Background: Topologically knotted tRNA methyltransferases specifically recognize substrate tRNA.
Results: Site-directed mutagenesis studies, chimeric protein analysis, and pre-steady state kinetics clarify the tRNA recognition sites of TrmH.
Conclusion: The N- and C-terminal regions function in the initial binding process, and substrate tRNA is discriminated by the catalytic domain in an induced-fit process.
Significance: Study of how proteins recognize RNA is crucial for understanding RNA maturation processes.
Keywords: Protein Structure, RNA Methylation, RNA Methyltransferase, RNA Modification, RNA-binding Proteins, RNA-Protein Interaction, Transfer RNA (tRNA), SPOUT Superfamily, SpoU Family, Pre-steady State Kinetics
Abstract
A conserved guanosine at position 18 (G18) in the D-loop of tRNAs is often modified to 2′-O-methylguanosine (Gm). Formation of Gm18 in eubacterial tRNA is catalyzed by tRNA (Gm18) methyltransferase (TrmH). TrmH enzymes can be divided into two types based on their substrate tRNA specificity. Type I TrmH, including Thermus thermophilus TrmH, can modify all tRNA species, whereas type II TrmH, for example Escherichia coli TrmH, modifies only a subset of tRNA species. Our previous crystal study showed that T. thermophilus TrmH is a class IV S-adenosyl-l-methionine-dependent methyltransferase, which maintains a topological knot structure in the catalytic domain. Because TrmH enzymes have short stretches at the N and C termini instead of a clear RNA binding domain, these stretches are believed to be involved in tRNA recognition. In this study, we demonstrate by site-directed mutagenesis that both N- and C-terminal regions function in tRNA binding. However, in vitro and in vivo chimera protein studies, in which four chimeric proteins of type I and II TrmHs were used, demonstrated that the catalytic domain discriminates substrate tRNAs from nonsubstrate tRNAs. Thus, the N- and C-terminal regions do not function in the substrate tRNA discrimination process. Pre-steady state analysis of complex formation between mutant TrmH proteins and tRNA by stopped-flow fluorescence measurement revealed that the C-terminal region works in the initial binding process, in which nonsubstrate tRNA is not excluded, and that structural movement of the motif 2 region of the catalytic domain in an induced-fit process is involved in substrate tRNA discrimination.
Introduction
Methylation is one of the most common chemical modifications that occur in a broad range of biomolecules, including nucleic acids, proteins, lipids, and small compounds. It is implicated in a variety of cellular processes, such as translation, transcription, processing of RNA, epigenetics, development, carcinogenesis, neurotransmission, cellular transport, infection, immunity, and bacterial host defense. Among RNA species, methylated nucleotides appear in most noncoding RNAs, constituting more than half of the post-transcriptional modifications identified so far. In particular, tRNA contains abundant methylated nucleotides, which stabilize the L-shaped tRNA structure and improve their molecular recognition (1–3).
A conserved guanosine at position 18 (G18) in the D-loop of tRNAs (1, 2) is often modified to 2′-O-methylguanosine (Gm). Methylation of G18 to Gm18 stabilizes the L-shaped three-dimensional structure of tRNA by interacting with pseudouridine 55 (4, 5). Recently, two groups, one with whom we collaborated (6), independently found that Gm18 modification in Escherichia coli tRNA suppresses immunostimulation via the Toll-like receptor 7 (7). Thus, enterobacteria utilize Gm18 modification in tRNA to avoid the host immune system. Therefore, the Gm18 study raises the importance from the viewpoint of control of infectious bacteria. In addition, because the Gm18-modified tRNA acts as a Toll-like receptor 7 antagonist (6), the Gm18-modified tRNA may be utilized as an anti-inflammatory drug.
Formation of Gm18 is catalyzed by tRNA (Gm18) methyltransferase (8–10) (tRNA (guanosine-2′)-methyltransferase; TrMet (Gm18) (2); EC 2.1.1.34). In the reaction, S-adenosyl-l-methionine (AdoMet)6 functions as a methyl group donor and is converted to S-adenosyl-l-homocysteine (AdoHcy). The tRNA (Gm18) methyltransferase gene in E. coli (8, 9), Thermus thermophilus (10, 11), and Aquifex aeolicus (12) is experimentally identified as trmH (classical name, spoU), and the homolog in Saccharomyces cerevisiae is trm3 (13). In accordance with this nomenclature, hereafter, we refer to the eubacterial tRNA (Gm18) methyltransferase as TrmH. TrmH enzymes can be divided to two types based on their substrate tRNA specificity. Type I TrmH, including T. thermophilus TrmH, can modify all tRNA species, whereas type II TrmH, for example E. coli TrmH, modifies only a subset tRNA species. In fact, all tRNA species reported from T. thermophilus have a Gm18 modification (2, 3), whereas of the 47 tRNA species from E. coli, only 14 tRNA species have a Gm18 modification (2, 3).
We reported identification of the trmH gene from T. thermophilus in 2002 (11). In extensive further studies, we then solved the crystal structures of apo- and AdoMet-bound forms (14). Our crystal study showed that TrmH is a class IV AdoMet-dependent methyltransferase (14). AdoMet-dependent methyltransferases are sorted into five classes based on the type of fold present in the catalytic domain (15). The majority of RNA methyltransferases are class I enzymes, which maintain a Rossmann fold in the catalytic domain. In contrast, class IV enzymes maintain a topological knot (so-called “trefoil knot”; a knot with three passes of the protein backbone in and out of the loop) structure in the catalytic domain (14, 16). Functional analysis of structure-based mutant TrmH proteins enabled us to propose a hypothetical reaction mechanism (14, 17) to clarify roles of the amino acid residues in the conserved motifs (17) and to predict the tRNA-binding site (18). Furthermore, in our recent study, we found that the tRNA-TrmH complex formation includes at least two processes, namely the initial tRNA binding and induced-fit processes (19). Moreover, we found that TrmH recognizes the position of the target guanosine with some flexibility (i.e. the modification position is changeable from 18 to neighboring positions according to the nucleotide sequence in tRNA) and that the 6-oxygen atom of guanosine is a positive determinant for enzyme recognition (19).
However, some important questions remain. What part of the protein structure of TrmH contributes to the initial tRNA binding and induced-fit processes? When is the substrate tRNA discriminated from nonsubstrate tRNA (for example, methylated tRNA)? Does this process occur in the initial tRNA binding or during the induced-fit process? In this study, we have focused on solving these questions.
EXPERIMENTAL PROCEDURES
Materials
[methyl-14C]AdoMet (1.95 GBq/mmol) and [methyl-3H]AdoMet (2.47 TBq/mmol) were purchased from ICN. Nonradioisotope-labeled AdoMet and AdoHcy were obtained from Sigma, and DE52 was purchased from Whatman. We produced the AdoHcy affinity column by coupling to a HiTrap N-hydroxysuccinimide-activated HP column (Amersham Biosciences) according to manufacturer's instructions. CM-Toyopearl 650 m was purchased from Tosoh. DNA oligomers were bought from Invitrogen. T7 RNA polymerase was from Toyobo. All other chemical reagents were of analytical grade.
Preparation of TrmH Proteins
T. thermophilus wild-type TrmH was expressed in the E. coli BL21 (DE3) Rosetta2 strain (Takara) and purified as reported previously (11). Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene). The mutant proteins were purified by the same method for the wild-type enzyme. The E. coli wild-type TrmH expression system was constructed as follows. E. coli trmH gene was amplified by polymerase chain reaction from E. coli BL21 (DE3) genomic DNA using the following primers: EcoTrmHN1 5′-GGG CAT ATG AAC CCA ACA CGT TAT GCA-3′; EcoTrmHC1 5′-CCC CCT CGA GTT ACC CTG CAG CCT GCA TAG T-3′. The underlined regions show the restriction enzyme sites, NdeI and XhoI, respectively. The amplified DNA was digested with NdeI and XhoI and then ligated into the multicloning linker of pET30a expression vector (Novagen). The expression of E. coli TrmH was performed according to the manufacturer's manual. Chimeric protein genes were generated by the fusion polymerase chain reaction. The resultant genes were inserted into pET30a vector. The chimeric proteins were purified by Q-Sepharose Fast Flow (GE Healthcare) and HiTrap Heparin HP (GE Healthcare) column chromatography without heat treatment. Protein concentration was measured using the Bio-Rad protein assay kit with bovine serum albumin as the standard. Purified proteins were diluted into 50% glycerol and stored at −30 °C.
TrmH Activity Measurement
Incorporation of a 14C-methyl group from AdoMet to yeast tRNAPhe transcript was used to assay TrmH activity (10). To obtain kinetic parameters of TrmH activity for tRNA, concentrations of TrmH and AdoMet were fixed at 0.03–0.06 and 37 μm, respectively, while incubation times varied from 2 to 30 min depending on the methyl group acceptance activity.
Gel Mobility Shift Assay
Gel mobility shift assays were performed as described in previous reports (18–20). In brief, purified protein and 0.05 A260 units of yeast tRNAPhe transcript were incubated in 20 μl of buffer A (50 mm Tris base, 50 mm acetic acid, and 5 mm Mg(OAc)2) at 37 °C for 20 min. Four microliters of loading solution (0.25% bromphenol blue and 30% glycerol) were then added to each sample, and samples were resolved on a 6% polyacrylamide gel (width, 90 mm; length, 90 mm; thickness, 1 mm) prepared with 1× buffer A. Electrophoresis was performed at room temperature for 1 h at 100 V. To detect protein, the gel was stained with Coomassie Brilliant Blue. Methylene blue was used for RNA detection.
Nucleoside Analysis by HPLC
Transfer RNA fractions were purified from E. coli BL21 (DE3) Rosetta2 cells with or without the expression vector. Briefly, total RNA fractions were prepared by phenol/chloroform extraction. Subsequently, the total RNA fraction was loaded on a Q-Sepharose Fast Flow column equilibrated with buffer B (20 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 400 mm NaCl), and then a small RNA fraction (mainly tRNA and 5 S rRNA) was eluted with buffer B containing 600 mm NaCl. The tRNA fraction was further purified by 10% PAGE (7 m urea). The tRNA fraction (0.2 A260) was digested with 2 μg of snake venom phosphodiesterase (Sigma), 20 μg of RNase A (Invitrogen), and 0.125 units of bacterial alkaline phosphatase (Takara) in 20 μl of 50 mm Tris-HCl (pH 8.0) at 37 °C overnight. Nucleosides were analyzed on an HPLC (Hitachi L-2000 system) equipped with a reverse phase C18 column (NUCLEOSIL 100 C18; 25 cm × 4.6 mm, 7 μm; GL Sciences, Inc). The solvent system consisted of buffer C (50 mm sodium phosphate (pH 5.1)) and buffer D (buffer C containing 70% methanol). The nucleoside samples (20 μl) were chromatographed using a flow rate of 1 ml/min with a multistep linear gradient as follows: 3% buffer D from 0 to 10 min; 3–35% D from 10 to 50 min; 35–98% D from 50–65 min; 98–100% D from 65 to 75 min, and 100% buffer D from 75 to 85 min. The elution positions of m5U and Gm were determined by enzymatic reactions with yeast tRNAPhe transcript and E. coli TrmA (tRNA (m5U54) methyltransferase (21)) and TrmH, respectively. The purification procedure for E. coli TrmA was described in our previous report (22). The content of Gm in each tRNA fraction was calculated based on the content of m5U which is 1.00 per one tRNA molecule.
Stopped-flow Fluorescence Measurements
To investigate the pre-steady state kinetics of the binding process between TrmH and tRNA, we performed stopped-flow fluorescence measurements using an Applied Photophysics SX20 stopped-flow apparatus (Applied Photophysics Ltd., Leatherhead, UK). We monitored changes in the fluorescence intensity of tryptophan residues in TrmH during the complex formation with tRNA. The excitation wavelength was 295 nm, and the fluorescence emission above 320 nm was monitored using a long pass UV cutoff filter. TrmH dissolved in buffer B containing 10 μm AdoMet was rapidly mixed with equimolar yeast tRNAPhe transcript in buffer B at 25 °C. The mixing experiments were carried out at two different TrmH and tRNA concentrations, 1.93 and 7.7 μm, after the mixing. The kinetic data were acquired in a split time base mode, in which data points were distributed in two linear intervals, one from 0 to 0.1 s and the other from 0.1 to 1.1 s. Within 1.1 s, the first catalytic cycle of TrmH is incomplete because the turnover time of the methyl transfer to yeast tRNAPhe transcript at 55 °C is around 10 s (10). Therefore, these measurements monitored the TrmH-tRNA complex formation processes. The scan was repeated 20 times for 7.7 μm TrmH and tRNA, 40 times for 3.85 μm TrmH and tRNA, and 80 times for 1.925 μm TrmH and tRNA. The dead time of the stopped-flow mixing, determined by the method of Brissette et al. (23), was 3 ms, and the optical path length was 1 mm.
RESULTS
Domain and Subdomain Structures of SpoU Family Enzymes
The majority of topological knot RNA methyltransferases reported thus far belong to either the SpoU (8, 9) or TrmD (24–27) families, and these two families constitute the SPOUT (SpoU-TrmD) superfamily (28). It should be mentioned that SpoU (a gene of unknown function in the SpoT operon) is the classical name of TrmH (8, 9) and remains in the protein family name. Fig. 1 shows the schematic drawing of domain and subdomain structures of representative SpoU family enzymes. This figure is based on crystal structures (14, 29–32), bioinformatics studies (8, 28, 33), and amino acid sequences (34–36). All of the SpoU family enzymes catalyze methyl transfer from AdoMet to 2′-OH of ribose in RNA. TrmJ (classical name, YfhQ) and TrmL (classical name, YibK) are eubacterial tRNA (Cm32/Um32) methyltransferases (34) and a tRNA (Um34/Cm34) methyltransferase (31, 32, 35), respectively. aTrm56 is an archaeal tRNA (Cm56) methyltransferase (29, 36). RlmB and AviRb are a 23 S rRNA (Gm2251) methyltransferase (37, 38) and a 23 S rRNA (Um2479) methyltransferase (39, 40), respectively. During the course of this study, two tRNA methyltransferases, which belong to the SPOUT superfamily, have been reported as follows: a dual base specificity enzyme for m1A9 and m1G9 (41) and a methyltransferase for m1Ψ54 (42–44). Because these new enzymes do not modify the 2′-OH of ribose, they are not included in the SpoU family. Furthermore, because the crystal structure of E. coli TrmH has not been solved, its C-terminal structure was predicted by the computational program, LOOPS (45). In Fig. 1, the catalytic domain is schematically represented as “SPOUT.” As shown in Fig. 1, rRNA methyltransferases have an obvious N-terminal domain, which shares homology with RNA-binding proteins. In contrast, tRNA methyltransferases generally have short stretches (or subdomains) at their N and/or C termini. On the basis of this comparison, the N-terminal domain of rRNA methyltransferases and short stretches in tRNA methyltransferases have been predicted to be involved in substrate RNA recognition (33). In particular, in the case of TrmH proteins, type II enzymes possess a relatively long stretch in their C termini as compared with the type I enzymes. Therefore, the C-terminal region of TrmH enzymes has been believed to include a key sequence(s) involved in substrate RNA recognition (18, 33).
FIGURE 1.
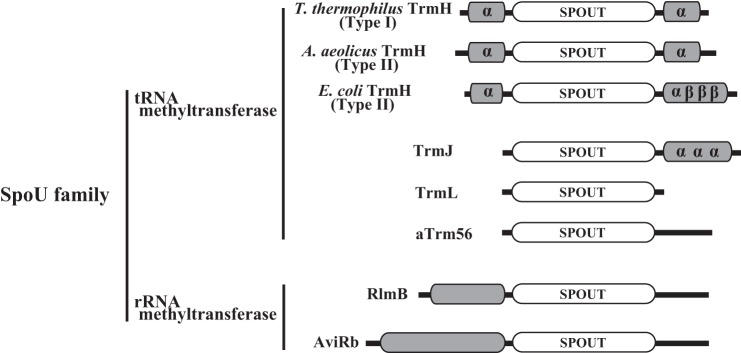
Schematic structure comparison of SpoU family proteins. Crystal structures of seven proteins have been reported, the one exception is E. coli TrmH. The topology of E. coli TrmH is predicted from a computational structural model (32). The catalytic domain is represented as SPOUT. α and β represent α-helix and β-strand, respectively.
C-terminal Region Is Involved in tRNA Recognition
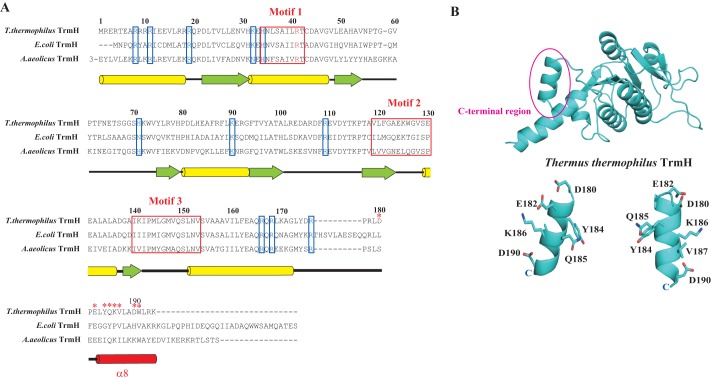
Fig. 2A shows the amino acid sequence alignment of TrmH proteins. The secondary structure of T. thermophilus TrmH is annotated at the bottom of the alignment. The amino acid residues from Asp-22 to Arg-176 of T. thermophilus TrmH form the catalytic domain of typical class IV AdoMet-dependent methyltransferases. The basic amino acid residues boxed (Fig. 2A, blue) are the residues for which alanine substitution decreases the affinity for tRNA (18). Thus, these residues are involved in the tRNA binding. The main part of the C-terminal region from Pro-181 to Lys-194 forms the α8-helix (14). As shown in Fig. 2A, the amino acid residues in the C-terminal region are not conserved in the TrmH proteins. To verify whether these residues are involved in tRNA recognition, seven amino acid residues (Asp-180, Glu-182, Tyr-184, Gln-185, Lys-186, Val-187, and Asp-190) were individually replaced by alanine. The mutant proteins were purified to homogeneity as judged by 15% SDS-PAGE and then used for methyl transfer kinetic assays. In this study, yeast tRNAPhe was used as substrate because the structure of this tRNA is well established and is a good substrate tRNA of T. thermophilus TrmH (10). As shown in Table 1, alanine substitution of Asp-180, Glu-182, Tyr-184, and Gln-185 did not cause any noteworthy effect on the methyl transfer activity. In contrast, alanine substitution of Lys-186, Val-187, and Asp-190 produced a slight decrease in the methyl transfer activity. We measured the kinetic parameters (Table 1) and found that the differences in Km values except for Asp-190 were very small. Furthermore, gel mobility shift assays with these mutant proteins did not demonstrate clear differences in shifted band intensities (data not shown). Indeed, only the shifted band for D190A was slightly weaker compared with that of the wild-type enzyme (Table 1). Thus, although these results suggest that the tail end of the C-terminal region is involved in tRNA binding, we could not confirm whether the C-terminal region is essential for tRNA binding. Therefore, we additionally prepared two deletion mutant proteins in which stop codons were introduced at either Lys-179 or Gln-185. The mutant proteins were purified as shown in Fig. 3A. The methyl transfer activity of Q185Stop mutant protein is clearly decreased via an increase in the Km value (Fig. 3, B and C). Furthermore, the introduction of a stop codon at Lys-179 (resulting in deletion of the entire C-terminal region) produced a complete loss of the methyl transfer activity (Fig. 3, B and C). To confirm the affinity of these mutant proteins for tRNA, the gel mobility shift assay was performed (Fig. 4D). The wild-type enzyme clearly captures tRNA under the tested conditions. In contrast, two mutant proteins did not form a clear band corresponding to the enzyme-tRNA complex. These results clearly show that the C-terminal region is involved in tRNA binding. Because the L179Stop mutant protein has a decreased methyl transfer activity and both mutant proteins have weak affinity for tRNA, the catalytic domain of these proteins probably folds correctly.
FIGURE 2.
A, amino acid sequence alignment of TrmH proteins. Three conserved motifs are enclosed in red boxes. Basic amino acid residues enclosed by blue boxes are residues for which alanine substitution decreases the affinity for tRNA as demonstrated in our previous study (18). The secondary structure of T. thermophilus TrmH is illustrated at the bottom of the alignment: yellow bars and green arrows show α-helices and β-strands, respectively. The α8-helix in the C-terminal region is highlighted in red. The red asterisks mark the alanine-substituted residues in this study. B, structure of T. thermophilus TrmH. The C-terminal region (α8-helix) is highlighted. The amino acid residues, substituted by alanine, are shown as stick models.
TABLE 1.
Kinetic parameters for yeast tRNAPhe transcript
Although the results of gel mobility shift assays were evaluated by 4° by the intensities of shift bands, the differences except for D190A were small as described in the text.
| Variant name | Km | Vmax | Vmax/Km | Gel mobility shift assay |
|---|---|---|---|---|
| nm | μmol mg−1 h−1 | |||
| Wild type | 150 | 1.1 | 1.00 | ++ |
| D180A | 120 | 0.4 | 0.42 | ++ |
| E182A | 110 | 0.2 | 0.29 | ++ |
| Y184A | 260 | 0.2 | 0.10 | + |
| Q185A | 120 | 0.1 | 0.16 | ± |
| K186A | 160 | 0.1 | 0.07 | ± |
| V187A | 140 | 0.1 | 0.11 | ± |
| D190A | 1300 | 0.2 | 0.05 | − |
FIGURE 3.
A, 15% SDS-polyacrylamide gel analysis of wild-type TrmH and two C-terminal deletion mutant proteins. The gel was stained with Coomassie Brilliant Blue. B, methyl transfer activities of wild-type TrmH and two C-terminal deletion mutant proteins. Yeast tRNAPhe transcript was used as substrate tRNA. The L179Stop mutant protein, in which the C-terminal region was deleted, completely lost methyl transfer activity. C, kinetic parameters of C-terminal deletion mutant proteins for yeast tRNAPhe. The Q185Stop mutant protein has decreased activity with a relatively large Km value, suggesting that amino acid residues from Gln-185 to Lys-194 are involved in the initial binding process. D, gel mobility shift assay with wild-type and mutant TrmH proteins. The gel was double-stained with Coomassie Brilliant Blue and methylene blue for detections of protein and RNA, respectively. The two mutant proteins (L179Stop and Q185Stop) did not show a clear band corresponding to the protein-tRNA complex. However, these proteins have weakened affinity for tRNA, because the bands derived from tRNA were smeared.
FIGURE 4.

Methyl transfer activity of N-terminal region deletion mutant protein. The deletion of amino acid residues from Met-1 to Arg-18 causes complete loss of methyl transfer activity.
N-terminal Region Is Also Involved in tRNA Recognition
In a previous study (18), we found that alanine substitution of conserved basic amino acid residues (Arg-8, Arg-11, and Arg-19) produced decreases in the methyl transfer activity via increases in the Km values. In the case of the R11A mutant enzyme, the enzymatic activity was completely lost, and the affinity for AdoMet was also lost in addition to the loss of affinity for tRNA (18). The crystal structure analysis revealed that Arg-11 hydrophobically interacts with Met-147 in another subunit. This interaction seems to stabilize the homodimer structure. Thus, the N-terminal region is required not only for tRNA binding but also for protein stability. In this study, we have made one mutant protein, in which the N-terminal region (Met-1–Arg-19) is deleted. This mutant protein is heavily proteolyzed in E. coli cells (data not shown), consistent with the importance of the Arg-11 residue for protein stability. Although we attempted purification of the mutant protein, we could not remove the proteolyzed fragments from the sample (data not shown). It should be mentioned that the intact mutant protein remained in the sample, and protease activity was not detected in the partially purified sample. As shown in Fig. 4, the deletion of the N-terminal region produced complete loss of methyl transfer activity. Taking previous and current results together, we concluded that the N-terminal region is involved in tRNA recognition and is required for protein stability.
N- and C-terminal Regions Do Not Work in the Substrate tRNA Selection Process
As described above, the N- and C-terminal regions are involved in substrate tRNA recognition. These results pose one important question. Does the N- and C-terminal regions work in substrate tRNA discrimination? To address this question, we utilized chimeric proteins of T. thermophilus and E. coli TrmHs (Fig. 5). T. thermophilus TrmH methylates all tRNA species and is classified as a type I TrmH (10–12), whereas E. coli TrmH methylates only 14 of the 47 E. coli tRNA species and is classified as a type II TrmH (Fig. 5A). Thus, the two types of TrmH enzyme have different substrate tRNA specificities. We prepared two chimeric proteins, the first was T. thermophilus TrmH with the C-terminal region of E. coli TrmH, and the second had both the N- and C-terminal regions of E. coli TrmH (Fig. 5B). When the C-terminal region of T. thermophilus TrmH was replaced by that of E. coli TrmH, the chimeric protein had methyl transfer activity toward all E. coli tRNA transcripts (Fig. 5C). It should be mentioned that native E. coli tRNAPhe, tRNAGlu, and tRNALys do not have the Gm18 modification, consistent with the results with E. coli TrmH in Fig. 5C. Thus, the substrate tRNA specificity of chimeric protein in which there is E. coli TrmH sequence is type I, demonstrating that the C-terminal region does not work in substrate tRNA discrimination. Next, we made a chimeric protein, in which both N- and C-terminal regions of T. thermophilus TrmH were replaced by those of E. coli TrmH (Fig. 5B). To our surprise, this chimeric protein also had the methyl transfer activity toward all E. coli tRNA transcripts (Fig. 5C). Thus, this result showed that the N- and C-terminal regions of TrmH do not work in substrate tRNA discrimination, although these regions are involved in tRNA recognition as described in the above sections. To confirm the affinities of both chimeric proteins for tRNA, gel mobility shift assays were performed. As shown in Fig. 5D, both chimeric proteins have affinity for yeast tRNAPhe transcript (this tRNA transcript is not methylated by E. coli TrmH), consistent with their broad methyl transfer specificities. Furthermore, because the affinities of these chimeric proteins for yeast tRNAPhe are weaker than that of T. thermophilus wild-type TrmH, the N- and C-terminal regions of E. coli TrmH may have weak affinity for tRNA.
FIGURE 5.
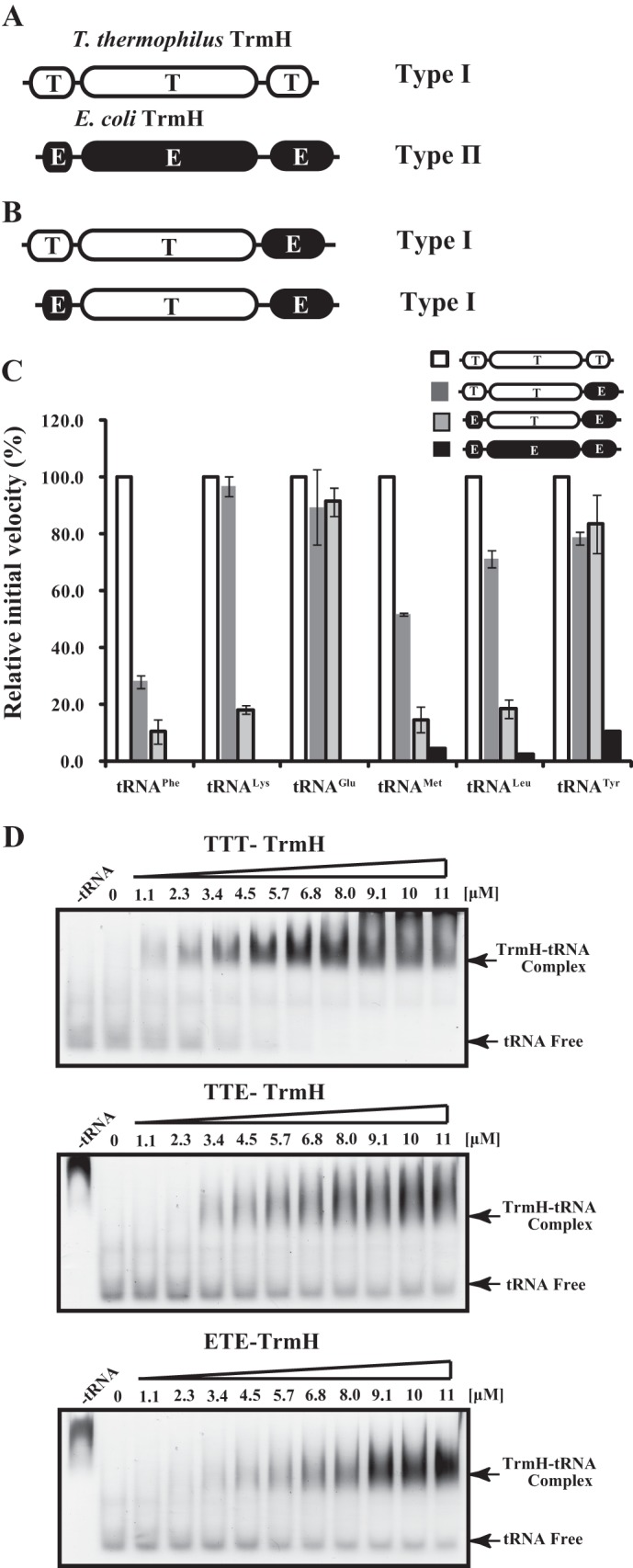
A, schematic drawing of T. thermophilus and E. coli TrmH proteins. Because T. thermophilus TrmH methylates all tRNA species, it is a member of the type I TrmH enzymes. In contrast, E. coli TrmH methylates a subset tRNA species and is categorized as a type II TrmH enzyme. B, schematic drawing of chimeric proteins. The C-terminal region of T. thermophilus TrmH was replaced by that of E. coli TrmH. Furthermore, both the N- and C-terminal regions of T. thermophilus TrmH were replaced by those of E. coli TrmH. C, methyl transfer activities of chimeric proteins. We tested six E. coli tRNA transcripts as substrate. As reported previously (1–3), in E. coli cells, tRNAPhe, tRNALys, and tRNAGlu are not methylated by E. coli TrmH. In contrast, tRNAmMet, tRNALeu, and tRNATyr are methylated by E. coli TrmH, although the Gm18 methylation of tRNAmMet has been reported to be a partial modification (33, 34). T. thermophilus TrmH methylates all these tRNA transcripts (white bars), consistent with the concept that T. thermophilus TrmH is a type I enzyme. In contrast, E. coli TrmH methylates only three tRNA transcripts (black bars; tRNAmMet, tRNALeu, and tRNATyr), showing that E. coli TrmH is a type II enzyme. The two chimeric proteins methylate all six tRNA transcripts akin to a type I enzyme (dark and light gray bars). The experiments were performed at 37 °C. The bars show the relative initial velocities, with the velocity of wild-type T. thermophilus TrmH expressed as 100.0. The averages of three independent experiments are shown. D, gel shift assay with chimeric TrmH proteins. The affinities of chimeric proteins for yeast tRNAPhe transcript were compared with that of the wild-type T. thermophilus TrmH (TTT).
Catalytic Domain Is Responsible for Substrate tRNA Selectivity
The N- and C-terminal regions are involved in tRNA recognition by TrmH. However, replacement of these regions did not change the substrate tRNA discrimination (selectivity). These results suggest that the catalytic domain is responsible for the substrate tRNA selectivity. Because this conclusion was very different from the previous concept in which it was proposed that the C-terminal region of TrmH proteins contains a key sequence(s) for substrate RNA recognition (11, 18, 33), we suspected that these results might be an artifact of the in vitro experiments. Therefore, we constructed an in vivo assay system. T. thermophilus and E. coli TrmHs and their chimeric proteins were expressed in E. coli cells, and then tRNA fractions were purified. In this experiment, we prepared a new chimeric protein, in which the catalytic domain of T. thermophilus TrmH was replaced by that of E. coli TrmH. It should be mentioned that endogenous E. coli TrmH is expressed both in the presence and absence of plasmid vector. The modified nucleosides in each tRNA fraction were analyzed by HPLC (Fig. 6). Because the m5U modification exists only at position 54 in tRNA and the content of m5U54 is near 1.00 per one tRNA molecule, we calculated the Gm content from the peak area of m5U as a control. The elution positions of m5U and Gm were determined using enzymatic reactions with yeast tRNAPhe transcript and E. coli TrmA (tRNA (m5U54) methyltransferase) and T. thermophilus TrmH, respectively (data not shown). The tRNA fractions were scarcely methylated by E. coli TrmA (data not shown). Thus, we confirmed that the m5U content in each tRNA fraction was near 100%. In the absence of expression vector, the ratio of Gm/m5U was calculated as 0.33 (Fig. 6A). When E. coli TrmH was overexpressed by the introduction of the expression plasmid, the ratio of Gm/m5U was raised to 0.57 (Fig. 6B). This result showed that Gm18 was not fully modified in vivo despite the tRNA species containing Gm18 modification. This idea is in line with mass spectrometric analysis of some tRNA species in E. coli; for example, tRNAmMet, which has a Gm18 modification, shows mass diversity due to the ratio of modifications (46, 47). When the chimeric protein, which has the catalytic domain of E. coli TrmH and the N- and C-terminal regions of T. thermophilus TrmH, was expressed, a similar result was obtained (Fig. 6C); the ratio of Gm/m5U was calculated as 0.64. In contrast, when the chimeric protein, which has the catalytic domain of T. thermophilus TrmH and the N- and C-terminal regions of E. coli TrmH, was expressed, the ratio of Gm/m5U was significantly raised to 0.91 (Fig. 6D). This ratio of Gm/m5U was near to full Gm18 modification, as was observed by the expression of T. thermophilus TrmH (Fig. 6E). Thus, these in vivo experiments demonstrate that the catalytic domain is responsible for substrate tRNA selectivity; this is in good agreement with the in vitro experiments.
FIGURE 6.
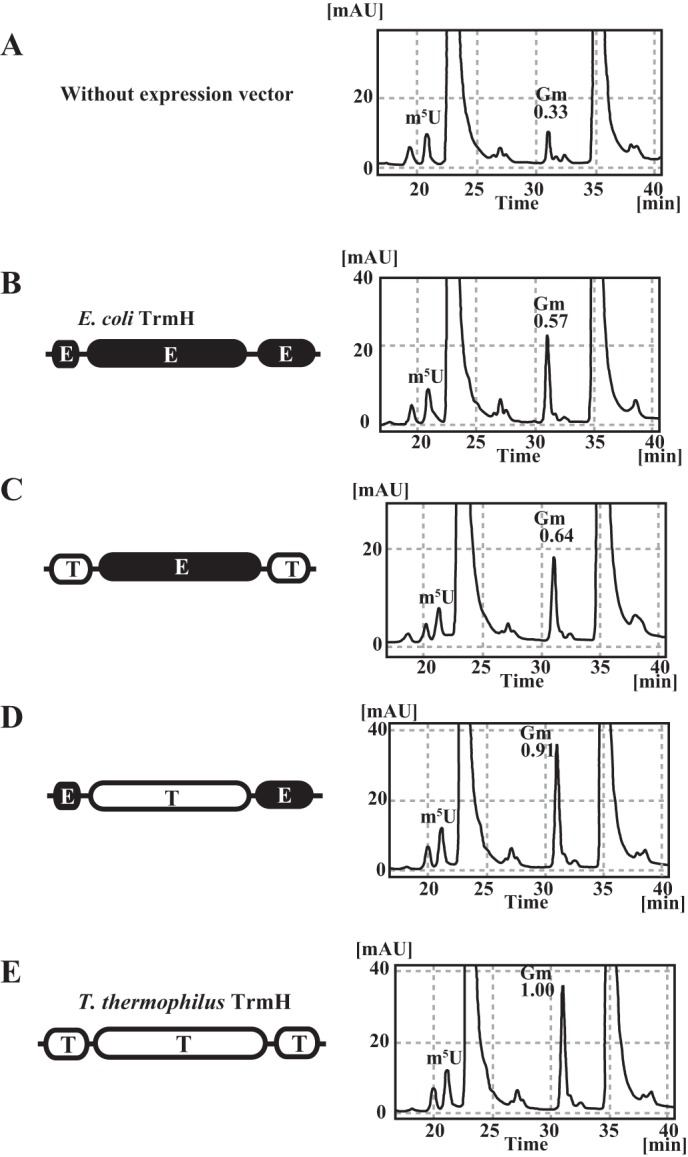
In vivo tRNA methylation activities of chimeric proteins. In the presence of endogenous E. coli TrmH (without expression vector), the Gm/m5U ratio in E. coli tRNA is 0.33 (A). When the wild-type E. coli TrmH was overexpressed, the ratio is raised to 0.57 but does not reach 1.00 (B). This result shows that limited tRNA species are methylated by E. coli TrmH and that the Gm18 modification in E. coli tRNA in natural conditions is a partial modification. When the chimeric protein, in which the catalytic domain is derived from E. coli TrmH, was expressed, the Gm/m5U ratio was close to the value from E. coli TrmH expression (C). Thus, this chimera protein methylates the limited tRNA species. In contrast, when the chimeric protein, which has the catalytic domain of T. thermophilus TrmH, was expressed, the Gm/m5U ratio was increased to 0.91, showing that this chimeric protein has broad substrate tRNA selectivity as compared with E. coli TrmH (D). When the wild-type T. thermophilus TrmH was expressed, tRNA was nearly fully modified (E). These in vivo experimental results suggest that the catalytic domain of E. coli TrmH distinguishes the substrate and nonsubstrate tRNAs, consistent with the in vitro results shown in Fig. 5.
Methylated tRNA Does Not Induce the Structural Change Process
In our previous report, we investigated the initial binding and structural change (induced fit) processes involved in the formation of the TrmH-tRNA complex by stopped-flow pre-steady state kinetic fluorescence measurements (Fig. 7) (19). Briefly, TrmH has three tryptophan residues (Fig. 7A), and their fluorescence intensities change according to the progress of TrmH-tRNA complex formation. As shown in Fig. 7B, a very fast decrease of fluorescence was observed within the first 10 ms, and then relatively slow increases were observed from 10 to 40 ms. In general, the decrease of tryptophan fluorescence intensity suggests an increase in accessibility of the residues to solvent water if the tryptophan fluorescence is not quenched in the initial state. The obtained data could be fitted to an equation (see Scheme 1 in Fig. 7B), where the first term represents the bi-molecular binding reaction of TrmH and tRNA, and the second and third terms represent conformational changes. The kinetic data after 3 ms were used for calculating the kinetic parameters (relative amplitudes, a1, a2, and a3, and apparent rate constants, k1, k2, and k3), and they are given in Table 2. As shown in Table 2, the first decrease of fluorescence can be well fitted to a second-order reaction. The very large k1 value of the order of 107–108 means that the complex formation of TrmH and tRNA is caused by a diffusion-limited association (48, 49). In contrast, the subsequent slow increase of fluorescence can be fitted as first-order reactions, in which two phases with different rate constants (k2 and k3) exist. Thus, the increase of fluorescence is derived from structural changes in the TrmH and tRNA complex, suggesting that these phases are involved in the induced fit processes. The activity measurement revealed that only 2% of substrate tRNA was methylated during the 8-s period of the test conditions, suggesting that the majority of complexes do not complete the methyl transfer during the 50-ms period (19).
FIGURE 7.
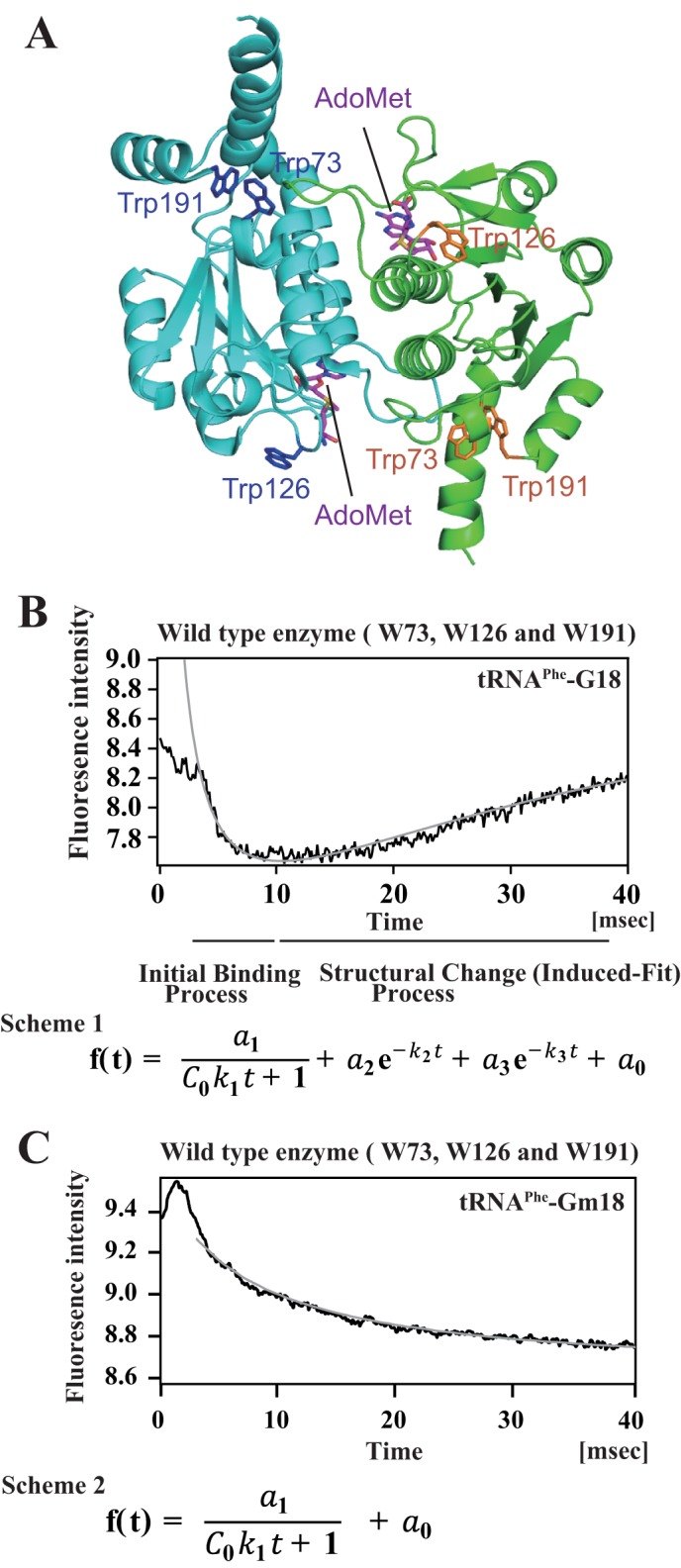
Methylated tRNA is excluded before the structural change (induced-fit) process. A, location of tryptophan residues. T. thermophilus TrmH has three tryptophan residues per one subunit. Bound AdoHcy and tryptophan residues are highlighted as stick models in the dimer TrmH structure. B, pre-steady state analysis of TrmH and tRNA complex formation by stopped-flow fluorescence measurements. This figure is derived from our previous report (19) with slight modifications. When TrmH and tRNA were mixed, the rapid decrease of fluorescence derived from the three tryptophan residues was observed during the first 10 ms. In general, the decrease of tryptophan fluorescence shows that the environment of the tryptophan residue(s) changes to become hydrophilic. This initial binding process could be approximated by a second-order reaction. After the initial binding process, the fluorescence intensity gradually increased at least 40 ms, suggesting that the environment of the tryptophan residue(s) changed to be hydrophobic after the structural change (induced-fit). Curve fitting of this process suggested that at least two types of structural changes are involved in this process. The parameters are given in Table 2. Probably, disruption of the D- and T-loops of tRNA, recognition of G18 base, and introduction of ribose into the catalytic pocket are included in this process. It should be mentioned that the methyl transfer reaction is not caused during the 40-ms period because only 2% of substrate tRNA was methylated after 8 s under the tested condition (19). C, pre-steady state kinetic analysis of TrmH and methylated tRNA complex formation. In this study, we analyzed the complex formation of TrmH with methylated tRNA by the same methods. The methylated tRNA caused the initial binding process within 10 ms as observed with unmodified tRNA. However, the methylated tRNA did not induce the structural change process. Two methyl groups (one in Gm18 and another in AdoMet) probably cause steric hindrance in the catalytic pocket. The curve could be fitted by a two molecule reaction (scheme 2).
TABLE 2.
Kinetic parameters of pre-steady state kinetics

In our previous study, an inhibition experiment with Gm18-methylated tRNA suggested that the methylated tRNA is excluded before the structural change process due to steric hindrance between the two methyl groups of Gm18 and AdoMet (19). Initially, we tested this phenomenon using pre-steady state kinetic analysis by stopped-flow fluorescence measurements. As shown in Fig. 7C, the methylated tRNAPhe did not induce the structural change process, consistent with the result of our previous inhibition experiment (19). This initial binding process was slow as compared with that of the unmodified tRNA transcript, suggesting that the methyl group of Gm18 ribose disturbs the efficient complex formation. However, the Gm18-tRNA clearly forms the complex with TrmH. In fact, the curve could be fitted to Fig. 7C, Scheme 2, which represents a bi-molecular binding reaction. Thus, the methylated tRNA is excluded before the structural change process.
Motif 2 in the Catalytic Domain Is Involved in the Structural Change Process
What part(s) of the enzyme is involved in the initial binding and structural change processes? To solve this question, we employed pre-steady state kinetics by stopped-flow fluorescence measurements. To distinguish the fluorescence intensities of the three tryptophan residues, we prepared three mutant TrmH proteins (W73, W126, and W191) (Fig. 7A). The W73 mutant has only one tryptophan residue at position 73, and the other two residues (Trp-126 and Trp-191) are substituted to Ala. Similarly, the W126 and W191 mutants have only one tryptophan residue (Trp-126 and Trp-191, respectively). Trp-73 is located in loop 2 in the catalytic domain, and this loop contacts the connecting loop between the catalytic domain and the C-terminal region (knotted loop) in the other subunit (Fig. 7A). Trp-126 is located in loop 4 (Fig. 7A), which corresponds to motif 2 of the catalytic domain (Fig. 2A). In our previous study, we found that Glu-124 in motif 2 is required for the formation of the AdoMet-binding site; the substitution of Glu-124 by Gln causes complete loss of affinity for AdoMet via structural disruption as observed in the CD spectrum (17). Thus, motif 2 is involved in the formation of the AdoMet-binding site in the topological knotted structure. Trp-191 is located in the C-terminal region, which contributes to tRNA binding (Fig. 3 and Table 1). Measurements of the methyl transfer activities showed that these mutant proteins have enough activities, although the W191 mutant had a slightly decreased velocity (Fig. 8A).
FIGURE 8.
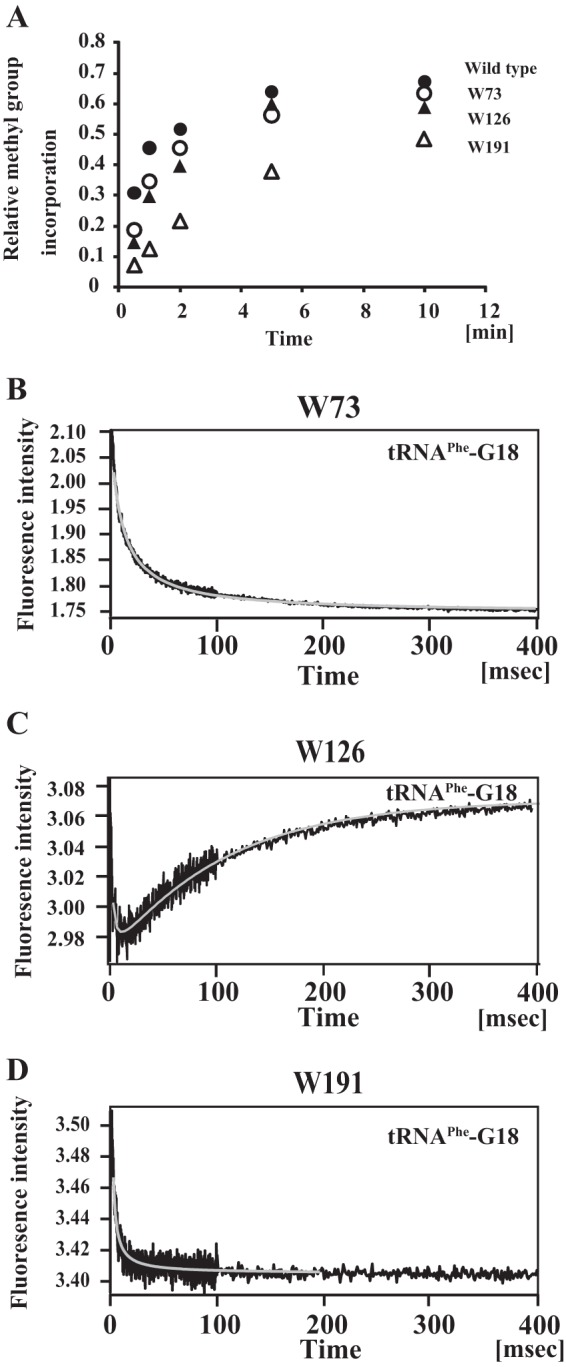
Pre-steady state kinetic analysis of complex formation of mutant TrmH proteins and tRNA by stopped-flow fluorescence measurements. We prepared three types of mutant TrmH proteins. The W73 mutant protein has only one tryptophan residue at position 73 with the two other tryptophan residues at 126 and 191 substituted by alanine. Similarly, the W126 and W191 mutant proteins were prepared. These three mutant proteins have sufficient methyl transfer activity toward yeast tRNAPhe transcript (A). The pre-steady state complex formation of W73 (B), W126 (C), and W191 (D) with tRNA were analyzed by stopped-flow fluorescence measurements. When W73 and W191 were used as the enzyme, the recovery of fluorescence intensity during the structural change process was not observed. It should be mentioned that the structural change process does progress in these mutant enzymes because these mutant proteins have methyl transfer activity. Thus, in the cases of W73 and W191 mutant enzymes, the structural change process could not be monitored by the recovery of tryptophan fluorescence intensity. In contrast, when the W126 mutant protein was used, the recovery of the fluorescence intensity derived from Trp-126 could be observed clearly, demonstrating that the structural change is around the Trp-126 residue (i.e. in motif 2 in the catalytic domain).
As shown in Fig. 8B, the fluorescence from Trp-73 is decreased in the initial binding process. In general, the decrease of fluorescence of tryptophan means that the environment of the residue (i.e. Trp-73) is changed to be hydrophilic. In contrast to the methylated tRNA (Fig. 7C), the W73 mutant protein undergoes the structural change process (the bound tRNA is not excluded from the enzyme-tRNA complex) because this mutant enzyme has high enough methylation activity (Fig. 8A). Thus, this result shows that the protein structure around Trp-73 is involved in the initial binding process but is not involved in the structural change process. In contrast, the fluorescence from Trp-126 gradually increases for 400 ms. Therefore, the profile means that the environment of Trp-126 was gradually changed to be hydrophobic during the structural change process (Fig. 8C, Table 3, and Scheme 3).
 |
This result reveals that the increase of fluorescence intensity during the structural change process observed with the wild-type enzyme is a reflection of the structural change around Trp-126 in motif 2. The fluorescence from Trp-191 gradually decreases in the initial binding process, demonstrating that the C-terminal region is involved in the initial binding process and is not involved in the structural change process (Fig. 8D).
TABLE 3.
Kinetic parameters of presteady state kinetics
The curves of W73 and W191 mutants were fitted to Scheme 2 (see Fig. 7). In the case of W126 mutant, the curve was fitted to Scheme 3.
| Variant name | a0 | a1 | a2 | k1 | k2 | C0 |
|---|---|---|---|---|---|---|
| μm | m−1s−1 | s−1 | m | |||
| W73 | 1.75 ± 0.0003 | 0.35 ± 0.002 | 2.38 × 107 ± 2.49 × 105 | 3.85 × 10−6 | ||
| W126 | 3.07 ± 0.0003 | 12,000 ± 4.65 × 105 | −0.10 ± 0.00065 | 3.06 × 107 ± 0.31 × 107 | 9.50 ± 0.12 | 3.85 × 10−6 |
| W191 | 3.41 ± 0.0002 | 120 ± 5.60 × 103 | 1.72 × 1011 ± 7.87 × 1012 | 3.85 × 10−6 |
Taking these experimental results together, we conclude that structural movement around motif 2 in the catalytic domain occurs during the structural change process and that the C-terminal region and a part of the catalytic domain function in the initial binding process. The final substrate tRNA discrimination (selection) by the catalytic domain is probably performed during the structural change process.
DISCUSSION
The majority of SpoU tRNA methyltransferases do not have a clear RNA binding domain in contrast to rRNA methyltransferases in which the RNA binding domain is more defined (Fig. 1). The question is therefore raised as to how SpoU tRNA methyltransferases specifically recognize the substrate tRNA and methylation target site? In this study, we have addressed this problem. Amino acid sequence alignments (Figs. 1 and 2), bioinformatics studies (8, 27, 28), partial digestion with protease (11), crystal structure studies (14, 24–26), and mutagenesis research (17, 18) suggested that small region(s) within the N- and/or C-terminal region(s) of SpoU family members is responsible for tRNA binding. Such an assumption has been common in the field. However, this study reveals that although this idea is mostly true, it is insufficient in the case of at least TrmH.
Our site-directed mutagenesis studies demonstrated that both N- and C-terminal regions are required for the methyl transfer reaction (Figs. 3 and 4). However, the chimeric protein, in which both N- and C-terminal regions are replaced by those of E. coli TrmH (type II enzyme), shows type I enzyme activity in vitro (Fig. 5) and in vivo (Fig. 6). Thus, these results demonstrate that the catalytic domain discriminates between substrate and nonsubstrate tRNAs. Stopped-flow fluorescence measurement experiments revealed that the methylated tRNA is excluded before the structural change process and that structural change during the induced-fit process includes the movement of motif 2 in the catalytic domain (Figs. 7 and 8). Furthermore, it was also demonstrated that the C-terminal region works in the initial binding process and does not discriminate between substrate and nonsubstrate tRNAs. Taking these experimental results together, nonsubstrate tRNA for E. coli TrmH (type II enzyme) is probably excluded in the structural change process by the catalytic domain. In this study, we focused on T. thermophilus TrmH (type I enzyme). Therefore, to understand the precise exclusion mechanism of nonsubstrate tRNA by E. coli TrmH, crystal structural studies of E. coli TrmH are required.
For a decade, it has been believed that short stretches of the N- and/or C-terminal regions of SpoU tRNA methyltransferases include a key sequence(s), which recognizes substrate RNAs. This study revealed that this idea is true but incomplete. In the case of TrmH, the N- and C-terminal regions recognize the L-shaped tRNA structure. These regions do not distinguish between substrate and nonsubstrate tRNAs. Rather, substrate discrimination takes place before and/or during the induced-fit process. This mechanism explains the methylation of truncated tRNA by TrmH; if the catalytic domain can capture the target site with some affinity, truncated tRNAs are methylated with decreased velocity (12, 50, 51). Our current findings seem to be applicable to the majority of other SpoU tRNA methyltransferases, because the protein structures share homology to each other. During the revision of this manuscript, the tRNA recognition mechanism of TrmL (tRNA (Cm34/Um34) methyltransferase) was reported; TrmL does not require another protein subunit for tRNA methylation (32). TrmL is composed only of the catalytic domain (Fig. 1) and thus discriminates substrate tRNA via the catalytic domain in its dimeric form. It has also been reported that TrmL requires other tRNA modification(s) for tRNA recognition (35). Thus, TrmL seems to recognize the conformation of the anticodon arm structure formed by other modified nucleotide(s) in substrate tRNA all via the catalytic domain alone. To understand the substrate RNA recognition mechanism of the other SpoU family proteins, further study is necessary.
The SPOUT superfamily includes another methyltransferase family (TrmD family) (28). TrmD catalyzes methyl transfer to the N1 atom of G37 in eubacterial tRNA (24). The m1G37 modification in archaea (54–56), eukaryotes (57, 58), and mitochondrial (59, 60) tRNA is conferred by Trm5, which belongs to the class I enzymes. Thus, the same modification at the same position in tRNA is conferred by different enzymes, namely TrmD (class IV) and Trm5 (class I), according to the organism. Therefore, these two enzymes have been compared from the viewpoints of the evolutionary process, protein structure, reaction mechanism, and substrate recognition (57, 58, 60–64). In 2003, Holmes and co-workers (26) reported that the deletion of the C-terminal domain of E. coli TrmD causes complete loss of activity. Thus, the C-terminal regions of both TrmH and TrmD may have the same function in the tRNA recognition mechanism. Recently, two groups independently identified new SPOUT tRNA methyltransferases (Mja 1640 from Methanocaldococcus jannaschii and Hvo 1989 from Haloferax volcanii) producing the m1Ψ54 modification in archaeal tRNA (43, 44). These proteins, which share homology with the catalytic domain of 18 S rRNA methyltransferase (Nep1), which gives rise to m1acp3Ψ (52, 53), are mainly formed by the catalytic domain (42–44). This observation is in line with our current findings that a small part(s) of the catalytic domain discriminates the substrate RNA. Moreover, the final discrimination of substrate RNA by the catalytic domain may be common with some other tRNA modification enzymes. For example, the RNA binding PUA domain of archaeal tRNA guanine transglycosylase is not required for archaeosine formation at the correct position (G15) in tRNA (65), suggesting that the PUA domain supports only the formation of the enzyme-tRNA complex (66), and the catalytic domain discriminates the modification site.
Recently, the study of Gm18 formation mechanism has increased in importance because the human Toll-like receptor 7 recognizes the Gm18 modification as a marker of intrinsic tRNA. Thus, although Gm18 modification in thermophilic bacteria is one of the tRNA stabilization factors, and is a response to environmental temperatures (22, 67), this tRNA modification of enteric bacteria is required for escape from the immune system in the animal gut (6, 7). Therefore, if we can control the Gm18 modification in infectious bacteria, the method may have application as a powerful anti-bacterial system. Furthermore, because the Gm18-modified tRNA acts as a Toll-like receptor 7 antagonist (6), the Gm18-modified tRNA may be utilized as an anti-inflammatory drug. Thus, this study will contribute not only to the RNA modification field but also to the medical and drug design fields.
This work was supported in part by Research Fellowship for Young Scientists 21-10011 (to A. O.) and a Grant-in-aid for Science Research 23350081 (to H. H.) from the Japan Society for the Promotion Science.
- AdoMet
- S-adenosyl-l-methionine
- AdoHcy
- S-adenosyl-l-homocysteine.
REFERENCES
- 1. Dunin-Horkawicz S., Czerwoniec A., Gajda M. J., Feder M., Grosjean H., Bujnicki J. M. (2006) MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 34, D145–D149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rozenski J., Crain P. F., McCloskey J. A. (1999) The RNA modification database: 1999 update. Nucleic Acids Res. 27, 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jühling F., Mörl M., Hartmann R. K., Sprinzl M., Stadler P. F., Pütz J. (2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250, 546–551 [DOI] [PubMed] [Google Scholar]
- 5. Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., Rich A. (1974) The general structure of transfer RNA molecules. Proc. Natl. Acad. Sci. U.S.A. 71, 4970–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jöckel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H., Ehm G., Schnare M., Nain M., Kaufmann A., Bauer S. (2012) The 2′-O-methylation status of a single guanosine controls transfer RNA mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gehrig S., Eberle M.-E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W. M., Erdmann V. A., Sprinzl M., Bec G., Keith G., Dalpke A. H., Helm M. (2012) Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gustafsson C., Reid R., Greene P. J., Santi D. V. (1996) Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 24, 3756–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Persson B. C., Jäger G., Gustafsson C. (1997) The spoU gene of Escherichia coli, the fourth gene of the spot operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 25, 3969–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hori H., Yamazaki N., Matsumoto T., Watanabe Y., Ueda T., Nishikawa K., Kumagai I., Watanabe K. (1998) Substrate recognition of tRNA (guanosine-2′)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem., 273, 25721–25727 [DOI] [PubMed] [Google Scholar]
- 11. Hori H., Suzuki T., Sugawara K., Inoue Y., Shibata T., Kuramitsu S., Yokoyama S., Oshima T., Watanabe K. (2002) Identification and characterization of tRNA (Gm18) methyltransferase from Thermus thermophilus HB8: domain structure and conserved amino acid sequence motifs. Genes Cells 7, 259–272 [DOI] [PubMed] [Google Scholar]
- 12. Hori H., Kubota S., Watanabe K., Kim J. M., Ogasawara T., Sawasaki T., Endo Y. (2003) Aquifex aeolicus tRNA (Gm18) methyltransferase has unique substrate specificity. J. Biol. Chem. 278, 25081–25090 [DOI] [PubMed] [Google Scholar]
- 13. Cavaillé J., Chetouani F., Bachellerie J.-P. (1999) The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA 5, 66–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nureki O., Watanabe K., Fukai S., Ishii R., Endo Y., Hori H., Yokoyama S. (2004) Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure 12, 593–602 [DOI] [PubMed] [Google Scholar]
- 15. Schubert H. L., Blumenthal R. M., Cheng X. (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nureki O., Shirouzu M., Hashimoto K., Ishitani R., Terada T., Tamakoshi M., Oshima T., Chijimatsu M., Takio K., Vassylyev D. G., Shibata T., Inoue Y., Kuramitsu S., Yokoyama S. (2002) An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr. D Biol. Crystallogr. 58, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 17. Watanabe K., Nureki O., Fukai S., Ishii R., Okamoto H., Yokoyama S., Endo Y., Hori H. (2005) Roles of conserved amino acid sequence motifs in the SpoU (TrmH) RNA methyltransferase family. J. Biol. Chem. 280, 10368–10377 [DOI] [PubMed] [Google Scholar]
- 18. Watanabe K., Nureki O., Fukai S., Endo Y., Hori H. (2006) Functional categorization of the conserved basic amino acid residues in TrmH (tRNA(Gm18)methyltransferase) enzymes. J. Biol. Chem. 281, 34630–34639 [DOI] [PubMed] [Google Scholar]
- 19. Ochi A., Makabe K., Kuwajima K., Hori H. (2010) Flexible recognition of the tRNA G18 methylation target site by TrmH methyltransferase through first binding and induced fit processes. J. Biol. Chem. 285, 9018–9029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Awai T., Ochi A., Ihsanawati, Sengoku T., Hirata A., Bessho Y., Yokoyama S., Hori H. (2011) Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure. J. Biol. Chem. 286, 35236–35246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ny T., Björk G. R. (1980) Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J. Bacteriol. 142, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishida K., Kunibayashi T., Tomikawa C., Ochi A., Kanai T., Hirata A., Iwashita C., Hori H. (2011) Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39, 2304–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brissette P., Ballou D. P., Massey V. (1989) Determination of the dead time of a stopped-flow fluorometer. Anal. Biochem. 181, 234–238 [DOI] [PubMed] [Google Scholar]
- 24. Byström A. S., Björk G. R. (1982) Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol. Gen. Genet. 188, 440–446 [DOI] [PubMed] [Google Scholar]
- 25. Ahn H. J., Kim H. W., Yoon H. J., Lee B. I., Suh S. W., Yang J. K. (2003) Crystal structure of tRNA (m1G37) methyltransferase: insights into tRNA recognition. EMBO J. 22, 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elkins P. A., Watts J. M., Zalacain M., van Thiel A., Vitazka P. R., Redlak M., Andraos-Selim C., Rastinejad F., Holmes W. M. (2003) Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 333, 931–949 [DOI] [PubMed] [Google Scholar]
- 27. Liu J., Wang W., Shin D. H., Yokota H., Kim R., Kim S.-H. (2003) Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 Å resolution: a novel methyltransferase fold. Proteins 53, 326–328 [DOI] [PubMed] [Google Scholar]
- 28. Anantharaman V., Koonin E. V., Aravind L. (2002) SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4, 71–75 [PubMed] [Google Scholar]
- 29. Kuratani M., Bessho Y., Nishimoto M., Grosjean H., Yokoyama S. (2008) Crystal structure and mutational study of a unique SpoU family archaeal methylase that forms 2′-O-methylcytidine at position 56 of tRNA. J. Mol. Biol. 375, 1064–1075 [DOI] [PubMed] [Google Scholar]
- 30. Pleshe E., Truesdell J., Batey R. T. (2005) Structure of a class II TrmH tRNA-modifying enzyme from Aquifex aeolicus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 722–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim K., Zhang H., Tempczyk A., Krajewski W., Bonander N., Toedt J., Howard A., Eisenstein E., Herzberg O. (2003) Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot. Proteins 51, 56–67 [DOI] [PubMed] [Google Scholar]
- 32. Liu R. J., Zhou M., Fang Z. P., Wang M., Zhou X. L., Wang E. D. (2013) The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tkaczuk K. L., Dunin-Horkawicz S., Purta E., Bujnicki J. M. (2007) Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Purta E., van Vliet F., Tkaczuk K. L., Dunin-Horkawicz S., Mori H., Droogmans L., Bujnicki J. M. (2006) The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol. Biol. 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Benítez-Páez A., Villarroya M., Douthwaite S., Gabaldón T., Armengod M. E. (2010) YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA (Leu) isoacceptors. RNA 16, 2131–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Renalier M. H., Joseph N., Gaspin C., Thebault P., Mougin A. (2005) The Cm56 tRNA modification in archaea is catalyzed either by a specific 2′-O-methylase or a C/D sRNP. RNA 11, 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lövgren J. M., Wikström P. M. (2001) The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 183, 6957–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michel G., Sauvé V., Larocque R., Li Y., Matte A., Cygler M. (2002) The structure of the RlmB 23S rRNA methyltransferase reveals a new methyltransferase fold with a unique knot. Structure 10, 1303–1315 [DOI] [PubMed] [Google Scholar]
- 39. Treede I., Jakobsen L., Kirpekar F., Vester B., Weitnauer G., Bechthold A., Douthwaite S. (2003) The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol. Microbiol. 49, 309–318 [DOI] [PubMed] [Google Scholar]
- 40. Mosbacher T. G., Bechthold A., Schulz G. E. (2005) Structure and function of the antibiotic resistance-mediating methyltransferase AviRb from Streptomyces viridochromogenes. J. Mol. Biol. 345, 535–545 [DOI] [PubMed] [Google Scholar]
- 41. Kempenaers M., Roovers M., Oudjama Y., Tkaczuk K. L., Bujnicki J. M., Droogmans L. (2010) New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res. 38, 6533–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen H. Y., Yuan Y. A. (2010) Crystal structure of Mj1640/DUF358 protein reveals a putative SPOUT-class RNA methyltransferase. J. Mol. Cell Biol. 2, 366–374 [DOI] [PubMed] [Google Scholar]
- 43. Chatterjee K., Blaby I. K., Thiaville P. C., Majumder M., Grosjean H., Yuan Y. A., Gupta R., de Crécy-Lagard V. (2012) The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 18, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wurm J. P., Griese M., Bahr U., Held M., Heckel A., Karas M., Soppa J., Wöhnert J. (2012) Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs. RNA 18, 412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vallat B. K., Pillardy J., Májek P., Meller J., Blom T., Cao B., Elber R. (2009) Building and assessing atomic models of proteins from structural templates: learning and benchmarks. Proteins 76, 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seno T., Kobayashi M., Nishimura S. (1968) Purification of Escherichia coli methionine tRNAF and methionine tRNAM and studies on their biophysical and biochemical properties. Biochim. Biophys. Acta 169, 80–94 [DOI] [PubMed] [Google Scholar]
- 47. Taniguchi H., Hayashi N. (1998) A liquid chromatography/electrospray mass spectrometric study on the post-transcriptional modification of tRNA. Nucleic Acids Res. 26, 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koren R., Hammes G. G. (1976) A kinetic study of protein-protein interactions. Biochemistry 15, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 49. Alsallaq R., Zhou H. X. (2008) Electrostatic rate enhancement and transient complex of protein-protein association. Proteins 71, 320–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hori H., Saneyoshi M., Kumagai I., Miura K., Watanabe K. (1989) Effects of modification of 4-thiouridine in E. coli tRNA(fMet) on its methyl acceptance activity by thermostable Gm-methylases. J. Biochem. 106, 798–802 [DOI] [PubMed] [Google Scholar]
- 51. Matsumoto T., Nishikawa K., Hori H., Ohta T., Miura K., Watanabe K. (1990) Recognition sites of tRNA by a thermostable tRNA (guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biochem. 107, 331–338 [DOI] [PubMed] [Google Scholar]
- 52. Wurm J. P., Meyer B., Bahr U., Held M., Frolow O., Kötter P., Engels J. W., Heckel A., Karas M., Entian K. D., Wöhnert J. (2010) The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 38, 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meyer B., Wurm J. P., Kötter P., Leisegang M. S., Schilling V., Buchhaupt M., Held M., Bahr U., Karas M., Heckel A., Bohnsack M. T., Wöhnert J., Entian K. D. (2011) The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Ψ1191 in yeast 18S rRNA. Nucleic Acids Res. 39, 1526–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Crécy-Lagard V., Brochier-Armanet C., Urbonavicius J., Fernandez B., Phillips G., Lyons B., Noma A., Alvarez S., Droogmans L., Armengaud J., Grosjean H. (2010) Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 27, 2062–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goto-Ito S., Ito T., Ishii R., Muto Y., Bessho Y., Yokoyama S. (2008) Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins 72, 1274–1289 [DOI] [PubMed] [Google Scholar]
- 56. Goto-Ito S., Ito T., Kuratani M., Bessho Y., Yokoyama S. (2009) Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 16, 1109–1115 [DOI] [PubMed] [Google Scholar]
- 57. Björk G. R., Jacobsson K., Nilsson K., Johansson M. J., Byström A. S., Persson O. P. (2001) A primordial tRNA modification required for the evolution of life? EMBO J. 20, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brulé H., Elliott M., Redlak M., Zehner Z. E., Holmes W. M. (2004) Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43, 9243–9255 [DOI] [PubMed] [Google Scholar]
- 59. Lee C., Kramer G., Graham D. E., Appling D. R. (2007) Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 282, 27744–27753 [DOI] [PubMed] [Google Scholar]
- 60. Paris Z., Horáková E., Rubio M. A., Sample P., Fleming I. M., Armocida S., Lukes J., Alfonzo J. D. (2013) The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 19, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Christian T., Hou Y. M. (2007) Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J. Mol. Biol. 373, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Christian T., Lahoud G., Liu C., Hou Y. M. (2010) Control of catalytic cycle by a pair of analogous tRNA modification enzymes. J. Mol. Biol. 400, 204–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lahoud G., Goto-Ito S., Yoshida K., Ito T., Yokoyama S., Hou Y. M. (2011) Differentiating analogous tRNA methyltransferases by fragments of the methyl donor. RNA 17, 1236–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sakaguchi R., Giessing A., Dai Q., Lahoud G., Liutkeviciute Z., Klimasauskas S., Piccirilli J., Kirpekar F., Hou Y. M. (2012) Recognition of guanosine by dissimilar tRNA methyltransferases. RNA 18, 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sabina J., Söll D. (2006) The RNA-binding PUA domain of archaeal tRNA-guanine transglycosylase is not required for archaeosine formation. J. Biol. Chem. 281, 6993–7001 [DOI] [PubMed] [Google Scholar]
- 66. Ishitani R., Nureki O., Nameki N., Okada N., Nishimura S., Yokoyama S. (2003) Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113, 383–394 [DOI] [PubMed] [Google Scholar]
- 67. Tomikawa C., Yokogawa T., Kanai T., Hori H. (2010) N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability through a tRNA modification network. Nucleic Acids Res. 38, 942–957 [DOI] [PMC free article] [PubMed] [Google Scholar]