Background: Copper nitrite reductases convert nitrite to nitric oxide in microorganisms.
Results: The copper nitrite reductase from an ammonia oxidizing bacterium is unusually oxygen tolerant and has several unique structural features.
Conclusion: The structure of this copper nitrite reductase enables it to function in an aerobic environment.
Significance: This enzyme plays a key role in an environmentally important pathway.
Keywords: Copper, Crystal Structure, Electron Transfer, Nitrogen Metabolism, Oxygen Binding, Nitrosomonas europaea, Denitrification, Nitrifier, Nitrite Reductase
Abstract
Nitrifier denitrification is the conversion of nitrite to nitrous oxide by ammonia-oxidizing organisms. This process, which is distinct from denitrification, is active under aerobic conditions in the model nitrifier Nitrosomonas europaea. The central enzyme of the nitrifier dentrification pathway is a copper nitrite reductase (CuNIR). To understand how a CuNIR, typically inactivated by oxygen, functions in this pathway, the enzyme isolated directly from N. europaea (NeNIR) was biochemically and structurally characterized. NeNIR reduces nitrite at a similar rate to other CuNIRs but appears to be oxygen tolerant. Crystal structures of oxidized and reduced NeNIR reveal a substrate channel to the active site that is much more restricted than channels in typical CuNIRs. In addition, there is a second fully hydrated channel leading to the active site that likely acts a water exit pathway. The structure is minimally affected by changes in pH. Taken together, these findings provide insight into the molecular basis for NeNIR oxygen tolerance.
Introduction
Nitrifier denitrification is the conversion of NO2− via NO to N2O, a potent greenhouse gas, by ammonia-oxidizing organisms (1). This biochemical pathway has been well documented in ammonia-oxidizing bacteria, and there is evidence that ammonia-oxidizing archaea may harbor a similar pathway (2–4). In ammonia oxidizing bacteria, NO2− is produced from the aerobic oxidation of ammonia via hydroxylamine (5, 6). The NO2− is reduced to NO resulting in the consumption of one electron by a copper nitrite reductase (CuNIR)2 encoded by the nirK gene. NO is then reduced to N2O using a variety of NO reductases (7–11). Nitrifier denitrification is distinct from denitrification because it does not appear to be inhibited by oxygen, and the final product is N2O rather than N2 (4, 9). This pathway also differs from a physiological perspective: typical denitrification supports oxidative phosphorylation in the absence of oxygen, whereas the function of nitrifier denitrification remains unresolved.
Three functions of the nitrifier denitrification pathway have been proposed. First, it may be a strategy to conserve oxygen for ammonia oxidation under oxygen-limiting conditions. Second, it may confer resistance to nitrite toxicity. Third, it could improve the kinetics of hydroxylamine oxidation by providing additional terminal electron acceptors (9, 12, 13). All three of these functions require tolerance to fluxes of oxygen. The nitrifier denitrification pathway may fulfill some or all of these interrelated functions depending on the growth environment. Moreover, the physiological function of the pathway likely varies among different ammonia oxidizing bacteria. This notion is supported by phylogenetic analysis of nirK sequences from different nitrifying bacteria. There are three separate sequence clusters, each with distinct characteristics. Cluster 1 nirKs are in multigene operons under regulation of a nitrite/nitric oxide sensor, cluster 2 nirKs have a fused N-terminal plastocyanin-like domain, and cluster 3 nirKs resemble typical CuNIRs from α-proteobacterial denitrifiers (7). In addition, the responses of different ammonia-oxidizing bacteria strains to nitrite addition are variable, consistent with divergent physiologies of the pathway (14).
The best characterized nitrifier denitrification pathway is that of Nitrosomonas europaea, a model nitrifying organism. The N. europaea CuNIR (NeNIR) falls within nirK phylogenetic cluster 1 and is part of an operon under control of the nitrite/nitric oxide sensor NsrR (15, 16). The multigene operon containing nirK also encodes two uncharacterized cytochromes, NcgB and NcgC, and a well characterized multicopper oxidase (MCO) (NcgA, also known as blue copper oxidase or NeMCO), of which the physiological function remains unknown (17–19). Most of the research on NeNIR has focused on strains deficient in the nirK gene as well as knock-outs of ncgA, ncgB, and ncgC (9, 20–22). Knock-outs of each gene within the operon result in increased sensitivity to nitrite. In addition, extensive analysis of the nirK-deficient strains combined with growth and metabolic studies (13, 23) indicate a close relationship between ammonia oxidation and nitrite reduction.
Despite its central role in nitrifier denitrification, NeNIR has not been isolated and characterized. Early work resulted in partially purified samples (24–26) that were likely a mixture of NeNIR, NeMCO, and hydroxylamine oxidoreductase. Sequence alignments indicate that NeNIR is similar to other CuNIRs typically found in denitrifiers (7). It possesses conserved ligands to the type 1 (T1) copper center, which is responsible for oxidizing proteinaceous electron donors, and to the type 2 (T2) copper center, which is the site of nitrite binding and reduction. NeNIR also contains conserved histidine and aspartic acid residues, which position nitrite adjacent to the T2 copper center (27, 28). Comparison of the sequence to those of structurally characterized CuNIRs indicates that it is most similar to the CuNIR from Neisseria gonorrhoeae (NgNIR) (29) with 37% identity.
Of particular interest is the observation that the N. europaea nitrifier denitrification pathway is active under aerobic conditions (4), suggesting that NeNIR might have an unusual ability to avoid inactivation by oxygen. CuNIRs are generally believed to be inactivated under aerobic conditions by generation of hydrogen peroxide. It has been demonstrated that CuNIRs can reduce oxygen at a fairly significant rate (∼3% of the rate of nitrite reduction) and prolonged exposure to oxygen in the presence of reduced azurin leads to enzyme inactivation, which can be prevented by addition of catalase (30, 31). In addition, CuNIR activity is inhibited when Alacaligenes faecalis cultures are subjected to rapid increases in oxygen concentration (31). To understand how NeNIR resists inactivation under aerobic conditions and to gain further insight into its role in nitrifier denitrification, we have purified native NeNIR, investigated its activity properties, and determined its crystal structure at two pH values and in the oxidized and reduced forms.
EXPERIMENTAL PROCEDURES
Cell Growth and Purification
Cultures of N. europaea were grown as described previously and flash frozen in liquid nitrogen for later use (17). Frozen cells were suspended to ∼25 mg (wet weight) ml−1 in a buffer containing 20 mm NaCl, 10% glycerol, 1 mm MgCl2, and 20 mm Pipes, pH 7.0, and then lysed by two passes through a 15,000 psi microfluidizer. The lysed cells were ultracentrifuged at 125,000 × g for 1 h to remove insoluble material. NH4SO4 was added to the supernatant of the 125,000 × g spin to a final concentration of 0.4 g ml−1 protein solution and allowed to stir for ∼10 min. The protein solution was then ultracentrifuged at 125,000 × g for 30 min to remove precipitated proteins. The resultant supernatant was applied to a 20-ml phenyl-Sepharose column (GE Healthcare) equilibrated with a buffer containing 2.5 m NH4SO4, 1 mm MgSO4, 50 μm CuSO4, 10% glycerol, and 20 mm Tris, pH 8.0. The protein was eluted using a 400-ml gradient with buffer containing 1 mm MgSO4, 50 μm CuSO4, 10% glycerol, and 20 mm Tris, pH 8.0. NeNIR typically elutes ∼250 ml into the gradient.
Fractions containing NeNIR were concentrated to <10 ml using an Amicon Ultra-15 centrifugal device and dialyzed against 1 liter of buffer containing 1 mm MgSO4, 50 μm CuSO4, 10% glycerol, and 5 mm Tris, pH 8.0, for at least 2 h. The dialysis buffer was changed two times. The desalted NeNIR fractions were loaded onto a 5-ml Source 15Q column (GE Healthcare) equilibrated with 1 mm MgSO4, 50 μm CuSO4, 10% glycerol and 5 mm Tris, pH 8.0, and eluted over a 200-ml gradient with a buffer containing 2.5 m NH4SO4, 1 mm MgSO4, 50 μm CuSO4, 10% glycerol, and 20 mm Tris, pH 8.0. NeNIR typically elutes 15 ml into gradient. Fractions containing NeNIR were concentrated to <1 ml using an Amicon Ultra-15 and further purified by size exclusion chromatography on a Sephacryl-200 column equilibrated with 20 mm NaCl, 1 mm MgCl2, 10% glycerol, and 20 mm Pipes, pH 7.0.
Activity Assays
Nitrite reductase activity was determined by monitoring the rate of nitrite-dependent methyl viologen oxidation (32) and by monitoring the rate of o-dianisidine oxidation (30). To measure activity using methyl viologen, the following protocol was performed inside of a Coy anaerobic chamber: 10 μl of 50 mm sodium dithionite was added to a screw top cuvette containing 980 μl of solution composed of 1.5 mm methyl viologen, 1 μm catalase, and 50 mm Pipes, pH 7.0. This typically yielded ∼650 μm reduced methyl viologen. NaNO2 (5 μl) was placed on the side wall of the cuvette to prevent mixing. The screw top cuvette was then sealed with a threaded lid containing a rubber septum. Prior to NeNIR addition, the vial was inverted to mix the NaNO2 with the methyl viologen solution, resulting in a final concentration of 2 mm NaNO2. The solution was allowed to stabilize for several minutes until a reliable background rate could be obtained (typically 2–5 min). The reaction was then initiated by the addition of 695 pmol of NeNIR. Methyl viologen oxidation was monitored at 730 nm (ϵ = 2,143 m cm−1) using a Cary 5000 UV-VIS spectrometer, and the data were fit using a linear regression with the program Origin (version 8) over the course of 1 min. Reported data are from an average of four replicates.
For o-dianisidine dependent activity, a saturated solution of o-dianisidine was prepared by stirring excess o-dianisidine in 50 mm Pipes, pH 7.0, overnight followed by filtering with a 0.22-μm filter. For a typical assay, NaNO2 and catalase were added to 950 μl of o-dianisidine-saturated buffer and brought to a final volume of 1 ml to give a final concentration of 10 mm NaNO2 and 1 μm catalase. Reactions were also performed without catalase and without NaNO2. Reactions containing NaNO2 were initiated by the addition of 17.3 pmol NeNIR, whereas reactions without NaNO2 were initiated by the addition of 2.5 nmol NeNIR. For anaerobic experiments, all manipulations were performed in a Coy chamber except for addition of NeNIR, which was performed using a gastight syringe. All reactions were performed in at least triplicate, and o-dianisidine oxidation was monitored at 460 nm (ϵ = 11,600 m cm−1).
Crystallization and Data Collection
NeNIR was crystallized by the sitting drop vapor diffusion method at 4 °C by mixing 1 μl of 10 mg ml−1 NeNIR with 1 μl of a well solution containing 5% glycerol, 20% PEG 3350, 0.7–1.05 mm LiCl, and either 75 mm Tris, pH 8.5, or 100 mm sodium cacodylate, pH 6.5. Rod-shaped crystals (pH 8.5) and hexagon-shaped crystals (pH 6.5) grew within 3 to 4 days. Reduced crystals were obtained by soaking crystals grown at pH 6.5 in well solution containing 100 mm ascorbate. All crystals were cryoprotected in well solution containing 30% glycerol and flash frozen in liquid nitrogen. Data collection was performed at sector 21 (the Life Sciences Collaborative Access Team) of the Advanced Photon Source at Argonne National Laboratory. HKL2000 was used to process and integrate all data sets (Table 1) (33).
TABLE 1.
Data collection and refinement statistics
| Data set (PDB code) | Phasing | pH 8.5 (4KNT) | pH 6.5 (4KNU) | pH 6.5 reduced (4KNS) |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2 | C2 | P21 | P21 |
| Cell dimensions | a = 100, b = 123, and c = 88 Å; γ = 97° | a = 100, b = 123, and c = 88 Å; γ = 97° | a = 51, a = 137, and c = 121 Å; γ = 101° | a = 51, b = 138, and c = 120 Å; γ = 101° |
| Resolution (Å)a | 2.4 (2.49–2.40) | 1.9 (1.90–1.95) | 1.8 (1.80–1.85) | 2.2 (2.20–2.26) |
| Rsyma,b | 0.191 (0.387) | 0.096 (0.443) | 0.120 (0.463) | 0.112 (0.530) |
| I/σIa | 11.7 (3.4) | 15.2 (2.8) | 11.5 (2.3) | 11.6 (2.1) |
| Completeness (%)a | 100 (100) | 98.6 (94.5) | 99.6 (98.1) | 91.0 (91.7) |
| Refinement | ||||
| Rworkc/Rfreed | 16.9/20.0 | 17.1/20.9 | 18.5/22.9 | |
| Ramachandran outliers (%) | 0 | 0 | 0 | |
| Average B-factor | 19.6 | 14.4 | 27.1 | |
| r.m.s.d.e | ||||
| Bond lengths (Å) | 0.006 | 0.008 | 0.005 | |
| Bond angles | 0.609° | 1.244° | 0.923° | |
a Values in parentheses refer to the highest resolution shell.
b Rsym = Σ|Iobs − Iav|/ΣFobs, where the summation is over all reflections.
c Rwork = Σ|Fobs − Fcalc|/ΣFobs.
d For calculation of Rfree, 5% of the reflections were reserved.
e r.m.s.d., root mean square deviation.
Structure Determination, Refinement, and Analysis
Copper single wavelength anomalous dispersion was used to determine phases for data sets collected on crystals grown at pH 8.5 (Table 1). Low resolution models built using the program COOT (34) for the pH 8.5 structure were subsequently used as molecular replacement search models for all other data sets. All data sets were collected at a wavelength of 1.3776 Å. Phaser was used to obtain molecular replacement solutions, and all structures were refined using REFMAC (35, 36). Three different crystal structures were determined: oxidized NeNIR, pH 8.5, to 1.9 Å resolution, oxidized NeNIR at pH 6.5 to 1.8 Å resolution, and reduced NeNIR pH at 6.5 to 2.2 Å resolution (Table 1). All regions of NeNIR were well ordered, and all of the residues following the signal peptide (residue numbers 22–309) were modeled. This very complete model allowed for an extensive comparison of nonconserved loop regions and termini with similar regions of other NIRs. The three structures are virtually identical with only minor differences in solvent molecules present in the substrate channel and metal-to-ligand distances. The copper sites were fully occupied in both structures at pH 6.5, whereas the metal centers in the pH 8.5 structure were best modeled with occupancies of 0.8–0.9. In the reduced structure, 2Fo − Fc density for a water molecule at the T2 copper center was not observed, although the presence of some density in the Fo − Fc difference map suggests that the crystal was still slightly oxidized. PDBeFold was used to perform three-dimensional structural alignments (37). Volume-filling analysis was performed using the program HOLLOW (38). Figures were generated with PyMOL (39), and the PyMOL plug-in for APBS was used to calculate electrostatic potential surfaces (40).
RESULTS AND DISCUSSION
Activity
Purified NeNIR was green in color and exhibited an optical spectrum with features at 450 and 597 nm, which are typical for a green CuNIR (Fig. 1) (27). The rate of nitrite reduction using methyl viologen as an electron donor was 288 ± 14.5 s−1. This rate is similar to other CuNIRs (30, 41) and suggests that differences in the substrate channel (see below) do not significantly affect its ability to reduce nitrite. The rate of nitrite reduction using o-dianisidine was measured aerobically (atmospheric oxygen concentrations) and anaerobically as well as with and without catalase (Table 2). Similar to the CuNIR from Alcaligenes faecalis (AfNIR) (30), the NeNIR o-dianisidine-dependent rate of NO2− reduction was significantly lower than the methyl viologen-dependent rate. This difference is likely due to the fact o-dianisidine has a much higher reduction potential than methyl viologen. No o-dianisidine oxidation was detected in assays performed aerobically without NO2−.
FIGURE 1.
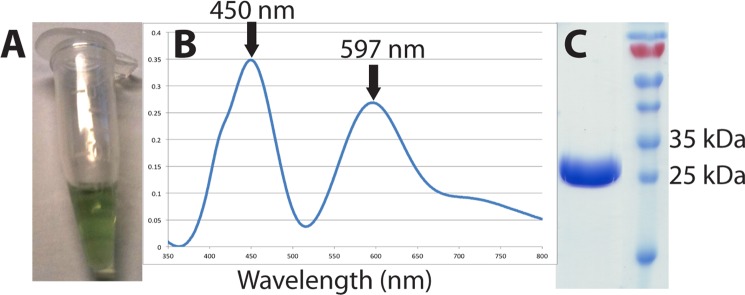
Purification of NeNIR from N. europaea. A, purified material is green. B, optical spectrum (7 mg/ml). C, SDS-PAGE analysis.
TABLE 2.
Activity of NeNIR measured using o-dianisidine
| Sample | Turnover |
|---|---|
| s−1 | |
| Ambienta | 1.42 ± 0.06 |
| Ambient + catalase | 1.23 ± 0.05 |
| Anaerobic | 0.78 ± 0.11 |
| Anaerobic + catalase | 0.72 ± 0.05 |
a Atmospheric oxygen concentrations.
The o-dianisidine-dependent activity assays reveal that oxygen does not inhibit nitrite reduction, and consistent with this observation, the presence of catalase has no effect on activity under aerobic or anaerobic conditions. Activity is actually enhanced in the presence of oxygen. The chemical basis for this enhancement is not immediately clear, but it is most likely due to an oxygen-dependent abiotic reaction between NO produced by the enzyme and o-dianisidine. Notably, NeNIR does not reduce oxygen. Even when the concentration of NeNIR is increased 144-fold, oxygen reduction is not detectable. By contrast, AfNIR reduces O2 at a rate of ∼3% of the NO2− reduction rate using either o-dianisidine or pseudoazurin as the electron donor (30). Taken together, these findings suggest that NeNIR is oxygen-tolerant. It would be informative to perform similar experiments using the native NeNIR electron donor rather than o-dianisidine, but its identity remains unknown.
Overall Structure and Metal Centers
NeNIR is a cylindrical homotrimer (Fig. 2) that closely resembles other CuNIRs despite the low sequence similarity (∼40% identity). The coordination environment and copper-to-ligand distances (Table 3) are also very similar to those in other structurally characterized CuNIRs and are consistent with NeNIR following the same catalytic cycle. However, the structure reveals several important differences (see below) that provide insight into the observed oxygen tolerance of NeNIR.
FIGURE 2.
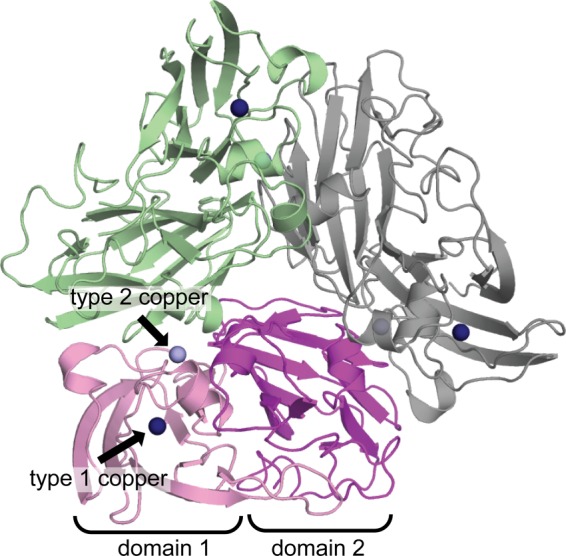
Overall architecture of NeNIR. Domains 1 and 2 of one monomer are shown in pink and purple, and the other two monomers are shown in green and gray. Type 1 copper centers are shown as dark blue spheres, and type 2 copper centers are shown as light blue spheres.
TABLE 3.
Metal-ligand distances in NeNIR structures
Distance and S.D. calculated from averaging the individual molecules in the asymmetric unit.
| Data set | pH 6.5 | pH 8.5 | Reduced |
|---|---|---|---|
| Molecules per asymmetric unit | 6 | 3 | 6 |
| Type 1 copper | Distance (Å) | Distance (Å) | Distance (Å) |
| His-83 | 2.10 ± 0.02 | 2.14 ± 0.01 | 2.21 ± 0.06 |
| His-131 | 2.10 ± 0.04 | 2.14 ± 0.02 | 2.20 ± 0.07 |
| Cys-123 | 2.23 ± 0.03 | 2.26 ± 0.04 | 2.30 ± 0.05 |
| Met-136 | 2.48 ± 0.03 | 2.47 ± 0.02 | 2.42 ± 0.10 |
| Type 2 copper | |||
| His-88 | 2.01 ± 0.04 | 2.03 ± 0.02 | 2.11 ± 0.04 |
| His-122 | 2.05 ± 0.05 | 2.07 ± 0.02 | 2.12 ± 0.08 |
| His-277 | 2.08 ± 0.03 | 2.12 ± 0.02 | 2.06 ± 0.08 |
| H2O/OH | 2.44 ± 0.06 | 2.31 ± 0.07 | |
Channels to the Type 2 Copper Center
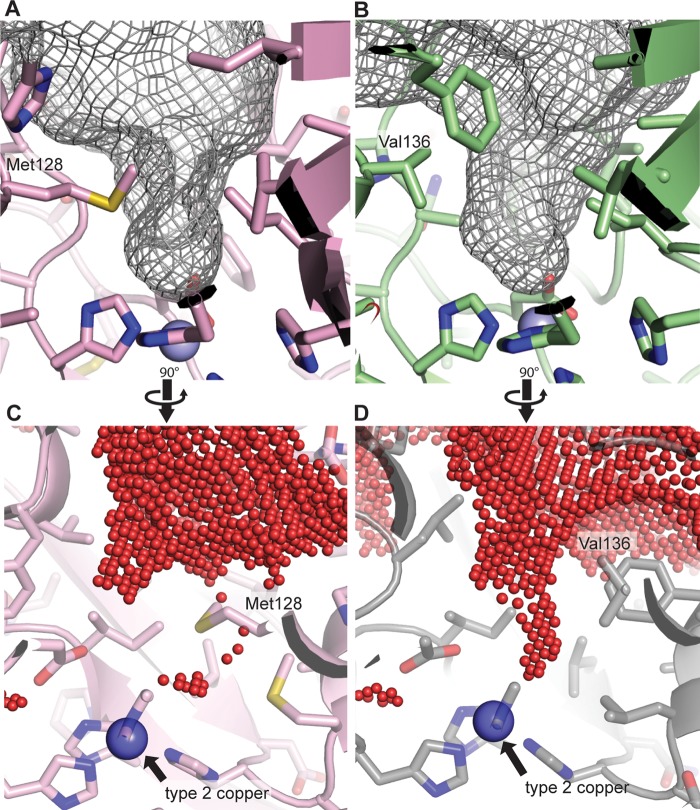
In typical CuNIRs, the T2 copper center is accessible by a single channel, which is responsible for delivering substrates, releasing product, and transferring protons (42–45). The channel roughly resembles a funnel with the opening formed by four different surface loops and β-strands 7 and 8 of the second domain. Even though the channel is funnel shaped, it is still relatively wide, and the T2 site is readily accessible (Fig. 3 and Fig. 4, B and D). By contrast, access to the T2 site in NeNIR is much more restricted due to two structural features. First, the backbone of one of the channel forming loops is longer and extends deeply into the channel. This loop is six amino acids longer than the equivalent loop in Alcaligenes xylosoxidans (AxNIR) (Fig. 3, extended loop), whereas the loop on the other side of the channel is one amino acid shorter than that AxNIR (Fig. 3, truncated loop). The net result is a more confined opening than that in typical CuNIRs (43).
FIGURE 3.
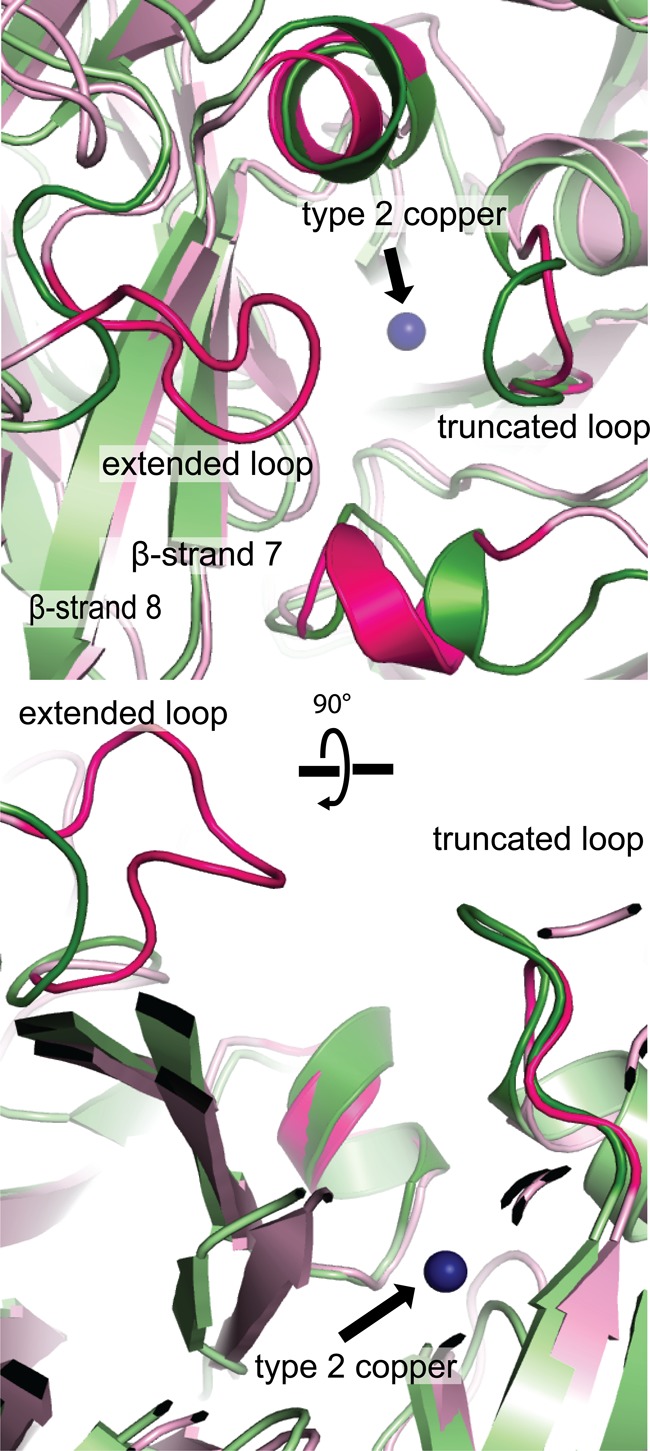
Comparison of channels to type 2 copper center. NeNIR is shown in pink, and AxNIR (PDB code 1OE1) is shown in green. Important loops in NeNIR are highlighted in magenta.
FIGURE 4.
Structural features defining the channel to the type 2 site. A and C, results of volume-filling analysis of NeNIR shown as gray mesh (A) and red spheres (C). B and D, results of volume-filling analysis of AxNIR (PDB code 2VN3) shown as a gray mesh (B) and red spheres (D).
The second key structural feature of the NeNIR channel is the presence of residue Met-128. This residue constricts the end of the channel such that the channel is no longer funnel-shaped, and only a single solvent molecule can be accommodated (Fig. 4, A and C, and Fig. 5A). This well ordered water molecule lies within 3.49 ± 0.06 Å of the sulfur atom of Met-128 and 2.72 Å ± 0.09 Å of the side chain oxygen of Asp-86, which is also hydrogen bonded to a coordinated water molecule (Fig. 5A). The distance between the sulfur atom of Met-128 and the water molecule is consistent with a sulfur-to-water hydrogen bond (46). In addition, the sulfur atom is 5.80 ± 0.03 Å from the T2 copper ion, and it is conceivable that this residue could interact directly with bound substrate. Importantly, structural alignments with other CuNIRs indicate that NeNIR is the only one with methionine at this position; the analogous location in most other CuNIRs is occupied by valine (e.g. AfNIR and AxNIR, Fig. 4, B and D) or isoleucine (e.g. Rhodobacter sphaeroides (RsNIR)).
FIGURE 5.
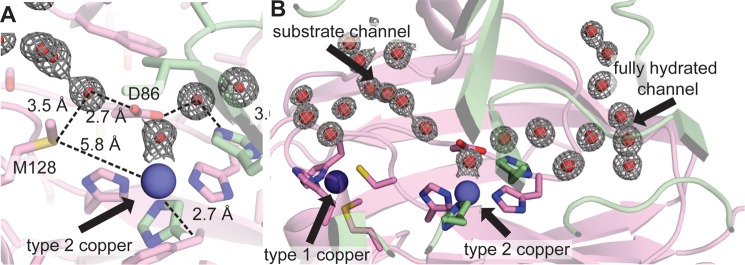
Solvent structure of NeNIR channels. Two NeNIR monomers are shown in green and pink with waters shown as red spheres with the corresponding 2Fo− Fc electron density superimposed at a contour level of 1σ. A, type 2 site showing residue Met-128 and the hydrogen-bonding network involving Asp-86 and the coordinated water molecule. B, locations of substrate channel and second water channel.
Unlike other CuNIRs, the NeNIR T2 copper center is also accessible by a second fully hydrated channel (Fig. 5B). This water channel is located at the monomer-monomer interface and extends in the opposite direction from the substrate channel. Its location is structurally analogous to a region in AxNIR that consists of several waters gated by a histidine residue and was thought to be a proton channel (termed the primary proton channel) (43–45) as well as to the proposed water exit channel found in MCOs (47). The channel contains eight well ordered solvent molecules connected by an elaborate hydrogen bonding network that extends all the way from the T2 copper center to the surface of the protein. Interactions of solvent molecules within the channel involve backbone hydrogen bonds, whereas interactions at the channel mouth engage the side chains of several residues, including Arg-91, Thr-112, and Asp-114. It should be noted that a glycerol molecule is hydrogen bonded to Arg-91, Asp-114, Asn-115, and Asp-234 on the protein surface and may influence the precise location of the channel opening.
The Effects of pH
To further understand the physical properties of the water channel and the confined space around the T2 copper center, structures of oxidized NeNIR at pH 8.5 and 6.5 as well as reduced NeNIR at pH 6.5 were compared with one another and to the structures of RsNIR at pH 6.0 and pH 8.4 (Fig. 6) (48, 49). RsNIR was selected for analysis because structures are available in the same pH range as those obtained for NeNIR. The hydrogen bonding network and the positions of solvent molecules within the water channel are not affected by lowering the pH of NeNIR to 6.5. In RsNIR at pH 6.0, the water molecule coordinated to the T2 site shifts by 1.2 Å, whereas only a slight elongation in bonding distance is observed in NeNIR at pH 6.5 (Fig. 6). Although the less pronounced effect could be due in part to the slightly higher pH of the NeNIR structure (NeNIR cannot be crystallized below pH 6.5), the lack of perturbation of the hydrogen-bonding network and water channel structure suggests that NeNIR is less sensitive to pH than other CuNIRs. The relatively minor effect of pH on the structure might be due to the presence of Met-128, which shields the T2 site from bulk solvent via its hydrogen bonding interactions (Fig. 5A). This idea is supported by volume-filling analysis, which shows that Met-128 displaces a large volume of solvent in the substrate channel and alters the shape of the substrate channel near the T2 copper center (Fig. 4C).
FIGURE 6.
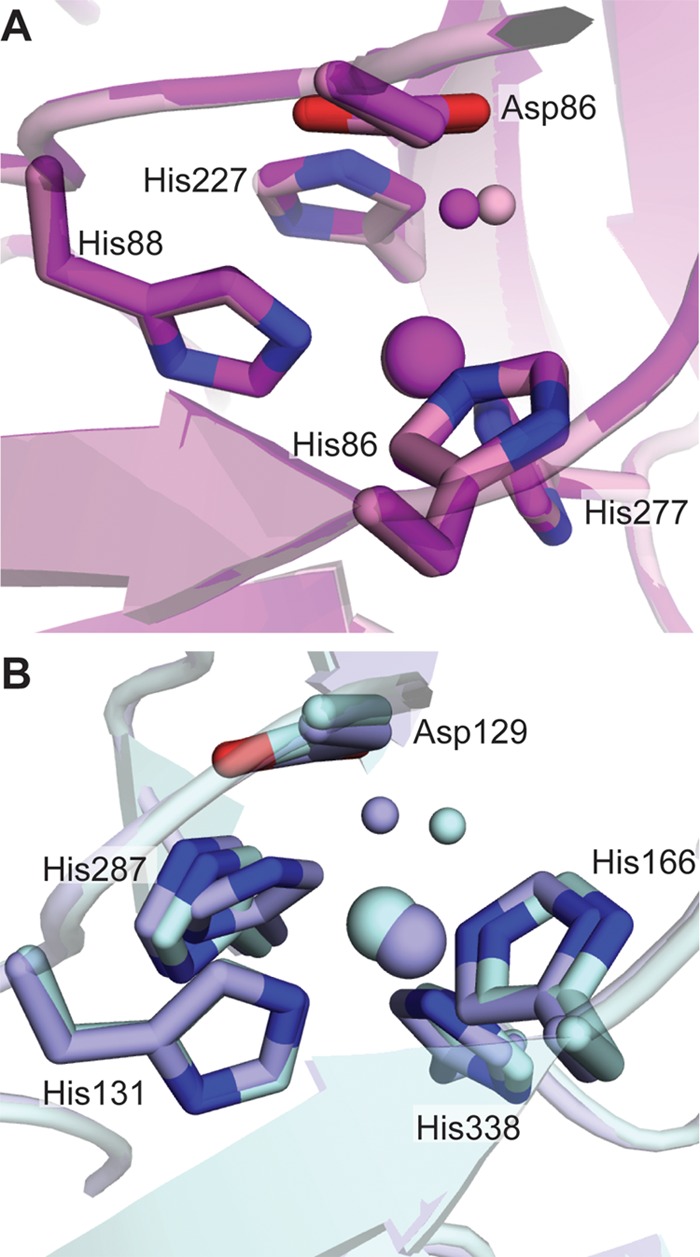
Effects of pH on the type 2 sites in NeNIR and RsNIR. A, NeNIR at pH 6.5 (purple) and pH 8.5 (pink). B, RsNIR at pH 6.0 (light blue, PDB code 2DY2) and pH 8.4 (dark blue, PDB code 1ZV2). Copper ions are shown as large spheres, and water molecules are shown as small spheres.
Electron Donor Docking Site
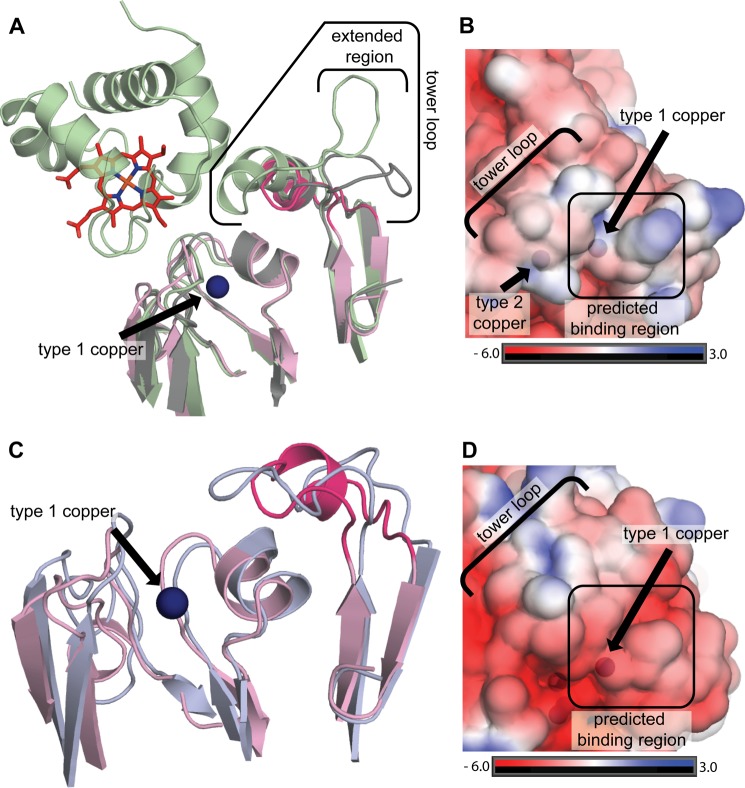
Docking of proteinaceous electron donors near the T1 copper center and intermolecular electron transfer from the T1 to the T2 site have been studied extensively (50, 51). X-ray and NMR structures of CuNIRs bound to their electron donors (cytochrome c analogs and pseudoazurin) combined with mutagenesis data have defined two specific regions critical for donor binding, the tower loop and the hydrophobic patch (50–53). In a typical CuNIR, the tower loop extends from the second domain over the T1 copper site located in the N-terminal domain (29) and helps position the proteinaceous electron donor on the hydrophobic patch directly above the T1 site, which lies between the hydrophobic patch and tower loop (Fig. 7A). In NeNIR, the tower loop is truncated by 17 residues as compared with AxNIR and by nine residues as compared with NgNIR (29, 50). As a result, the NeNIR tower loop does not extend nearly as far over the N-terminal domain as its AxNIR counterpart. The portion of the NeNIR tower loop facing the N-terminal domain is instead structurally similar to NgNIR (Fig. 7A). An extensive region of the NgNIR and AxNIR tower loops that resides on the top of the C-terminal domain is completely absent in NeNIR.
FIGURE 7.
Electron donor docking sites in CuNIRs. A, superposition of NeNIR (pink, tower loop in magenta) with the AxNIR-cytochrome c551 co-crystal structure (green, PDB code 2ZON) and NgNIR (gray, PDB code 1KBW). B, NeNIR surface colored by electrostatic potential. C, superpostion of NeNIR (pink and magenta) and NeMCO (blue, PDB code 3G5W) highlighting the tower loop. D, NeMCO (PDB code 3G5W) surface colored by electrostatic potential.
The size and conformation of the NeNIR tower loop is reminiscent of the tower loop in NeMCO, the two-domain MCO encoded by the same operon as NeNIR (Fig. 7C) (17). This MCO is unique because it has considerable oxidase activity toward proteinaceous electron donors, suggesting it is a terminal oxidase (18). The truncated tower loops found in NgNIR and NeMCO have been proposed to allow access of a larger electron donor or to decrease specificity of the electron donor (17, 29). The structural similarity between the NeNIR and NeMCO tower loops might suggest that they utilize the same electron donor. However, comparison of the hydrophobic patches in NeMCO and NeNIR indicates that the NeMCO surface is considerably more negatively charged (Fig. 7, B and D), which would be consistent with different donors. Thus, it may be that NeNIR and NeMCO either obtain electrons from two separate donors or that they interact with the same donor in different ways. The NcgB and NcgC proteins, also encoded by the nirK operon, are obvious candidates, but these two proteins form a complex, which is not typical of small electron carrier proteins. Other possible candidates include the tetraheme cytochrome c554 (the electron carrier from hydroxylamine oxidoreductase to the quinone oxidase), cytochrome c552 (cytochrome c analog), and nitrosocyanin (54, 55).
Mechanistic Implications
There are two proposed mechanisms for CuNIR. For many years, the prevailing mechanism involved displacement of the coordinated water molecule by nitrite at the oxidized T2 copper followed by electron transfer from the T1 to the T2 site. This mechanism is often referred to as the “ordered” or the “bind first mechanism” (56). However, there is strong kinetic evidence as well as single molecule data indicating that CuNIRs may follow a “random sequential mechanism” in which either nitrite can bind first or electron transfer from the T1 to the T2 copper center can occur prior to nitrite binding (57, 58). In the random sequential mechanism, the order of events is proposed to be dictated by substrate concentrations and pH. If the bind first mechanism is operational in NeNIR, the unique water channel may play a critical role. Because the NeNIR substrate channel narrows to width of one solvent molecule, the coordinated water molecule must be displaced by nitrite into the water channel rather than into the constrained substrate channel. The water channel may not be as important if the T2 site is reduced before nitrite binding because the coordinated water molecule becomes disordered upon reduction of NeNIR, similar to other CuNIRs (59). In reduced NeNIR, ordered solvent is not observed in the substrate channel near the T2 center, but following reduction, there is probably still solvent present that would need to be displaced into the water channel.
The water channel could also play a role in proton transfer. It has been demonstrated that AxNIR catalysis involves proton-coupled electron transfer and residue Asn-90, which is near the entrance of the substrate channel, is critical for proton transfer. The AxNIR mutant in which Asn-90 is replaced with serine has a 70% loss in activity, whereas mutations in the channel formerly known as the primary proton channel (analogous to the NeNIR water channel) do not affect activity (45, 60). Interestingly, the NeNR residue equivalent to AxNIR Asn-90 is a serine (Ser-84), implying that NeNIR probably utilizes a different proton transfer pathway.
Beyond constricting the substrate channel, residue Met-128 might play a direct role in preventing oxygen reduction. In particular, it has been proposed that O2 binds at the T2 site in an end-on manner and is reduced to peroxide (30). Peroxide formation in a CuNIR would occur via a stable one-electron reduced oxygen species. Presumably, the bound oxygen would be located in the same position as the bound water molecule, linked to the Met-128 hydrogen-bonding network (Fig. 5A). In a typical CuNIR, the open substrate channel and could likely accommodate any number of reactive oxygen species. In NeNIR, the proximity of the sulfur of Met-128 to the T2 copper site may prevent formation of a reactive oxygen species either by sterically blocking oxygen from binding in a reactive conformation (which is unlikely because NO2− readily binds) or by preventing formation of specific intermediates.
In sum, NeNIR is an oxygen tolerant CuNIR, consistent with its presence in a primarily aerobic organism. The NeNIR crystal structures reveal a constrained substrate channel due to the presence of Met-128 and a second water channel that may function as an egress for displaced water molecules. These two features are completely unprecedented in CuNIRs and suggest a new paradigm for functional aerobic CuNIR activity. Finally, the NeNIR electron donor docking site is structurally similar to that in NeMCO but differs in electrostatic properties, consistent with distinct electron donors. Functional expression of NeNIR in a recombinant host and site-directed mutagenesis as well as identification of the native electron donors will be required to further understand these unique characteristics.
Acknowledgments
We thank the staff of the Life Sciences Collaborative Access Team, especially Zdzislaw Wawrzak, for assistance in collecting data. We also thank A. Andrew Pacheco and Matthew Youngblut for helpful discussions.
This work was supported by Agriculture and Food Research Initiative Competitive Grant 2010-65115-20380 from the U. S. Department of Agriculture (to A. C. R.) and a grant from the Oregon Agricultural Experimental Station (to D. J. A).
The atomic coordinates and structure factors (codes 4KNU, 4KNT, and 4KNS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CuNIR
- copper nitrite reductase
- NeNIR
- Nitrosomonas europaea copper nitrite reductase
- PDB
- Protein Data Bank
- MCO
- multicopper oxidase
- NeMCO
- two domain multicopper oxidase from Nitrosomonas europaea
- T1
- type 1
- T2
- type 2
- NgNIR
- Neisseria gonorrhoeae copper nitrite reductase
- AfNIR
- Alcaligenes faecalis copper nitrite reductase
- AxNIR
- Alcaligenes xylosoxidans copper nitrite reductase
- RsNIR
- Rhodobacter sphaeroides copper nitrite reductase.
REFERENCES
- 1. Wrage N., Velthof G. L., van Beusichem M. L., Oenema O. (2001) Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 33, 1723–1732 [Google Scholar]
- 2. Santoro A. E., Buchwald C., McIlvin M. R., Casciotti K. L. (2011) Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333, 1282–1285 [DOI] [PubMed] [Google Scholar]
- 3. Shaw L. J., Nicol G. W., Smith Z., Fear J., Prosser J. I., Baggs E. M. (2006) Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8, 214–222 [DOI] [PubMed] [Google Scholar]
- 4. Dundee L., Hopkins D. W. (2001) Different sensitivities to oxygen of nitrous oxide production by Nitrosomonas europaea and Nitrosolobus multiformis. Soil Biol. Biochem. 33, 1563–1565 [Google Scholar]
- 5. Arp D. J., Stein L. Y. (2003) Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38, 471–495 [DOI] [PubMed] [Google Scholar]
- 6. Hooper A. B., Vannelli T., Bergmann D. J., Arciero D. M. (1997) Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie Leeuwenhoek 71, 59–67 [DOI] [PubMed] [Google Scholar]
- 7. Cantera J. J., Stein L. Y. (2007) Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 9, 765–776 [DOI] [PubMed] [Google Scholar]
- 8. Casciotti K. L., Ward B. B. (2001) Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 67, 2213–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beaumont H. J., Hommes N. G., Sayavedra-Soto L. A., Arp D. J., Arciero D. M., Hooper A. B., Westerhoff H. V., van Spanning R. J. (2002) Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J. Bacteriol. 184, 2557–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chandran K., Stein L. Y., Klotz M. G., van Loosdrecht M. C. (2011) Nitrous oxide production by lithotrophic ammonia-oxidizing bacteria and implications for engineered nitrogen-removal systems. Biochem. Soc. Trans. 39, 1832–1837 [DOI] [PubMed] [Google Scholar]
- 11. Klotz M. G., Stein L. Y. (2008) Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 278, 146–156 [DOI] [PubMed] [Google Scholar]
- 12. Poth M., Focht D. D. (1985) N kinetic snalysis of N2O Production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 49, 1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cantera J. J., Stein L. Y. (2007) Role of nitrite reductase in the ammonia-oxidizing pathway of Nitrosomonas europaea. Arch. Microbiol. 188, 349–354 [DOI] [PubMed] [Google Scholar]
- 14. Cua L. S., Stein L. Y. (2011) Effects of nitrite on ammonia-oxidizing activity and gene regulation in three ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 319, 169–175 [DOI] [PubMed] [Google Scholar]
- 15. Beaumont H. J., Lens S. I., Reijnders W. N., Westerhoff H. V., van Spanning R. J. (2004) Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54, 148–158 [DOI] [PubMed] [Google Scholar]
- 16. Chain P., Lamerdin J., Larimer F., Regala W., Lao V., Land M., Hauser L., Hooper A., Klotz M., Norton J., Sayavedra-Soto L., Arciero D., Hommes N., Whittaker M., Arp D. (2003) Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185, 2759–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawton T. J., Sayavedra-Soto L. A., Arp D. J., Rosenzweig A. C. (2009) Crystal structure of a two-domain multicopper oxidase: implications for the evolution of multicopper blue proteins. J. Biol. Chem. 284, 10174–10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DiSpirito A. A., Taaffe L. R., Lipscomb J. D., Hooper A. B. (1985) A ‘blue’ copper oxidase from Nitrosomonas europaea. Biochim. Biophys. Acta 827, 320–326 [Google Scholar]
- 19. Lawton T. J., Rosenzweig A. C. (2011) Detection and characterization of a multicopper oxidase from Nitrosomonas europaea. Methods Enzymol. 496, 423–433 [DOI] [PubMed] [Google Scholar]
- 20. Beaumont H. J., Lens S. I., Westerhoff H. V., van Spanning R. J. (2005) Novel nirK cluster genes in Nitrosomonas europaea are required for NirK-dependent tolerance to nitrite. J. Bacteriol. 187, 6849–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho C. M., Yan T., Liu X., Wu L., Zhou J., Stein L. Y. (2006) Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl. Environ. Microbiol. 72, 4450–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt I., van Spanning R. J., Jetten M. S. (2004) Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology 150, 4107–4114 [DOI] [PubMed] [Google Scholar]
- 23. Stein L. Y., Arp D. J. (1998) Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 64, 4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hooper A. B. (1968) A nitrite-reducing enzyme from Nitrosomonas europaea. Preliminary characterization with hydroxylamine as electron donor. Biochim. Biophys. Acta 162, 49–65 [DOI] [PubMed] [Google Scholar]
- 25. Miller D. J., Wood P. M. (1983) The soluble cytochrome oxidase of Nitrosomonas europaea. J. Gen. Microbiol. 129, 1645–1650 [Google Scholar]
- 26. Ritchie G. A., Nicholas D. J. (1974) Partial characterization of purified nitrite reductase and hydroxylamine oxidase from Nitrosomonas europaea. Biochem. J. 138, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zumft W. G. (1997) Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacPherson I. S., Murphy M. E. (2007) Type-2 copper-containing enzymes. Cell Mol. Life Sci. 64, 2887–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boulanger M. J., Murphy M. E. (2002) Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: a new class of copper-containing nitrite reductases. J. Mol. Biol. 315, 1111–1127 [DOI] [PubMed] [Google Scholar]
- 30. MacPherson I. S., Rosell F. I., Scofield M., Mauk A. G., Murphy M. E. (2010) Directed evolution of copper nitrite reductase to a chromogenic reductant. Protein Eng. Des. Sel. 23, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kakutani T., Watanabe H., Arima K., Beppu T. (1981) A blue protein as an inactivating factor for nitrite reductase from Alcaligenes faecalis Strain S-6. J. Biochem. 89, 463–472 [DOI] [PubMed] [Google Scholar]
- 32. Kakutani T., Watanabe H., Arima K., Beppu T. (1981) Purification and properties of a copper-containing nitrite reductase from a denitrifying bacterium, Alcaligenes faecalis strain S-6. J. Biochem. 89, 453–461 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 38. Ho B. K., Gruswitz F. (2008) HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.5.0.1, Schrödinger, LLC, New York [Google Scholar]
- 40. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kataoka K., Furusawa H., Takagi K., Yamaguchi K., Suzuki S. (2000) Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J. Biochem. 127, 345–350 [DOI] [PubMed] [Google Scholar]
- 42. Dodd F. E., Van Beeumen J., Eady R. R., Hasnain S. S. (1998) X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner. J. Mol. Biol. 282, 369–382 [DOI] [PubMed] [Google Scholar]
- 43. Ellis M. J., Dodd F. E., Sawers G., Eady R. R., Hasnain S. S. (2003) Atomic resolution structures of native copper nitrite reductase from Alcaligenes xylosoxidans and the active site mutant Asp92Glu. J. Mol. Biol. 328, 429–438 [DOI] [PubMed] [Google Scholar]
- 44. Hough M. A., Eady R. R., Hasnain S. S. (2008) Identification of the proton channel to the active site type 2 Cu center of nitrite reductase: structural and enzymatic properties of the His254Phe and Asn90Ser mutants. Biochemistry 47, 13547–13553 [DOI] [PubMed] [Google Scholar]
- 45. Leferink N. G., Han C., Antonyuk S. V., Heyes D. J., Rigby S. E., Hough M. A., Eady R. R., Scrutton N. S., Hasnain S. S. (2011) Proton-coupled electron transfer in the catalytic cycle of Alcaligenes xylosoxidans copper-dependent nitrite reductase. Biochemistry 50, 4121–4131 [DOI] [PubMed] [Google Scholar]
- 46. Zhou P., Tian F., Lv F., Shang Z. (2009) Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins 76, 151–163 [DOI] [PubMed] [Google Scholar]
- 47. Bento I., Carrondo M. A., Lindley P. F. (2006) Reduction of dioxygen by enzymes containing copper. J. Biol. Inorg. Chem. 11, 539–547 [DOI] [PubMed] [Google Scholar]
- 48. Jacobson F., Guo H., Olesen K., Okvist M., Neutze R., Sjölin L. (2005) Structures of the oxidized and reduced forms of nitrite reductase from Rhodobacter sphaeroides 2.4.3 at high pH: changes in the interactions of the type 2 copper. Acta Crystallogr. D Biol. Crystallogr. 61, 1190–1198 [DOI] [PubMed] [Google Scholar]
- 49. Jacobson F., Pistorius A., Farkas D., De Grip W., Hansson O., Sjölin L., Neutze R. (2007) pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase. J. Biol. Chem. 282, 6347–6355 [DOI] [PubMed] [Google Scholar]
- 50. Nojiri M., Koteishi H., Nakagami T., Kobayashi K., Inoue T., Yamaguchi K., Suzuki S. (2009) Structural basis of inter-protein electron transfer for nitrite reduction in denitrification. Nature 462, 117–120 [DOI] [PubMed] [Google Scholar]
- 51. Impagliazzo A., Ubbink M. (2004) Mapping of the binding site on pseudoazurin in the transient 152 kDa complex with nitrite reductase. J. Am. Chem. Soc. 126, 5658–5659 [DOI] [PubMed] [Google Scholar]
- 52. van de Kamp M., Silvestrini M. C., Brunori M., Van Beeumen J., Hali F. C., Canters G. W. (1990) Involvement of the hydrophobic patch of azurin in the electron-transfer reactions with cytochrome c551 and nitrite reductase. Eur. J. Biochem. 194, 109–118 [DOI] [PubMed] [Google Scholar]
- 53. Murphy L. M., Dodd F. E., Yousafzai F. K., Eady R. R., Hasnain S. S. (2002) Electron donation between copper containing nitrite reductases and cupredoxins: The nature of protein-protein interaction in complex formation. J. Mol. Biol. 315, 859–871 [DOI] [PubMed] [Google Scholar]
- 54. DiSpirito A. A., Taaffe L. R., Hooper A. B. (1985) Localization and concentration of hydroxylamine oxidoreductase and cytochrome c-552, cytochrome c-554, cytochrome c-m522, cytochrome cm-552, and cytochrome a in Nitrosomonas europaea. Biochim. Biophys. Acta 806, 320–330 [Google Scholar]
- 55. Lieberman R. L., Arciero D. M., Hooper A. B., Rosenzweig A. C. (2001) Crystal structure of a novel red copper protein from Nitrosomonas europaea. Biochemistry 40, 5674–5681 [DOI] [PubMed] [Google Scholar]
- 56. Hough M. A., Antonyuk S. V., Strange R. W., Eady R. R., Hasnain S. S. (2008) Crystallography with online optical and x-ray absorption spectroscopies demonstrates an ordered mechanism in copper nitrite reductase. J. Mol. Biol. 378, 353–361 [DOI] [PubMed] [Google Scholar]
- 57. Wijma H. J., Jeuken L. J., Verbeet M. P., Armstrong F. A., Canters G. W. (2006) A random-sequential mechanism for nitrite binding and active site reduction in copper-containing nitrite reductase. J. Biol. Chem. 281, 16340–16346 [DOI] [PubMed] [Google Scholar]
- 58. Goldsmith R. H., Tabares L. C., Kostrz D., Dennison C., Aartsma T. J., Canters G. W., Moerner W. E. (2011) Redox cycling and kinetic analysis of single molecules of solution-phase nitrite reductase. Proc. Natl. Acad. Sci. U.S.A. 108, 17269–17274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Murphy M. E., Turley S., Adman E. T. (1997) Structure of nitrite bound to copper-containing nitrite reductase from Alcaligenes faecalis - mechanistic implications. J. Biol. Chem. 272, 28455–28460 [DOI] [PubMed] [Google Scholar]
- 60. Leferink N. G., Eady R. R., Hasnain S. S., Scrutton N. S. (2012) Laser-flash photolysis indicates that internal electron transfer is triggered by proton uptake by Alcaligenes xylosoxidans copper-dependent nitrite reductase. FEBS J. 279, 2174–2181 [DOI] [PubMed] [Google Scholar]