saeR-saeS is transcribed from P1 and saeR-saeS is transcribed from P3 (13, 14). saeP and saeQ has an essential role in saeRS function (14, 15). SaeQ protein is assumed to stabilize SaeS protein (16). The P1 promoter is activated by hydrogen peroxide and α-defensin (17, 18), whereas it is inactivated at low pH and high salt concentrations (17). How these environmental signals alter the P1 activity of saeRS, however, is not clear.
S. aureus produces lipoteichoic acids (LTA)2 and wall teichoic acids (WTA) that have an important role in S. aureus virulence (19). S. aureus teichoic acids promote bacterial adherence to host cells (20, 21). An S. aureus gene-disrupted mutant of the tagO gene that encodes WTA synthetase decreases adherence activity to human epithelial cells (22, 23), biofilm forming ability (24), colony spreading ability (25), and virulence in mammals (22, 23). Both LTA and WTA are d-alanylated by enzymes encoded by the dlt operon. The gene-disrupted mutant of the dlt operon decreases resistance ability to host antimicrobial peptides (26, 27), biofilm-forming ability (24), colony-spreading ability (25), and virulence in mice (28). Whether teichoic acids are involved in bacterial gene expression, however, is unknown.
We investigated the interaction between S. aureus and host animals using silkworms, which are larvae of the moth, Bombyx mori (29–34). We previously reported that the silkworm hemolymph protein ApoLp inhibits the P1 activity of saeRS and decreased the expression of saePQRS and hemolysin genes (35). Administration of anti-ApoLp antibody sensitized silkworms to S. aureus, indicating that inhibition of hemolysin production by ApoLp contributes to silkworm resistance to S. aureus (35). How ApoLp inhibits the expression of S. aureus hemolysin genes was, however, not revealed. In the present study, we demonstrated that ApoLp binds LTA, a cell surface component of S. aureus, and LTA binding inhibits the expression of saePQRS and hemolysin genes.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
S. aureus strains were aerobically cultured in tryptic soy broth at 37 °C, and 10 μg/ml erythromycin, 100 μg/ml kanamycin, or 20 μg/ml phleomycin/ml was added to the medium if required. The JM109 strain of Escherichia coli was used as a host for plasmids. Details of the bacterial strains and plasmids used in this study are shown in Table 1. The ltaS deletion mutant (M0674N) and the conditional knockdown mutant of ltaS (M0674Nspac) were cultured at 30 °C in tryptic soy broth supplemented with 0.5 m NaCl to keep out damaging cells. M0674Nspac was cultured in the presence of 1 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG) or in the absence of IPTG.
TABLE 1.
List of bacterial strains and plasmids used
| Strain or plasmid | Genotypes or characteristics | Source or Ref. |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | NCTC8325-4, restriction mutant | Ref. 41 |
| NCTC8325–4 | NCTC8325 cured of ϕ11, ϕ12, and ϕ13 | Ref. 42 |
| M0661 | RN4220 ΔsaePQRS::Kanr | This work |
| M0661N | NCTC8325-4 ΔsaePQRS::Kanr | This work |
| M0674 | RN4220 ΔltaS::phleor | Ref. 36 |
| M0674N | NCTC8325-4 ΔltaS::phleor (transduction from M0674) | This work |
| M0674Nspac | NCTC8325-4 Pspac::ltaS; Ermr | This work |
| M0702 | RN4220 ΔtagO::Ermr | Ref. 25 |
| M0793 | RN4220 ΔdltABCD::Ermr | Ref. 25 |
| M0702N | NCTC8325-4 ΔtagO::Ermr (transduction from M0702) | This work |
| M0793N | NCTC8325-4 ΔdltABCD::Ermr (transduction from M0793) | This work |
| Plasmids | ||
| pMutinT3 | Suicidal vector for Gram-positive bacteria; Ampr, Ermr | Ref. 43 |
| pMutinT3-ltaS | pMutinT3 carrying 5′-partial region of ltaS | This work |
| pUC-Int | pUC19 carrying integration region for NCTC8325-4 | This work |
| pUC-Int-erm | pUC-Int carrying erythromycin-resistant gene | This work |
| pInt-sae | pUC-Int-erm carrying saePQRS gene | This work |
| pInt-I9Q-SaeS | pInt-sae carrying SaeS with I9Q substitution | This work |
| pInt-I9Q,L63Q-SaeS | pInt-sae carrying SaeS with I9Q and L63Q substitution | This work |
| pInt-Δ34–36-SaeS | pInt-sae carrying SaeS deleted with 34 to 36th amino acids | This work |
| pInt-Δ35–37-SaeS | pInt-sae carrying SaeS deleted with 35 to 37th amino acids | This work |
| pInt-Δ34–37-SaeS | pInt-sae carrying SaeS deleted with 34 to 37th amino acids | This work |
| pKOR3a | Vector for allelic replacement in S. aureus, Cmr | Ref. 38 |
| pluc | pND50 with luc + with a ribosomal binding site | Ref. 44 |
| pluc-hla | pluc with hla promoter from RN4220 | Ref. 44 |
| pluc-saeP1 | pluc with sae P1 promoter from RN4220 | Ref. 35 |
Purification of ApoLp
We purified ApoLp from the silkworm hemolymph according to our previously published method (35). We measured the activity required to inhibit S. aureus hemolysin production in each purification step. In this study, we used an ApoLp protein fraction purified by phosphocellulose chromatography using buffer A (50 mm MES (pH 6.9), 100 mm NaCl, 2 mm dithiothreitol, 5% glycerol).
Measurement of S. aureus Hemolysin Production
S. aureus culture supernatants were collected by centrifugation, and 2-fold serial dilutions were mixed with an equal volume of 5% sheep erythrocytes. Incubation was performed at 37 °C for 1 h and at 4 °C overnight. Hemolytic activity was defined as the reciprocal of dilutions that yielded 50% erythrocyte lysis.
Binding Assay of ApoLp to LTA
Lipoteichoic acids from S. aureus (Sigma) were poured into Immulon 1B ELISA plates (Dynatech Laboratories, Chantilly, VA) and incubated at 4 °C overnight. The plates were washed three times with 200 μl of PBS containing 0.5% Tween 20 (PBST) and treated with 200 μl of PBST containing 3% bovine serum albumin for 1 h at room temperature. The plates were washed three times with PBST and incubated with 100 μl of ApoLp solution at room temperature for 30 min. The plates were washed three times with PBST and incubated with anti-ApoLp IgG (35) at room temperature for 2 h. The plates were washed five times with PBST and incubated with anti-mouse IgG AP conjugate (Promega) at room temperature for 2 h. The plates were washed seven times with PBST and incubated with 200 μl of substrate buffer (50 mm Tris-HCl (pH 9.0), 1 mm MgCl2, 1 mg/ml p-nitrophenylphosphate (Sigma)). Absorbance of 450 nm was measured using a microplate reader (MTP-300, COLONA ELECTRONIC, Ibaraki, Japan). Binding of ApoLp to LTA was determined by subtracting the value without LTA.
Competitive Assay of ApoLp with LTA against S. aureus Hemolysin Production
Overnight culture of S. aureus NCTC8325–4 was inoculated into a 200-fold amount of fresh tryptic soy broth and cultured at 37 °C for 1 h. A 600-μl aliquot of the cultures was supplemented with 200 μl of ApoLp solution or a mixed solution of ApoLp and LTA and was cultured at 37 °C for 4 h. The culture supernatant was collected by centrifugation at 10,000 × g for 2 min. Hemolytic activity of the supernatant was measured using the hot-cold method described above.
Construction of the ltaS Deletion Mutant
Deletion of the ltaS gene in the S. aureus RN4220 strain was performed by Oku et al. (36). The deletion was transferred to NCTC8325-4 by phage transduction using phage 80α and confirmed by Southern blot analysis.
Construction of the ltaS Conditional Knockdown Mutant
The ltaS conditional knockdown mutant was constructed according to the previously published method (37). Briefly, the 5′ region of the ltaS gene was amplified by PCR and inserted into pMutinT3, resulting in pMutinT3-ltaS. The RN4220 strain was transformed with pMutinT3-ltaS, and the transformed strain was resistant against erythromycin. The mutation was transferred to the NCTC8325-4 strain by phage transduction using phage 80α. The desired chromosomal DNA mutation was confirmed by Southern blot analysis.
Construction of the saePQRS Deletion Mutant
Upstream and downstream regions of the saePQRS were amplified by PCR using the oligonucleotide primers listed in Table 2 and NCTC8325-4 genomic DNA as the template. The aph gene, conferring kanamycin resistance, was amplified by PCR using pSF151 as the template (38). Three DNA fragments of the upstream region, downstream region, and aph gene were spliced together using splicing by overlap extension PCR, resulting in an saePQRS-cassette. The saePQRS cassette was inserted into the SmaI site of pKOR3a, resulting in pKOR3a-saePQRS. S. aureus RN4220 was transformed with pKOR3a-saePQRS. The transformant was cultured in tryptic soy broth, and 103 cells were spread onto tryptic soy agar plates containing 12.5 μg/ml chloramphenicol. The plates were incubated at 43 °C overnight. The resulting colonies were cultured in tryptic soy broth at 37 °C and spread onto tryptic soy agar plates containing 1 μg/ml anhydrotetracycline and 50 μg/ml kanamycin. The resulting colonies were examined for sensitivity to chloramphenicol. The saePQRS deletion was confirmed by Southern blot analysis.
TABLE 2.
Primers used in this study
| Target | Sequence (5′-3′) | Ref. |
|---|---|---|
| saePQRS (upstream) | ||
| Forward | ATAACGAACAGGAGTCCATCA | This work |
| Reverse | ATCACCTCAAATGGTTCGCTTCAAAATAAGAGGAGGGCATTT | |
| saePQRS (downstream) | ||
| Forward | GTTCGCTAGATAGGGGTCCCATTACACAAATTAGACATTA | This work |
| Reverse | TGATTTGCAACATCCACCTG | |
| ltaS (5′-region) | ||
| Forward | AAGAAGCTTCTAAATAACGGGGGAAAGAATCATGAGTTC | Ref. 37 |
| Reverse | GGAGGATCCGACAGGAACAAATTTCTTACTAAATGCTTTTG | |
| saePQRS (complementation) | ||
| Forward | GGAGGATCCTTATTGTGGCAAAAGGTTT | This work |
| Reverse | AAGAAGCTTATTATTAGGCGGCATACAG | |
| Genome integration | ||
| Forward | GGAGGATCCTTACGCATCCAAACACTCC | Ref. 45 |
| Reverse | GGTGGTACCAACACAACACTGACACGTCATTTA | |
| Erythromycin-resistant gene | ||
| Forward | GAAGAATTCAACCTATAAAAATAGGCGTAT | Ref. 46 |
| Reverse | GGTGGTACCAACGTTCTTGCCATTGCTGCA | |
| SaeS | ||
| R-7S | ACTTCTAATTGATAACACCATTATCG | This work |
| F-I9Q | CAACAAATTATTGGCGTCGTTTCGAG | |
| R-62P | TGGATTAATAAAAATACTACATATTAATAAGG | |
| F-L63Q | CAAATACAAAAAATTAAGCAGTTTAATATAAAAACTAAGC | |
| R-33F | AAACCACATTAAAATATATGCAATTGC | |
| F-37M | ATGACACTAACTTTGACCTTAACG | |
| R-34N | GTTAAACCACATTAAAATATATGCAATTGC | |
| F-38T | ACACTAACTTTGACCTTAACG | |
Construction of Mutated saeS Genes
A DNA fragment containing saePQRS was amplified by PCR using the oligonucleotide primers listed in Table 2 and inserted into pUC-Int-erm, resulting in pInt-sae. Point mutations or deletions were introduced into pInt-sae by PCR using oligonucleotide primers. The mutations were confirmed by sequencing. The saePQRS-deleted mutant (M0661) was transformed with plasmids and the desired integration of the plasmids into chromosomal DNA was confirmed by Southern blot analysis. The integrated plasmids were transferred to M0661N by phage 80α. These strains were used as S. aureus strains expressing wild-type SaeS (WT) or mutated SaeS.
Measurement of Gene Expression by Quantitative Real-time PCR
The protocol and oligonucleotide primers used were essentially the same as those used previously (35). Briefly, S. aureus cells were collected by centrifugation, treated with RNAprotect bacteria reagent (Qiagen, Gaithersburg, MD) and lysed in a buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mg/ml lysostaphin). RNA was extracted using an RNeasy Mini kit (Qiagen). RNA was reverse transcribed to cDNA. Quantitative real-time PCR was performed using cDNA as a template and primers for target mRNAs. The signals were detected using a StepOnePlus real-time PCR System (Applied Biosystems). The data were normalized to 16S ribosomal RNA.
Reporter Assay
The RN4220 strain was transformed with reporter plasmids (Table 1). Plasmids were transferred into NCTC8325-4 and mutant strains by phage transduction using phage 80α. The strains were aerobically cultured at 30 °C in tryptic soy broth supplemented with 0.5 m NaCl. The S. aureus cultured solutions (600 μl, A600 = 0.1) were supplemented with ApoLp solution and cultured for 6 h at 30 °C. The cells were collected by centrifugation and lysed in a buffer (20 mm KH2PO4 (pH 7.8), 0.04% Triton X-100, 0.1 mm dithiothreitol, 10 μg/ml lysostaphin, and one tablet of protease inhibitor (Roche Applied Science)). Cell lysate supernatant was incubated with luciferase substrate, and luminescence was measured using a luminometer (Berthold Technologies, Bad Wildbad, Germany). The promoter activity was calculated as luminescence units per milligram of protein.
RESULTS
ApoLp Binds S. aureus Lipoteichoic Acids
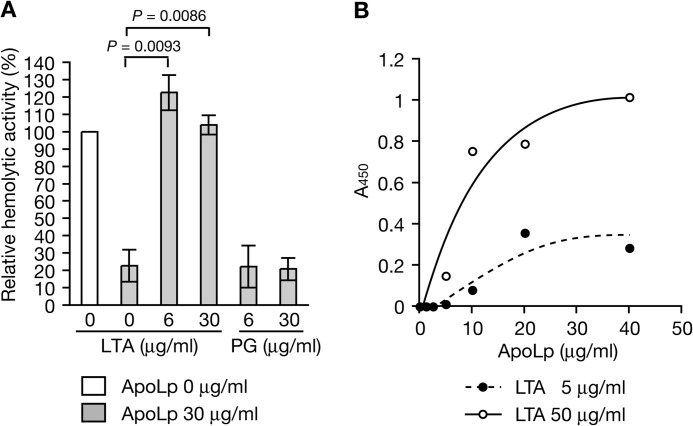
We previously reported that ApoLp binds S. aureus cell surfaces (35). To identify the S. aureus cell surface molecule that ApoLp binds, we examined whether LTA and peptidoglycan, major cell surface components, abolished the ApoLp-induced inhibition of S. aureus hemolysin production. Addition of LTA to S. aureus culture medium blocked the inhibitory effect of ApoLp on S. aureus hemolysin production (Fig. 1A). In contrast, the addition of peptidoglycan did not block the inhibitory effect of ApoLp (Fig. 1A). These findings indicate that ApoLp binds LTA of S. aureus cell surfaces. We then examined whether ApoLp directly binds LTA. We measured the amount of ApoLp bound to immobilized LTA by enzyme-linked immunosorbent assay. The amount of ApoLp bound to LTA was increased with increasing amounts of added ApoLp (Fig. 1B). The amount of ApoLp bound to LTA increased with increasing amounts of immobilized LTA (Fig. 1B). These findings indicate that ApoLp directly binds LTA.
FIGURE 1.
ApoLp binds LTA. A, ApoLp, ApoLp, and LTA (from S. aureus, Sigma), or ApoLp and peptidoglycan (PG, from S. aureus, Sigma) was added to S. aureus NCTC8325-4 culture (A600 of 0.1), and the mixtures were cultured further for 4 h. Hemolytic activity of the culture supernatant against sheep erythrocytes was measured. Means ± S.D. from two independent experiments are presented. Student's t test p values are presented. B, LTA (5 μg/ml or 50 μg/ml) was immobilized onto a microplate. ApoLp was added to the plate. The amount of ApoLp bound to the plate was measured using an ELISA with anti-ApoLp IgG. The vertical axis represents A450, which reflects the amount of ApoLp bound to LTA.
The Inhibitory Effect of ApoLp on S. aureus Hemolysin Production Is Attenuated in LTA Synthetase S. aureus Mutants
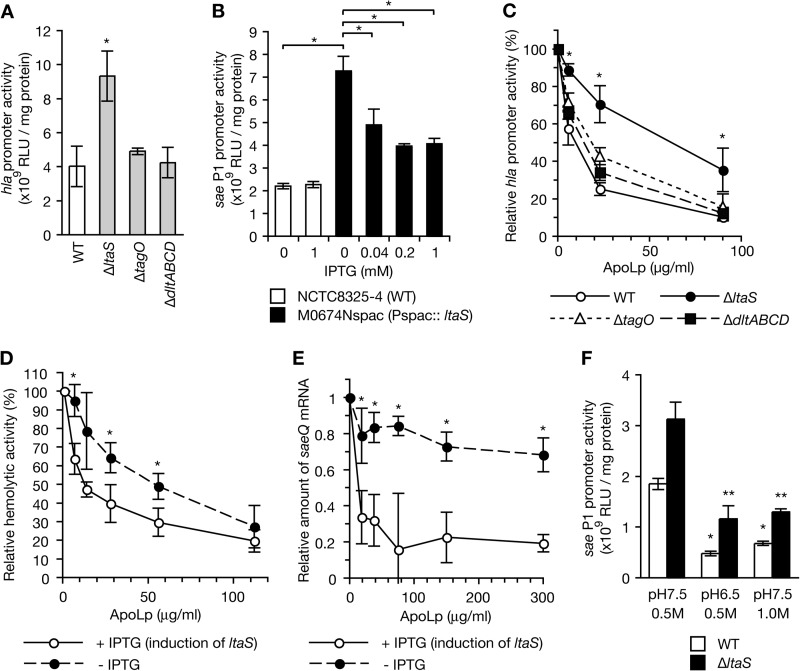
To determine whether ApoLp binding to LTA is required for the inhibitory effect of ApoLp on S. aureus hemolysin production and saeQ expression, we examined the inhibitory effect of ApoLp using gene deletion mutants of the ltaS gene, which encodes LTA synthetase, and an ltaS conditional knockdown mutant. Furthermore, we examined the effect of ApoLp in the deletion mutant of tagO encoding WTA synthetase or dltABCD, which functions in the d-alanylation of teichoic acids. First, we examined the activity of the hla promoter encoding α-hemolysin in the absence of ApoLp in these mutants. The promoter activity of hla was higher in the ltaS-deleted mutant than in the wild-type strain (Fig. 2A). The promoter activity of hla did not differ between the wild-type strain, the tagO-deleted mutant, and the dltABCD-deleted mutant (Fig. 2A). Thus, LTA has a role in decreasing hla expression, whereas WTA and d-alanylation of teichoic acids have no role in hla expression in the absence of ApoLp. In addition, in the ltaS conditional knockdown mutant that contains the IPTG-inducible promoter Pspac upstream of the ltaS gene, the activity of sae P1, which is the promoter for the saeQ gene, was higher in the absence of IPTG than in the wild-type strain (Fig. 2B). The addition of IPTG to the ltaS-conditional mutant decreased the activity of sae P1 (Fig. 2B). Thus, LTA has a negative role in sae P1 expression in the steady state.
FIGURE 2.
The inhibitory effect of ApoLp against hemolysin production was attenuated in the ltaS mutant encoding LTA synthetase. A, S. aureus NCTC8325-4 strain (WT), the ltaS mutant (ΔltaS), the tagO mutant (ΔtagO), and the dltABCD mutant (ΔdltABCD) were transformed with a reporter plasmid carrying the hla promoter and cultured at 30 °C for 6 h. Luciferase activities (relative light units (RLU)) of the cell lysates were measured. Means ± S.D. of three independent experiments are shown. The asterisk indicates Student's t test p value of <0.05 between the wild-type strain and the ltaS mutant. B, S. aureus NCTC8325–4 strain (WT) and the ltaS conditional knockdown mutant (Pspac::ltaS) were transformed with a reporter plasmid carrying sae P1 and cultured in the absence or presence of IPTG at 30 °C for 6 h. Luciferase activities of the cell lysates were measured. Means ± S.D. of three independent experiments are shown. The asterisk indicates Student's t test p value of <0.05. C, various amounts of ApoLp were added to S. aureus cultures (A600 of 0.1) of NCTC8325-4 (WT), M0674N (ΔltaS), M0702N (ΔtagO), and M0793N (ΔdltABCD) that were transformed with a reporter plasmid carrying the hla promoter, and the mixtures were cultured further at 30 °C for 6 h. Luciferase activity of the cell lysate was measured. The vertical axis represents relative luciferase activity against that without ApoLp. Asterisks indicate a Student's t test p value of <0.05 between NCTC8325-4 and the ltaS mutant. D, various amounts of ApoLp were added to S. aureus culture of M0674spac carrying IPTG-inducible ltaS (A600 of 0.1) in the presence or absence of IPTG, and the mixtures were cultured further at 30 °C for 6 h. Hemolytic activity of the culture supernatant against sheep erythrocytes was measured. The vertical axis represents relative hemolytic activity against that without ApoLp. Means ± S.D. from two independent experiments are shown. Asterisks indicate a Student's t test p value of <0.05 between +IPTG and −IPTG. E, various amounts of ApoLp were added to S. aureus culture of M0674spac carrying IPTG-inducible ltaS (A600 of 0.1) in the presence or absence of IPTG and further cultured at 30 °C for 6 h. Total RNA was extracted from the cultured cells. The amount of saeQ mRNA was measured by quantitative RT-PCR. Means ± S.D. from three independent experiments are shown. Asterisks indicate a Student's t test p value of <0.05 between +IPTG and −IPTG. F, S. aureus NCTC8325–4 strain (WT) and the ltaS mutant (ΔltaS) that were transformed with a reporter plasmid carrying sae P1 were cultured under normal conditions (pH 7.5, 0.5 m NaCl), a low pH condition (pH 6.5, 0.5 m NaCl), or a high salt condition (pH 7.5, 1.0 m NaCl). Luciferase activities of the cell lysates were measured. Means ± S.D. of three independent experiments are shown. The asterisks indicate Student's t test p value of <0.05 between the normal condition and the low pH condition or between the normal condition and the high salt condition.
We then examined the effect of ApoLp in the ltaS, tagO, and dltABCD mutants. In the wild-type strain, the tagO-deleted mutant, and the dltABCD-deleted mutant, the addition of 90 μg/ml ApoLp decreased the promoter activity of hla to <20% that of the non-treated sample, whereas in the ltaS-deleted strain, the addition of ApoLp decreased the promoter activity only to ∼40% that of the non-treated sample (Fig. 2C). In addition, in the ltaS conditional knockdown mutant, the inhibitory effect of ApoLp on hemolysin production was attenuated in the absence of IPTG compared with that in the presence of IPTG, which induces ltaS expression (Fig. 2D). Furthermore, in the presence of IPTG, the addition of ApoLp decreased saeQ expression to <20% that of the non-treated sample, whereas in the absence of IPTG, the addition of ApoLp decreased saeQ expression only to ∼70% that of the non-treated sample (Fig. 2E). These findings indicate that repression of ltaS expression reduced the inhibitory effects of ApoLp on hemolysin production and saeQ expression and thus indicated that ApoLp binding to LTA was required for the subsequent inhibition of hemolysin production and saeQ expression.
Low pH and high salt conditions decrease the activity of sae P1 (17). We examined whether LTA is required for the inactivation of sae P1 activity by a low pH or high salt condition. The sae P1 activity was lower at pH 6.5 than at pH 7.5 in both the wild-type and the ltaS-deleted mutant (Fig. 2F). In addition, the sae P1 activity was lower in 1.0 m NaCl than in 0.5 m NaCl in both the wild-type and the ltaS-deleted mutant (Fig. 2F). Thus, LTA is not involved in the regulation of sae P1 activity by low pH and high salt conditions.
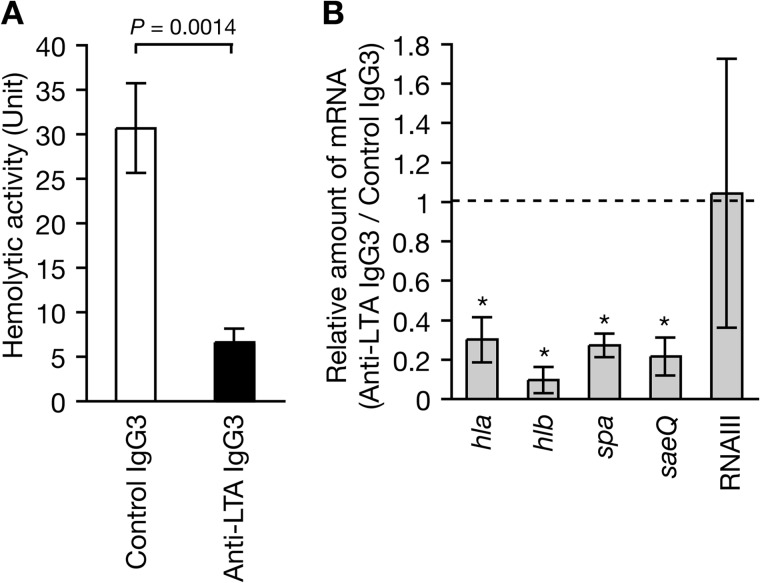
Addition of Anti-LTA Monoclonal Antibody Decreases S. aureus Hemolysin Production and saeQ Expression
Based on the findings that the ApoLp binding to LTA is necessary to inhibit the expression of the hemolysin genes and saeQ, we hypothesized that ApoLp binding to LTA triggers signals to inhibit the expression of hemolysin genes and saeQ. Studies of the signal transduction mechanism in mammals revealed that binding of an antibody to the receptor sometimes activates the signaling pathway in the similar way as the binding of true ligands (39, 40). We examined whether binding of the monoclonal antibody to LTA causes similar effects as ApoLp. The addition of anti-LTA IgG3 decreased S. aureus hemolysin production compared with the addition of control IgG3 (Fig. 3A). Furthermore, the addition of anti-LTA IgG3 decreased the expression of hla encoding α-hemolysin, hlb encoding β-hemolysin, spa encoding protein A, and saeQ compared with the addition of control IgG3 (Fig. 3B). The addition of anti-LTA IgG3 did not decrease the expression of RNAIII, which is a regulatory RNA on various S. aureus virulence genes. These findings indicate that IgG3 binding to LTA triggers signaling that leads to inhibition of the expression of hemolysin genes, spa, and saeQ, as observed for ApoLp, and that LTA acts as a receptor to transmit the signaling pathway that leads to down-regulation of the expression of virulence genes.
FIGURE 3.
Addition of anti-LTA antibody decreases S. aureus hemolysin production. A, anti-LTA mouse IgG3 monoclonal antibody (mAb 55, Hycult Biotech, The Netherlands) or control mouse IgG3 antibody (Bethyl Laboratories, Montgomery, TX) was added to S. aureus NCTC8325-4 culture (A600 = 0.1) at a final concentration of 250 μg/ml, and the mixture was cultured further for 4 h. Hemolytic activities of the culture supernatants against sheep erythrocytes was measured. Means ± S.D. from three independent experiments are shown. Student's t test p value is presented. B, anti-LTA mouse IgG3 monoclonal antibody or control mouse IgG3 antibody was added to S. aureus NCTC8325-4 culture (A600 = 0.1) at a final concentration of 250 μg/ml, and the mixture was cultured further for 4 h. Total RNA was extracted from the cultured cells. RNA amounts of hla, hlb, spa, saeQ, and RNAIII were measured by quantitative RT-PCR. The vertical axis shows the relative values against the amount of RNAs in the presence of control IgG3. Means ± S.D. of three independent experiments are shown. Asterisks indicate Student's t test p value of <0.05 between anti-LTA IgG3 and control IgG3.
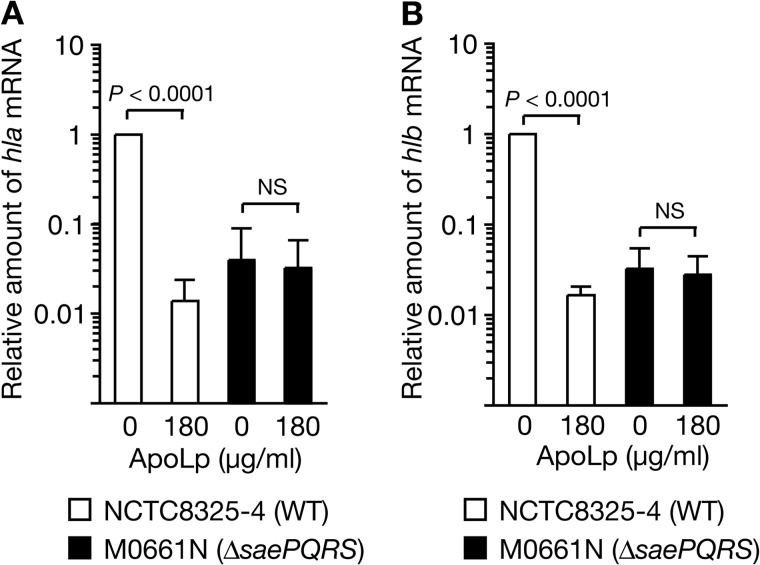
Inhibitory Effect of ApoLp on Hemolysin Expression Is Diminished in the saePQRS-deleted Mutant and the saeS Mutant with a Shortened Extracellular Domain
Based on our findings that ApoLp or IgG3 binding to LTA inhibits the expression of saeQ and hemolysin genes, together with the reports that the two-component SaeRS is required for the expression of hemolysin genes (10, 11), we hypothesized that ApoLp binding to LTA inactivates SaeRS and thus inhibits the expression of hemolysin genes. We examined whether saePQRS is required for the inhibitory effect of ApoLp on hemolysin genes. In a wild-type strain of S. aureus, as we reported previously, the addition of 180 μg/ml of ApoLp decreased the expression of hla and hlb (Fig. 4, A and B). In contrast, in the saePQRS-deleted mutant, the addition of 180 μg/ml of ApoLp did not decrease the expression of hla and hlb (Fig. 4, A and B). These findings indicate that saePQRS is required for the inhibitory effect of ApoLp on the expression of hemolysin genes.
FIGURE 4.
The inhibitory effect of ApoLp against S. aureus hemolysin expression was diminished in the saePQRS-deleted mutant. ApoLp (180 μg/ml) was added to S. aureus cultures (A600 of 0.1) of NCTC8325-4 (WT) and M0661N (ΔsaePQRS), and the mixture was cultured further for 4 h. Total RNA was extracted from the cultured cells. RNA amounts of hla (A) and hlb (B) were measured by quantitative RT-PCR. The vertical axis shows the relative value against the amount of RNA of NCTC8325-4 in the absence of ApoLp. Means ± S.D. of three independent experiments are shown. Student's t test p values are presented. NS, not significant.
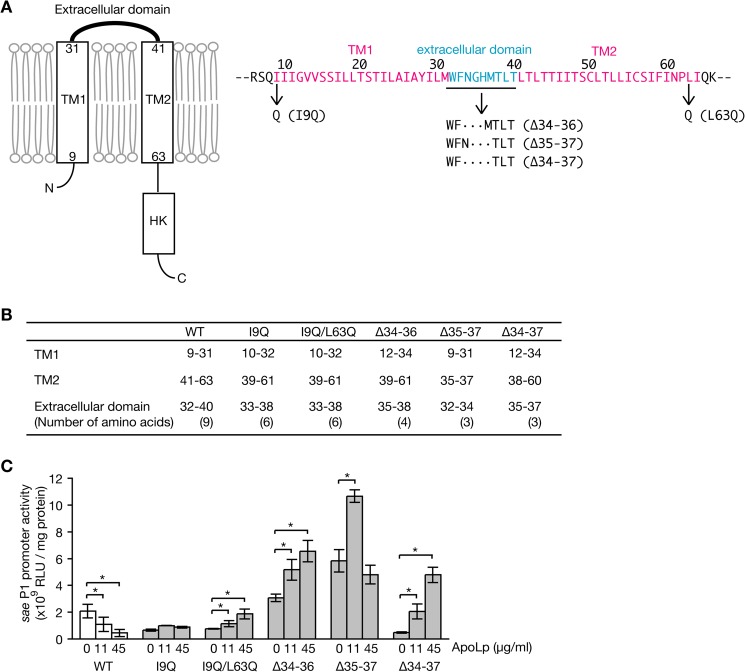
The requirement of LTA and saePQRS for the inhibitory effect of ApoLp on hemolysin expression suggests that the sensor protein SaeS senses the binding of ApoLp to LTA. SaeS is a transmembrane protein with two transmembrane helixes and a short extracellular region of 9 amino acids (Fig. 5A) (12). We hypothesized that the short extracellular region contributes to recognize ApoLp. To address this point, we constructed a deletion mutant of the extracellular domain or introduced an amino acid substitution in the transmembrane domain that theoretically shortens the extracellular domain. The lengths of the extracellular domains of the SaeS mutant proteins were predicted to be short by the in silico program TMHMM (version 2.0; Fig. 5B). The addition of ApoLp to an S. aureus strain expressing wild-type SaeS decreased sae P1 activity in a dose-dependent manner, whereas the addition of ApoLp to S. aureus strains expressing SaeS mutant proteins with a shortened extracellular domain did not decrease sae P1 activity (Fig. 5C). Rather, some mutants with a shortened extracellular domain increased sae P1 activity in the presence of ApoLp. These findings indicate that the extracellular domain of SaeS is required for the recognition of ApoLp binding to LTA and the subsequent down-regulation of sae P1 activity.
FIGURE 5.
The inhibitory effect of ApoLp against S. aureus sae P1 activity was abolished in S. aureus strains expressing mutated SaeS with a shortened extracellular domain. A, schematic representation of SaeS. TM1 and TM2 are two-transmembrane domains. HK, histidine kinase domain. Amino acid sequences of TM1, TM2, and the extracellular domain are presented in the right panel. Amino acid substitutions and deletions in mutated SaeS proteins are shown. B, regions of TM1, TM2, and the extracellular domain in wild-type SaeS (WT) and mutated SaeS were predicted by the in silico program TMHMM (version 2.0). The number of extracellular amino acids is presented in parentheses. C, S. aureus strains expressing wild-type SaeS (WT) or mutated SaeS were transformed with a reporter plasmid carrying sae P1 and cultured in the presence of ApoLp. S. aureus cells were collected at A600 of 1.5, and luciferase activities of the cell lysates were measured. Means ± S.D. of three independent experiments are shown. The asterisks indicate Student's t test p value of <0.05.
DISCUSSION
The findings of the present study revealed that ApoLp binds LTA. In addition, the inhibitory effects of ApoLp against the expression of hemolysin genes and saeQ were attenuated in LTA synthetase-deficient S. aureus mutants. Furthermore, anti-LTA monoclonal antibody decreased the expression of hemolysin genes and saeQ in a similar way as ApoLp. Thus, binding of ApoLp or antibody protein to LTA transmits a signal to decrease the expression of hemolysin genes and saeQ. This study first revealed that LTA, a non-protein bacterial cell surface component, acts as a receptor to recognize environmental signals.
This study revealed that the two-component system saeRS has an essential role in the inhibitory effect of ApoLp against hemolysin expression. Furthermore, the extracellular domain of the sensor protein of saeRS was required for the inhibitory effect of ApoLp against sae P1 activity. These findings suggest that the extracellular domain of SaeS recognizes ApoLp binding to LTA and transmits signals to decrease the expression of hemolysin genes. Because ApoLp is a large molecule of 294 kDa (35), the binding of ApoLp to LTA will occur in a milieu outside the peptidoglycan layer (Fig. 6). The conformational change of LTA by ApoLp binding might alter the lipid fluidity to transmit signals to SaeS or it might allow direct access to the extracellular region of SaeS (Fig. 6). Furthermore, this study demonstrated that LTA has a negative role in the sae P1 expression in the absence of ApoLp. This suggests that the presence of LTA is recognized by SaeS in the steady state. Additional studies are needed to elucidate how the extracellular domain of SaeS recognizes LTA and its binding to ApoLp.
FIGURE 6.
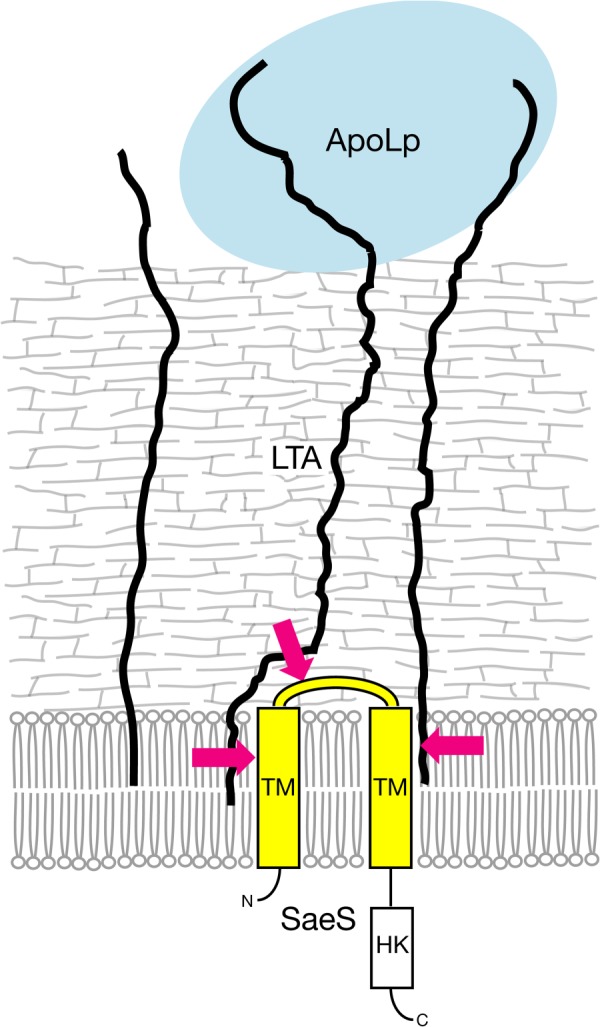
Model of the inhibitory mechanism of ApoLp against SaeS through binding to LTA. Binding of ApoLp to LTA inactivates the SaeS sensor kinase. The extracellular domain of SaeS is essential for recognizing the binding of ApoLp to LTA. The magenta arrows indicate possible pathways for the physical alteration of LTA that leads to inactivation of SaeS.
Low pH and high salt conditions inactivate the P1 activity of saePQRS (17, 18). The inhibitory effects of low pH and high salt conditions against sae P1 activity were not abolished in the ltaS mutant (Fig. 2F). This finding suggests that LTA does not act as a receptor for SaeS to recognize low pH and high salt conditions and that there is an LTA-independent pathway to inactivate the SaeRS two-component system.
Teichoic acids have been thought to have an important role in S. aureus virulence by promoting S. aureus adherence to host cells or as a required component for S. aureus resistance against host antimicrobial peptides. The present study revealed a novel function of LTA to bind host proteins and transfer signals to repress the expression of S. aureus virulence genes. Thus, LTA acts as a receptor in bacteria to facilitate recognition of the host environment. The biologic meaning of the novel function of LTA in S. aureus infectious processes requires further study.
This work was supported by grants-in-aid for scientific research. This work was supported in part by the Naito Foundation and the Genome Pharmaceuticals Institute.
- LTA
- lipoteichoic acid(s)
- WTA
- wall teichoic acid(s)
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1. Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P. (2002) An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561 [DOI] [PubMed] [Google Scholar]
- 2. Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., Kennedy A. D., Dorward D. W., Klebanoff S. J., Peschel A., DeLeo F. R., Otto M. (2007) Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514 [DOI] [PubMed] [Google Scholar]
- 3. Bubeck Wardenburg J., Bae T., Otto M., Deleo F. R., Schneewind O. (2007) Poring over pores: α-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13, 1405–1406 [DOI] [PubMed] [Google Scholar]
- 4. Ji Y., Yu C., Liang X. (2007) Transcriptomic analysis of ArlRS two-component signaling regulon, a global regulator, in Staphylococcus aureus. Methods Enzymol. 423, 502–513 [DOI] [PubMed] [Google Scholar]
- 5. Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P. (1988) Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunman P. M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., Brown E. L., Zagursky R. J., Shlaes D., Projan S. J. (2001) Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fournier B., Klier A., Rapoport G. (2001) The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41, 247–261 [DOI] [PubMed] [Google Scholar]
- 8. Liang X., Zheng L., Landwehr C., Lunsford D., Holmes D., Ji Y. (2005) Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187, 5486–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rogasch K., Rühmling V., Pané-Farré J., Höper D., Weinberg C., Fuchs S., Schmudde M., Bröker B. M., Wolz C., Hecker M., Engelmann S. (2006) Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188, 7742–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giraudo A. T., Cheung A. L., Nagel R. (1997) The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168, 53–58 [DOI] [PubMed] [Google Scholar]
- 11. Giraudo A. T., Calzolari A., Cataldi A. A., Bogni C., Nagel R. (1999) The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177, 15–22 [DOI] [PubMed] [Google Scholar]
- 12. Mascher T. (2006) Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264, 133–144 [DOI] [PubMed] [Google Scholar]
- 13. Steinhuber A., Goerke C., Bayer M. G., Döring G., Wolz C. (2003) Molecular architecture of the regulatory Locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185, 6278–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adhikari R. P., Novick R. P. (2008) Regulatory organization of the staphylococcal sae locus. Microbiology 154, 949–959 [DOI] [PubMed] [Google Scholar]
- 15. Jeong D. W., Cho H., Jones M. B., Shatzkes K., Sun F., Ji Q., Liu Q., Peterson S. N., He C., Bae T. (2012) The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 86, 331–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeong D. W., Cho H., Lee H., Li C., Garza J., Fried M., Bae T. (2011) Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J. Bacteriol. 193, 4672–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geiger T., Goerke C., Mainiero M., Kraus D., Wolz C. (2008) The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190, 3419–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palazzolo-Ballance A. M., Reniere M. L., Braughton K. R., Sturdevant D. E., Otto M., Kreiswirth B. N., Skaar E. P., DeLeo F. R. (2008) Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180, 500–509 [DOI] [PubMed] [Google Scholar]
- 19. Neuhaus F. C., Baddiley J. (2003) A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aly R., Shinefield H. R., Litz C., Maibach H. I. (1980) Role of teichoic acid in the binding of Staphylococcus aureus to nasal epithelial cells. J. Infect. Dis. 141, 463–465 [DOI] [PubMed] [Google Scholar]
- 21. Carruthers M. M., Kabat W. J. (1983) Mediation of staphylococcal adherence to mucosal cells by lipoteichoic acid. Infect. Immun. 40, 444–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weidenmaier C., Peschel A., Xiong Y. Q., Kristian S. A., Dietz K., Yeaman M. R., Bayer A. S. (2005) Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 191, 1771–1777 [DOI] [PubMed] [Google Scholar]
- 23. Weidenmaier C., Kokai-Kun J. F., Kristian S. A., Chanturiya T., Kalbacher H., Gross M., Nicholson G., Neumeister B., Mond J. J., Peschel A. (2004) Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10, 243–245 [DOI] [PubMed] [Google Scholar]
- 24. Gross M., Cramton S. E., Götz F., Peschel A. (2001) Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69, 3423–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaito C., Sekimizu K. (2007) Colony spreading in Staphylococcus aureus. J. Bacteriol. 189, 2553–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peschel A. (2002) How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10, 179–186 [DOI] [PubMed] [Google Scholar]
- 27. Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., Götz F. (1999) Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 [DOI] [PubMed] [Google Scholar]
- 28. Collins L. V., Kristian S. A., Weidenmaier C., Faigle M., Van Kessel K. P., Van Strijp J. A., Götz F., Neumeister B., Peschel A. (2002) Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186, 214–219 [DOI] [PubMed] [Google Scholar]
- 29. Kaito C., Akimitsu N., Watanabe H., Sekimizu K. (2002) Silkworm larvae as an animal model of bacterial infection pathogenic to humans. Microb. Pathog. 32, 183–190 [DOI] [PubMed] [Google Scholar]
- 30. Kaito C., Kurokawa K., Matsumoto Y., Terao Y., Kawabata S., Hamada S., Sekimizu K. (2005) Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56, 934–944 [DOI] [PubMed] [Google Scholar]
- 31. Kaito C., Morishita D., Matsumoto Y., Kurokawa K., Sekimizu K. (2006) Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol. Microbiol. 62, 1601–1617 [DOI] [PubMed] [Google Scholar]
- 32. Kaito C., Sekimizu K. (2007) A silkworm model of pathogenic bacterial infection. Drug Discov. Ther. 1, 89–93 [PubMed] [Google Scholar]
- 33. Nagata M., Kaito C., Sekimizu K. (2008) Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J. Biol. Chem. 283, 2176–2184 [DOI] [PubMed] [Google Scholar]
- 34. Miyazaki S., Matsumoto Y., Sekimizu K., Kaito C. (2012) Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol. Lett. 326, 116–124 [DOI] [PubMed] [Google Scholar]
- 35. Hanada Y., Sekimizu K., Kaito C. (2011) Silkworm apolipophorin protein inhibits Staphylococcus aureus virulence. J. Biol. Chem. 286, 39360–39369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oku Y., Kurokawa K., Matsuo M., Yamada S., Lee B. L., Sekimizu K. (2009) Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J. Bacteriol. 191, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gründling A., Schneewind O. (2007) Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaito C., Hirano T., Omae Y., Sekimizu K. (2011) Digestion of extracellular DNA is required for giant colony formation of Staphylococcus aureus. Microb. Pathog. 51, 142–148 [DOI] [PubMed] [Google Scholar]
- 39. Taub R., Greene M. I. (1992) Functional validation of ligand mimicry by anti-receptor antibodies: structural and therapeutic implications. Biochemistry 31, 7431–7435 [DOI] [PubMed] [Google Scholar]
- 40. Bona C. A., Kang C. Y., Kohler H., Monestier M. (1986) Epibody: the image of the network created by a single antibody. Immunol. Rev. 90, 115–127 [DOI] [PubMed] [Google Scholar]
- 41. Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 [DOI] [PubMed] [Google Scholar]
- 42. Novick R. (1967) Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33, 155–166 [DOI] [PubMed] [Google Scholar]
- 43. Moriya S., Tsujikawa E., Hassan A. K., Asai K., Kodama T., Ogasawara N. (1998) A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29, 179–187 [DOI] [PubMed] [Google Scholar]
- 44. Matsumoto Y., Kaito C., Morishita D., Kurokawa K., Sekimizu K. (2007) Regulation of exoprotein gene expression by the Staphylococcus aureus cvfB gene. Infect. Immun. 75, 1964–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Omae Y., Sekimizu K., Kaito C. (2012) Inhibition of colony-spreading activity of Staphylococcus aureus by secretion of δ-hemolysin. J. Biol. Chem. 287, 15570–15579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ueda T., Kaito C., Omae Y., Sekimizu K. (2011) Sugar-responsive gene expression and the agr system are required for colony spreading in Staphylococcus aureus. Microb. Pathog. 51, 178–185 [DOI] [PubMed] [Google Scholar]