Background: Transportin-SR2 (TRN-SR2) is a karyopherin implicated in nuclear import of the HIV-1 preintegration complex.
Results: RanGTP can displace HIV-1 integrase and induces large scale structural changes in TRN-SR2.
Conclusion: Structural and functional analysis of TRN-SR2 supports its role in nuclear import.
Significance: Characterization of TRN-SR2 in the nuclear and cytoplasmic states allows further insights into its function during nuclear import.
Keywords: GTPase, HIV-1, Homology Modeling, Host-Pathogen Interactions, Nuclear Transport, Protein-Protein Interactions, X-ray Scattering
Abstract
The human immunodeficiency virus type 1 (HIV-1) and other lentiviruses are capable of infecting non-dividing cells and, therefore, need to be imported into the nucleus before integration into the host cell chromatin. Transportin-SR2 (TRN-SR2, Transportin-3, TNPO3) is a cellular karyopherin implicated in nuclear import of HIV-1. A model in which TRN-SR2 imports the viral preintegration complex into the nucleus is supported by direct interaction between TRN-SR2 and HIV-1 integrase (IN). Residues in the C-terminal domain of HIV-1 IN that mediate binding to TRN-SR2 were recently delineated. As for most nuclear import cargoes, the driving force behind HIV-1 preintegration complex import is likely a gradient of the GDP- and GTP-bound forms of Ran, a small GTPase. In this study we offer biochemical and structural characterization of the interaction between TRN-SR2 and Ran. By size exclusion chromatography we demonstrate stable complex formation of TRN-SR2 and RanGTP in solution. Consistent with the behavior of normal nuclear import cargoes, HIV-1 IN is released from the complex with TRN-SR2 by RanGTP. Although in concentrated solutions TRN-SR2 by itself was predominantly present as a dimer, the TRN-SR2-RanGTP complex was significantly more compact. Further analysis supported a model wherein one monomer of TRN-SR2 is bound to one monomer of RanGTP. Finally, we present a homology model of the TRN-SR2-RanGTP complex that is in excellent agreement with the experimental small angle x-ray scattering data.
Introduction
The human immunodeficiency virus type 1 (HIV-1) and other lentiviruses have the capacity to infect non-dividing cells such as macrophages through an active nuclear import mechanism (1, 2). Nuclear import is particularly important in the pathogenesis of HIV-1 because non-dividing cells are a key reservoir of virus in infected individuals. After viral entry and partial uncoating the reverse transcriptase produces a double-stranded DNA copy of the viral RNA genome. During its migration to the nucleus, the reverse transcription complex is gradually transformed into the preintegration complex (PIC).7 Capsid (CA) proteins remain at least partially associated with the PIC during its journey to the nucleopore (3). Upon arrival at the nuclear membrane, the PIC has to overcome the formidable challenge of crossing the nuclear membrane (1, 2). The nuclear membrane is composed of a double lipid bilayer. The outer and the inner nuclear membrane are joined at nuclear pore complexes (NPCs) that serve as entry gates. It is believed that γ-retroviruses wait for the breakdown of the nuclear membrane to access the chromatin, whereas lentiviruses use cellular import pathways for the transport of the PIC through the nucleopore. Different viral signals have been implicated in HIV-1 nuclear import (DNA-flap, IN, Vpr, Matrix), but no single one is accepted as the dominant nuclear import factor (1, 2). The ability of HIV-1 PICs to cross an intact nuclear envelope during interphase implicates the involvement of active cellular transport machineries. Involvement of the classic importin α/β pathway and importin 7 has been proposed by several authors (4–9), and a role for nuclear pore proteins has been described as well (10–12). Also tRNA was proposed as potential nuclear import factor (13).
Using yeast-two-hybrid, we identified Transportin-SR2 (TRN-SR2, Transportin-3), encoded by the TNPO3 gene, as a cofactor of HIV-1 IN (14). TRN-SR2 was independently discovered as a host factor of HIV replication in two large scale siRNA screens (15, 16). The direct interaction between HIV IN and TRN-SR2 has been confirmed independently (17, 18). The karyopherin TRN-SR2 is known to shuttle essential splicing factors, serine/arginine-rich proteins (SR-proteins), between the nucleus and the cytoplasm and is involved in the regulation of mRNA splicing (19). Recognition of SR-proteins by TRN-SR2 mainly relies on the conserved RS-domain and requires phosphorylation. Alternative cargoes lacking an RS domain have been identified as well, indicating that other interactions are possible (20, 21). Transient (siRNA) as well as stable (shRNA) depletion of TRN-SR2 strongly hampers HIV-1 but not MLV infection. Knockdown of TRN-SR2 in primary macrophages likewise interferes with HIV-1 replication, demonstrating its requirement for productive infection of non-dividing cells. Using quantitative PCR we could pinpoint the block in replication to an event after reverse transcription but before integration and could exclude that TRN-SR2 knockdown affects later steps in the replication cycle. The reduction in the number of two long-terminal repeat circles (14) was confirmed by some (22–24) but not all groups (25–27). In any case the consistent reduction in integration is never accompanied by an increase in two long-terminal repeat circles, suggesting a defect in nuclear import. Interestingly, TRN-SR2 depletion appears to affect integration site selection (28). Using enhanced GFP-labeled IN, a defect in HIV nuclear import upon TRN-SR2 depletion was shown (14).
Some HIV capsid mutations (e.g. N74D CA) reduce the dependence of HIV replication on TRN-SR2 (18, 29). Whether this implies a direct and specific interaction between TRN-SR2 and capsid or capsid core particles remains controversial (29–34). Because many capsid mutations are known to affect uncoating, a plausible explanation for the observed phenotype is the requirement for capsid uncoating before direct interaction between TRN-SR2 and IN can take place (33). The N74D CA mutant was originally selected to overcome restriction by the C-terminally truncated fragment of cleavage and polyadenylation specificity factor subunit 6 (CPSF) (29). Full-length CPSF6, a cellular protein involved in splicing, contains an RS domain at its C terminus. Because CPSF6 binds TRN-SR2 and interacts with CA through its N-terminal domain, TRN-SR2 depletion may lead to cytoplasmic accumulation of CPSF6 that in turn may restrict HIV replication at the uncoating step (24, 35). In contrast, spreading replication of N74D CA HIV (as compared with single round transduction) remained highly sensitive to TRN-SR2 depletion (33), suggesting that CPSF6 accumulation does not explain the full phenotype of TRN-SR2 depletion.
Both a peptide-based approach and mass spectrometry-based protein footprinting revealed hot spots for the interaction with TRN-SR2 in the C terminus of HIV-1 IN (30, 36). The cargo domain of TRN-SR2 is required for nuclear import of HIV (22).
TRN-SR2 is a 923-amino acid protein consisting entirely of stacked HEAT repeats (two antiparallel α-helices connected by a small turn linker) (37). These create a curving structure with a high degree of flexibility, allowing binding to different types of cargo and regulatory proteins (38). Importins bind their cargo in the cytoplasm either directly or through the adaptor importin α (39). After docking at the NPC on the cytoplasmic side, the importin-cargo complex moves through the nucleopore channel via its interactions with nucleoporins (Nups) (40). RanGTP binding induces conformational changes in importins leading to the release of the cargo in the nucleus (41). The complex of the importin and RanGTP moves back through the NPC to the cytoplasm where GTP is hydrolyzed, and the import factor is available for a new round of nuclear transport (39). The 25-kDa Ran (Ras-related nuclear protein), a small GTPase, is a key modulator of protein interactions of importins and the motor behind nuclear transport (42). The direction of nuclear transport (import/export) is controlled by its gradient (GTP/GDP-bound forms) across the NPC. RanGTP is enriched in the nucleoplasm, and RanGDP is enriched in the cytoplasm. The nature of the bound nucleotide (GTP or GDP) modulates the interaction between Ran and importins (38, 39).
Because nuclear import is generally believed to be a bottleneck during HIV infection and precedes the integration of the proviral DNA into the host genome, the interaction of IN and TRN-SR2 holds promise as a potential target for anti-HIV therapy. Clear understanding and structural analysis of the interaction of TRN-SR2 with various cargoes is essential before efficient drug development. Here we present biochemical and structural biology studies on the interactions of TRN-SR2 with RanGTP.
EXPERIMENTAL PROCEDURES
Purification of His9-TRN-SR2
Cultures of Escherichia coli strain Rosetta (DE3) transformed with pET19b-TRN-SR2 in LB medium were induced with 0.5 mm isopropyl β-d-thiogalactoside at A600 nm ∼ 0.6 and incubated 6 h at 30 °C. The cultures were harvested by centrifugation for 15 min at 4000 rpm and 4 °C, and pellets were washed with STE buffer (100 mm NaCl, 10 mm Tris-HCl, pH 7.3, 0.1 mm EDTA), centrifuged again, and then stored at −20 °C until purification. The frozen cultures were thawed and resuspended in lysis buffer (50 mm Tris-HCl, pH 7.3, 0.5 m NaCl, protease inhibitors (Complete, EDTA-free, Roche Applied Science), 2 units DNase/10 ml, 5 mm dithiothreitol (DTT). Cells were lysed by a French press and sonication on ice, and the lysate was centrifuged at 15,000 rpm at 4 °C for 15 min. The soluble lysate containing His9-fused protein was loaded onto a Ni2+-affinity column equilibrated with binding buffer (20 mm Tris-HCl, pH 7.5, 250 mm NaCl, 5 mm DTT) using an AKTA purifier system. The protein was eluted with a linear gradient of imidazole (0–1 m), and 1-ml fractions were collected. Fractions with a peak absorption at 280 nm were pooled, concentrated by centrifugal concentrators (Vivaspin 6 50,000 MWCO PES, Sartorius Stedim Biotech), and dialyzed overnight against buffer A (20 mm Tris-HCl, pH 7.5, 5 mm DTT) before ion exchange chromatography. A gradient from 50 mm to 1 m NaCl in buffer A was run using a HiTrap Q HP 5- ml column. The 1-ml fractions containing His9-TRN-SR2 protein were pooled and concentrated with Vivaspin. The last step in the purification procedure was size-exclusion chromatography (SEC) using a HiLoad 16/60 Superdex 200 prep grade column. SEC was performed in 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT. The pooled fractions with His9-TRN-SR2 were stored at 4 °C.
Purification of GST-TRN-SR2
Cultures of E. coli strain BL21 transformed with pGEX-6P2-TRN-SR2 in LB medium substituted with 1 m d-sorbitol and 2.5 mm trimethylglycine were induced with 0.5 mm isopropyl β-d-thiogalactoside at A600 nm ∼ 0.6 and incubated overnight at 28 °C. The cultures were harvested by centrifugation for 10 min at 4000 rpm at 4 °C, and pellets were washed with STE buffer, centrifuged again, and stored at −20 °C until purification. The frozen cultures were thawed, resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, protease inhibitors, 2 units DNase/10 ml, 5 mm DTT), and lysed by a French press, and the lysate was centrifuged at 15,000 rpm at 4 °C for 30 min. The soluble lysate containing GST-fused protein was loaded onto a glutathione-Sepharose column equilibrated with binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT). The protein was eluted with the elution buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT, 20 mm reduced glutathione), and 1-ml fractions were collected. Fractions exhibiting a peak absorption at 280 nm were pooled and dialyzed overnight against 100× volume of dialysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT, 10% glycerol) and stored at −80 °C.
Purification of GST-ASF/SF2
Cultures of E. coli strain BL21 transformed with pGEX-2TK-ASF/SF2 in LB medium were induced with 0.5 mm isopropyl β-d-thiogalactoside at A600 nm ∼ 0.6 and incubated for 6 h at 25 °C. The cultures were harvested by centrifugation for 10 min at 4000 rpm at 4 °C, and pellets were washed with STE buffer, centrifuged again, and stored at −20 °C until purification. The frozen cultures were thawed, resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm PMSF, 2 units DNase/10 ml, 10 μg/ml RNase A, 5 mm DTT), and lysed by sonication, and the lysate was centrifuged at 15,000 rpm at 4 °C for 30 min. The soluble lysate containing GST-fused protein was loaded onto a glutathione-Sepharose column equilibrated with binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT). The protein was eluted with the elution buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT, 20 mm reduced glutathione), and 1-ml fractions were collected. Fractions exhibiting a peak absorption at 280 nm were pooled and dialyzed overnight against 100× volume of dialysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT, 10% glycerol) and stored at −80 °C.
Purification of His6-HIV-1 IN
N-terminally His6-tagged HIV-1 IN was purified as described previously (43).
Purification of His9-Ran and Ran
Cultures of E. coli strain BL21 (DE3) transformed with pET-3d-His9-HRV3C-RanQ69L in LB medium were induced with 0.5 mm isopropyl β-d-thiogalactoside at A600 nm ∼ 0.6 and incubated overnight at 28 °C. The cultures were harvested by centrifugation for 10 min at 6000 rpm and 4 °C, and pellets were washed with STE buffer, centrifuged again, and then stored at −20 °C until purification. The frozen cultures were thawed, resuspended in lysis buffer (20 mm Tris-HCl, pH 7.5, 0.25 m NaCl, 1 mm PMSF, 20 mm imidazole, 4 mm MgCl2, 2 units DNase/10 ml, 5 mm DTT), and lysed by sonication on ice, and the lysate was centrifuged at 15,000 rpm and 4 °C for 30 min. The soluble lysate containing His9-fused protein was loaded onto nickel column equilibrated with binding buffer (20 mm Tris-HCl, pH 7.5, 250 mm NaCl, 50 mm imidazole, 4 mm MgCl2, 5 mm DTT). The protein was eluted with elution buffer (20 mm Tris-HCl, pH 7.5, 250 mm NaCl, 250 mm imidazole, 4 mm MgCl2, 5 mm DTT), and 1 ml fractions were collected. Fractions exhibiting peak absorption at 280 nm were pooled and dialyzed overnight against 100× volume of dialysis buffer (20 mm Tris-HCl, pH 7.5, 250 mm NaCl, 4 mm MgCl2, 5 mm DTT, 10% glycerol) or used to cleave off the His tag. The dialyzed protein was stored at −80 or −20 °C. Cleavage of the His tag was performed during dialysis using GST-tagged Human Rhinovirus 3C Protease (1 unit for each 50 μg of target protein) for 12 h. The added protease was removed again by affinity purification over GSH-Sepharose as described by the manufacturer.
Nucleotide Loading of Ran
To load recombinant Ran with nucleotides (GDP or GTP), maximally 100 μm Ran was incubated for 30 min at 30 °C with 1 mm GDP or GTP and 20 mm EDTA. After 30 min, the reaction was stopped by adding 50 mm MgCl2. The buffer was exchanged on a PD10 desalting column to the Ran dialysis buffer (20 mm Tris-HCl, pH 7.5, 250 mm NaCl, 4 mm MgCl2, 5 mm DTT, 10% glycerol). The peak fractions were pooled and used immediately or stored for later use at −80 °C.
Complex Formation and Analytical Size Exclusion Chromatography
For complex formation, His-TRN-SR2 and RanGTP/GDP were mixed in a 1:3 molar ratio and incubated on ice for at least 1 h. SEC runs were performed on a Superdex 200 10/300 GL column attached to an AKTA purifier system (GE Healthcare) at a flow rate of 0.5 ml/min at 4 °C in SEC buffer containing 10 mm Tris-HCl, pH 7.3, 150 mm NaCl, 5 mm MgCl2, and 5 mm DTT. Proteins were detected by absorbance at 280 nm. The column was calibrated with the following proteins: ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa), and aprotinin (6.5 kDa) (Gel Filtration Calibration kits HMW & LMW; GE Healthcare). The fractions were analyzed by SDS-PAGE and silver-stained following the manufacturer's instructions.
AlphaScreen Interaction Assays
The AlphaScreen assays (Amplified Luminescent Proximity Homogeneous Assay, ALPHA; PerkinElmer Life Sciences) were optimized for use in 384-well OptiPlate microplates (PerkinElmer Life Sciences) with a final volume of 25 μl. Recombinant proteins (GST- or His9-tagged TRN-SR2, GST-ASF/SF2, His6-tagged or untagged Ran charged with GDP or GTP and His6-IN) were diluted to 5× working solutions in assay buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm DTT, 1 mm MgCl2, 0.1% (v/v) Tween 20, 0.1% (w/v) bovine serum albumin). First, 5 μl of buffer or diluted RanGTP was pipetted into the wells followed by 5 μl of each of the interacting protein dilutions. The plate was sealed and left to incubate for 1 h at 4 °C, allowing an equilibrium to be established. Next, 10 μl of a mix of Ni2+ chelate acceptor and glutathione donor AlphaScreen beads (PerkinElmer Life Sciences) was added, bringing the total volume to 25 μl and establishing final concentrations of 10 μg/ml for each of the beads. The plate was then placed at room temperature and incubated for one more hour before being read in an EnVision Multilabel Reader (PerkinElmer Life Sciences). Background AlphaScreen counts were subtracted, and data were analyzed in Prism 5.0 (GraphPad).
The cargo dissociation experiments were performed with final concentrations of 10 nm GST-TRN-SR2 or His9-TRN-SR2 and 40 nm His6-IN or 40 nm GST-ASF/SF2, respectively. All titrations (RanGDP/GDP) were performed against 10 nm GST-TRN-SR2.
Small Angle X-ray Scattering (SAXS) Measurements
SAXS data were collected using synchrotron radiation at the European Molecular Biology Laboratory X33 beamline of the DORIS III storage ring (DESY, Hamburg, Germany). SAXS curves were measured over the range of momentum transfer 0.006 < q = 4π sin(θ)/λ < 0.63 Å−1, where 2θ is the scattering angle, and λ = 1.5 Å is the x-ray wavelength. Protein samples were in 20 mm Tris-HCl buffer, pH 7.3, 150 mm NaCl, 5 mm DTT, and 5 mm MgCl2. The ATSAS program package (44, 45) was used for data processing. Guinier plots were used to evaluate the zero angle scattering (I0) and the radius of gyration (Rg). The particle distance distributions P(r) were calculated using GNOM (46); these distributions were used to estimate the maximal particle dimension (Dmax). Molecular weights were estimated from SAXS data for q < 0.3 Å−1 using SAXSMoW (47). The fits between the experimental scattering curves and the theoretical scattering from atomic models were calculated using CRYSOL (48).
Dynamic Light Scattering
Measurements were made in small droplets with the SpectroSize 300 instrument at the scattering angle of 150° and at 20 °C. As with the SAXS measurements, protein samples were in 20 mm Tris-HCl buffer, pH 7.3, 150 mm NaCl, 5 mm DTT, and 5 mm MgCl2.
Differential Scanning Fluorimetry
His9-TRN-SR2 WT or the E145Q,V149A,E152Q,E153Q mutant at 1 μm final concentration was mixed with 1× SYPRO Red dye (Invitrogen), and the corresponding dilution of Ran was loaded with either GDP or GTP (3-fold dilution series from 3 to 0.33 μm). Mixtures were left for 15 min at room temperature before 25 μl was transferred to two wells of a 96-well plate (Bio-Rad). The plate was sealed with optical flat 8-cap strips (Bio-Rad), and differential scanning fluorimetry (DSF) melting curves were obtained on a Bio-Rad iCycler equipped with an iQ5 real-time PCR detection system. The raw fluorescence data were analyzed with Excel (Microsoft), whereas Prism 5.0 (GraphPad) was used to fit the transitions with a Boltzmann sigmoidal equation and extract melting temperatures.
Homology Modeling
The TRN-SR2 amino acid sequence was retrieved from the UniProt Knowledge base (identifier: Q9Y5L0–2) and used as a query on the HHpred server (49). The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) was hence searched with the profile Hidden Markov Model-Hidden Markov Model (HMM-HMM) comparison through HHblits (8 iterations maximum, secondary structure scoring on), and the resulting query-template alignments were realigned with the Maximum ACcuracy alignment (MAC) algorithm. The best templates were selected, and a multiple alignment was generated. These alignments pointed to Importin-13 (Imp13) as the highest quality template for homology modeling of TRN-SR2 (E values of 0 for different HMMs). The relatively low sequence homology (23% identical, similarity around 38%), however, necessitates careful validation of the final model quality. The optimal alignment to Imp13, corresponding to a maximal TM-score of 0.9194, was subsequently used for model building in MODELLER 9.8 (50). Three crystal structures of Imp13 are available in the PDB, 2XWU, 2X19, and 2X1G, which represent two complexes of human Imp13 with UBC9 or RanGTP and one of Drosophila Imp13 with Mago and Y14, respectively. For the RanGTP-bound model of TRN-SR2, structure 2X19 was used as a template, and the RanGTP structure (Saccharomyces cerevisiae) from 2X19 was set as an environment for induced fit after it was modified to its human counterpart.
RESULTS
Functionality of the TRN-SR2-RanGTP Interaction
We first studied the direct interaction between TRN-SR2 and Ran loaded with either GDP or GTP (RanGDP or RanGTP, respectively). Ran is a crucial regulator of nuclear transport, and the regulation is executed through conformational changes in the Ran switch I and switch II loops depending on whether guanosine di- or triphosphate is bound (51, 52), altering the affinity for different binding partners. To ensure that GTP is not hydrolyzed to GDP during our experiments, a GTPase-deficient Ran mutant, RanQ69L (henceforth simply referred as Ran), was used that is characterized by a severely reduced hydrolysis of GTP to GDP (53). As expected, RanGDP and RanGTP bound TRN-SR2 to a different extent, as evidenced by AlphaScreen (Fig. 1). We determined an apparent KD = 4.7 ± 1.2 nm for the TRN-SR2-RanGTP interaction, whereas no binding to RanGDP could be detected under tested conditions.
FIGURE 1.
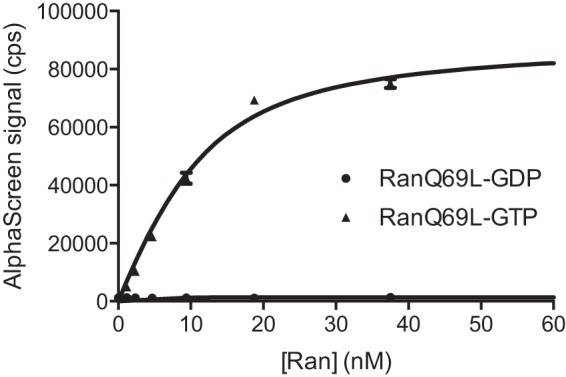
TRN-SR2 specifically interacts with RanGTP. GTPase-defective His6-RanQ69L was loaded with GDP (●) or GTP (▴) and titrated against 10 nm GST-TRN-SR in AlpaScreen. Data shown are the averages ± S.D. for triplicate measurements.
Using DSF, we provided further support for this specific interaction and assessed the thermostability of the established complex. The addition of RanGDP to TRN-SR2 did not significantly affect the observed melting temperature of TRN-SR2 (supplemental Fig. S1A). In contrast, the addition of increasing amounts of RanGTP markedly increased the melting temperature. In the presence of a 3× molar excess of RanGTP, the melting temperature of TRN-SR2 was 56.6 °C compared with 45.7 °C for TRN-SR2 alone (supplemental Fig. S1B). These data clearly point to the formation of a stable TRN-SR2-RanGTP complex in solution.
Structural analysis of proteins in vitro gains in relevance if the purified recombinant proteins are able to perform their physiological reactions. After evaluating the interaction with RanGTP, we determined whether recombinant TRN-SR2 binds its natural cellular cargo and, more importantly, whether cargo can be released from the complex upon the addition of RanGTP. 20 nm His9-TRN-SR2 (henceforth referred to as TRN-SR2) was incubated with 80 nm GST-tagged ASF/SF2 (SRSF1), one of the known cellular cargoes of TRN-SR2 (19) or 80 nm His6-HIV-1 IN and increasing amounts of RanGTP. When the TRN-SR2-ASF/SF2 interaction was probed with AlphaScreen, a clear RanGTP-dependent dissociation could be observed, demonstrating the functionality of the recombinant protein (Fig. 2A). Similarly, with HIV-1 IN, the viral cargo was efficiently released from TRN-SR2 by increasing the concentrations of RanGTP (Fig. 2B). Indirectly this finding indicates that the TRN-SR2 basic functionality is likely independent of eukaryotic post-translational modifications.
FIGURE 2.
Cargo dissociation from TRN-SR2. The cellular cargo GST-ASF/SF2 was dissociated from 10 nm His9-TRN-SR2 (A), and HIV-1 His6-IN was dissociated from 10 nm GST-TRN-SR2 (B) by adding increasing amounts of RanGTP, effectively inhibiting the AlphaScreen signal. Data shown are the averages ± S.D. for triplicate measurements in two experiments.
TRN-SR2 and Its Complex with Ran as Studied by Analytical SEC
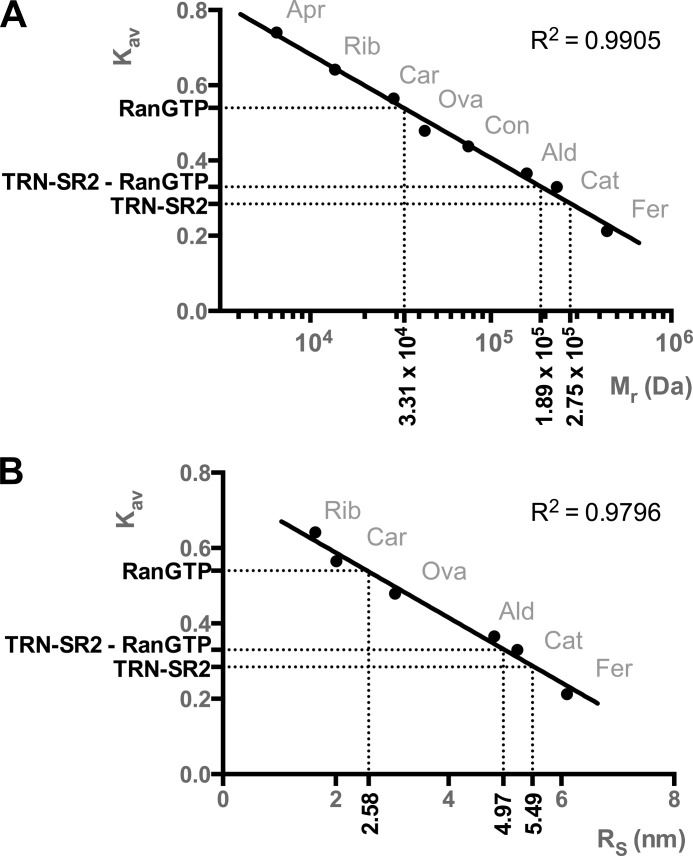
We applied analytical SEC to investigate the behavior of TRN-SR2 and its complexes in solution. Fig. 3A displays chromatograms from four independent runs for TRN-SR2, TRN-SR2 with RanGDP or RanGTP, and RanGDP alone. Chromatograms from analytical SEC and the corresponding SDS-PAGE (Fig. 3) confirm that TRN-SR2 and RanGTP establish a stable complex. In stark contrast, there is almost no complex formation detected after mixing TRN-SR2 with RanGDP (Fig. 3A). The silver-stained gels of equally loaded single fractions from the SEC runs (Fig. 3B) confirmed the higher affinity of TRN-SR2 for RanGTP than for RanGDP. Note the more pronounced band for unbound RanGDP in comparison with unbound RanGTP (Fig. 3B). Additionally, there are remarkable changes; TRN-SR2 in complex with RanGTP has a larger peak elution volume than free TRN-SR2 (Ve = 13.4 ml versus 12.7 ml, respectively), translating into a higher gel phase distribution coefficient (Kav) and hence a smaller apparent Stokes radius (RS = 4.97 nm versus 5.49 nm, respectively), which is unexpected for a larger protein complex (Figs. 3A and 4).
FIGURE 3.
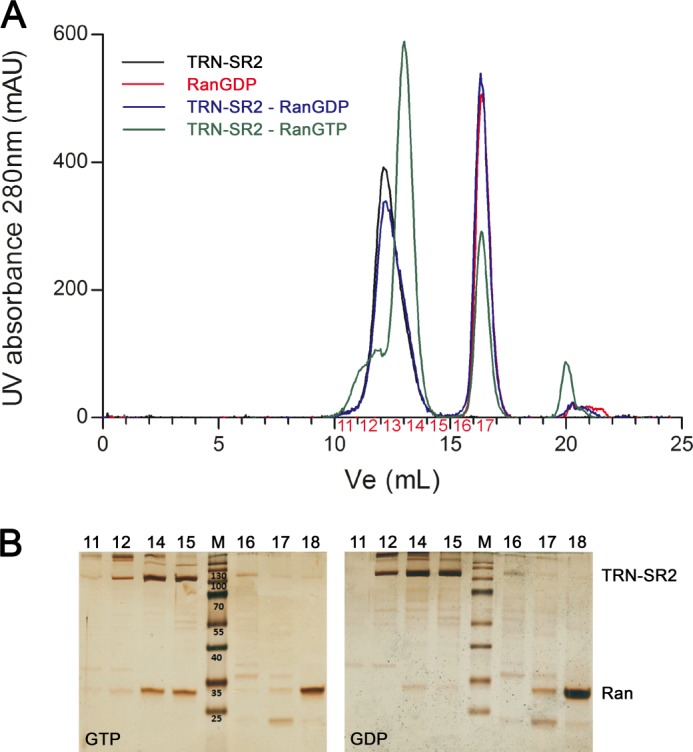
Impact of the Ran nucleotide state on size exclusion chromatography mobility of TRN-SR2. SEC of TRN-SR2 was carried out with Ran loaded either by GTP or GDP. A, shown are analytical SEC chromatograms TRN-SR2 (black), TRN-SR2 + RanGDP (blue), TRN-SR2 + RanGTP (green), or RanGDP (red). All runs were performed on a Superdex 200 10/300 GL column connected to an AKTA Purifier FPLC system. mAU, millabsorbance units. B, shown is a silver-stained SDS-PAGE of the fractions obtained from SEC runs of the complexes.
FIGURE 4.
Relative molecular mass (Mr) and Stokes radii (RS) as determined by size exclusion chromatography. Linear regression was based on calibration with ferritin (Fer, 440 kDa), catalase (Cat, 232 kDa), aldolase (Ald, 158 kDa), conalbumin (Con, 75 kDa), ovalbumin (Ova, 43 kDa), carbonic anhydrase (Car, 29 kDa), ribonuclease A (Rib, 13.7 kDa), and aprotinin (Apr, 6.5 kDa). Gel phase distribution coefficients (Kav) were calculated from the respective elution volumes (Ve) and plotted versus Mr or RS for the standard proteins. Unknown Mr and RS values for RanGTP, TRN-SR2, and the TRN-SR2-RanGTP complex were then interpolated from the calibration curves, yielding Mr values of 33.1 kDa, 275 kDa, and 189 kDa and RS values of 2.58, 5.49, and 4.97 nm respectively.
TRN-SR2 and Its Complex with Ran Characterized by DLS and SAXS
We next used DLS to obtain further size characteristics of the proteins. We measured samples of TRN-SR2 alone and the TRN-SR2-RanGTP complex at varying total protein concentrations (2–16 mg/ml). All samples were shown to be monodisperse. DLS data allowed determination of radii of hydration (Rh) of 3.92 ± 0.17 and 4.78 ± 0.10 nm for the TRN-SR2-RanGTP complex and TRN-SR2 alone, respectively (Table 1 and supplemental Fig. S2).
TABLE 1.
Solution species parameters derived from dynamic light scattering
Rh, hydrodynamic radius.
| Sample | Rh |
|---|---|
| nm | |
| TRN-SR2 | 4.78 ± 0.10 |
| TRN-SR2-RanGTP | 3.92 ± 0.17 |
Subsequently, we performed SAXS to characterize the solution structure of TRN-SR2 and its complex with RanGTP. The measurements were done on samples with varying total protein concentrations (2–16 mg/ml) (Fig. 5A). The SAXS data permitted the calculation of the radius of gyration (Rg), the maximum dimension (Dmax) (Table 2), and pair-distance distributions P(r) for each sample (Fig. 5B). All studied solutions of both TRN-SR2 alone and the complex yielded scattering with linearity in the Guinier region, indicating no considerable aggregation. All samples were monodisperse, with little dependence of the normalized SAXS curve on protein concentration (Table 2). For free TRN-SR2 at 8 mg/ml, the measured Rg was 4.5 nm, and the maximum particle dimension Dmax was 15.5 nm, which is close to the values reported earlier for TRN-SR2 dimers (30). Moreover, the apparent molecular mass estimated from the SAXS data at this concentration is 202 kDa, which is a close match to the theoretical mass of the dimer (214 kDa). Interestingly, comparison of the intraparticle distance distributions for TRN-SR2 alone and for the TRN-SR2-RanGTP complex (Fig. 5B) clearly suggests a smaller particle in the latter case. For the complex at 8.1 mg/ml, the Rg was 3.6 nm, and Dmax was 10.7 nm (Table 2). The apparent molecular mass for the complex was also clearly smaller (131 kDa) than for TRN-SR2 alone. The fact that the TRN-SR2-RanGTP complex in solution is smaller than TRN-SR2 alone is also evident from comparing the lowest q parts of the scaled scattering curves (Fig. 5A), which clearly point to smaller extrapolated zero angle scattering (I0) for the complex. Importantly, the measured mass of 131 kDa is very close to the expected mass of a complex containing one TRN-SR2 molecule and one RanGTP molecule (a total of 132 kDa). The values for Rg correspond well to the hydrodynamic radii (Rh) (Table 1) as obtained in the DLS experiments, considering the most ordered first solvation shell to be 0.3 nm thick (54).
FIGURE 5.
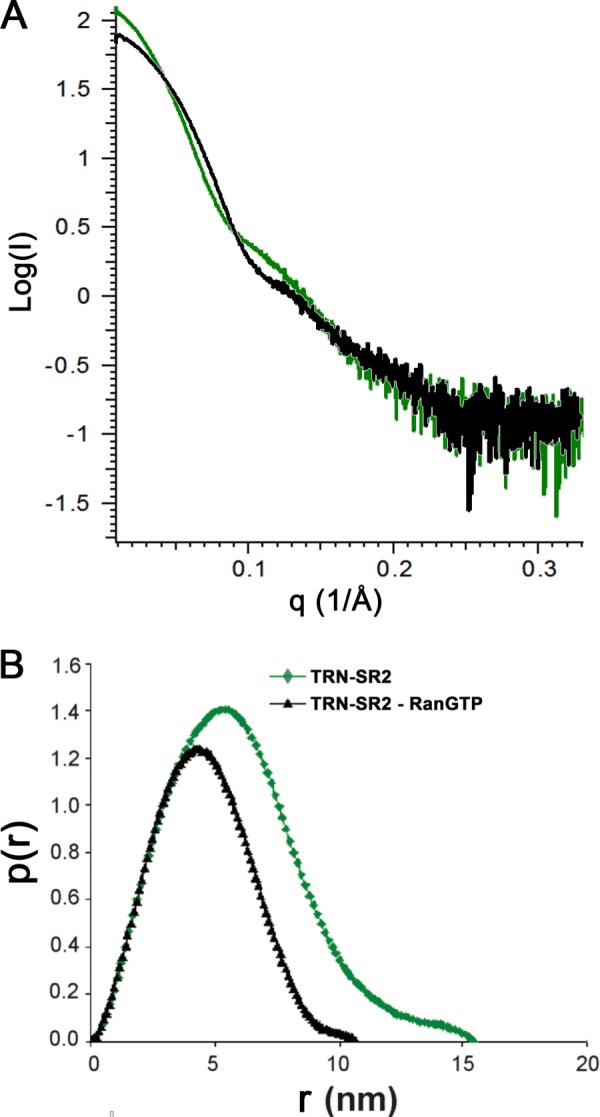
SAXS analysis of TRN-SR2-RanGTP complex. A, shown are experimental SAXS curves for TRN-SR2 alone (green) and for its complex with RanGTP (black). B, shown are SAXS-derived distance distributions P(r) for TRN-SR2 alone (green) and for the TRN-SR2-RanGTP complex (black); r represents the distances between all pairs of points within the particle, weighted by the respective electron densities.
TABLE 2.
Solution species parameters derived from small-angle x-ray scattering
Rh, hydrodynamic radius; I0, extrapolated scattering intensity at zero angle; Rg, radius of gyration; Dmax, maximal dimension; Mr, relative molecular mass. a.u., arbitrary units.
| Sample | Concentration | SAXS |
|||
|---|---|---|---|---|---|
| I0 | Rg | Dmax | Mr | ||
| mg/ml | a.u. | nm | nm | kDa | |
| TRN-SR2 | 1.8 | 118.4 | 4.37 | 15.3 | 181.0 |
| 4 | 115.2 | 4.37 | 15.3 | 200.5 | |
| 8 | 119.4 | 4.51 | 15.5 | 202.3 | |
| 16.3 | 133.0 | 4.86 | 17.2 | 219.8 | |
| TRN-SR2-RanGTP | 2.1 | 81.2 | 3.60 | 11.3 | 144.9 |
| 4 | 85.7 | 3.66 | 11.2 | 140.8 | |
| 8.1 | 80.7 | 3.61 | 10.7 | 130.5 | |
| 15.8 | 88.1 | 3.65 | 10.3 | 134.3 | |
Homology Model of the TRN-SR2-RanGTP Complex
At the moment there is no high resolution structure reported for the TRN-SR2 protein described. However, the structure of its closest paralogue, Imp13, has been solved. In 2010 Bono et al. (55) reported the structures of the complexes of human Imp13 with the S. cerevisiae Ran homologue, GTP binding nuclear protein GSP1/CNR1, and of the Drosophila melanogaster Imp13 (Cadmus) with Mago and Y14 homologues (Protein mago nashi and RNA-binding protein 8A, respectively, in fruit fly). Later the structure of the complex of human Imp13 with sumo-conjugating enzyme Ubc9 was published (56). Since the original duplication event, TRN-SR2 and Imp13 have diverged significantly, resulting in a present day low sequence identity. Nonetheless, we could produce alignments of human TRN-SR2 to both human and Drosophila Imp13 using HHPred, the most optimal of which is shown in supplemental Fig. S3 (22% sequence identity, probabilities = 100% and E values = 0) (49). This alignment was used to guide modeling of the human TRN-SR2-RanGTP complex (Fig. 6A).
FIGURE 6.
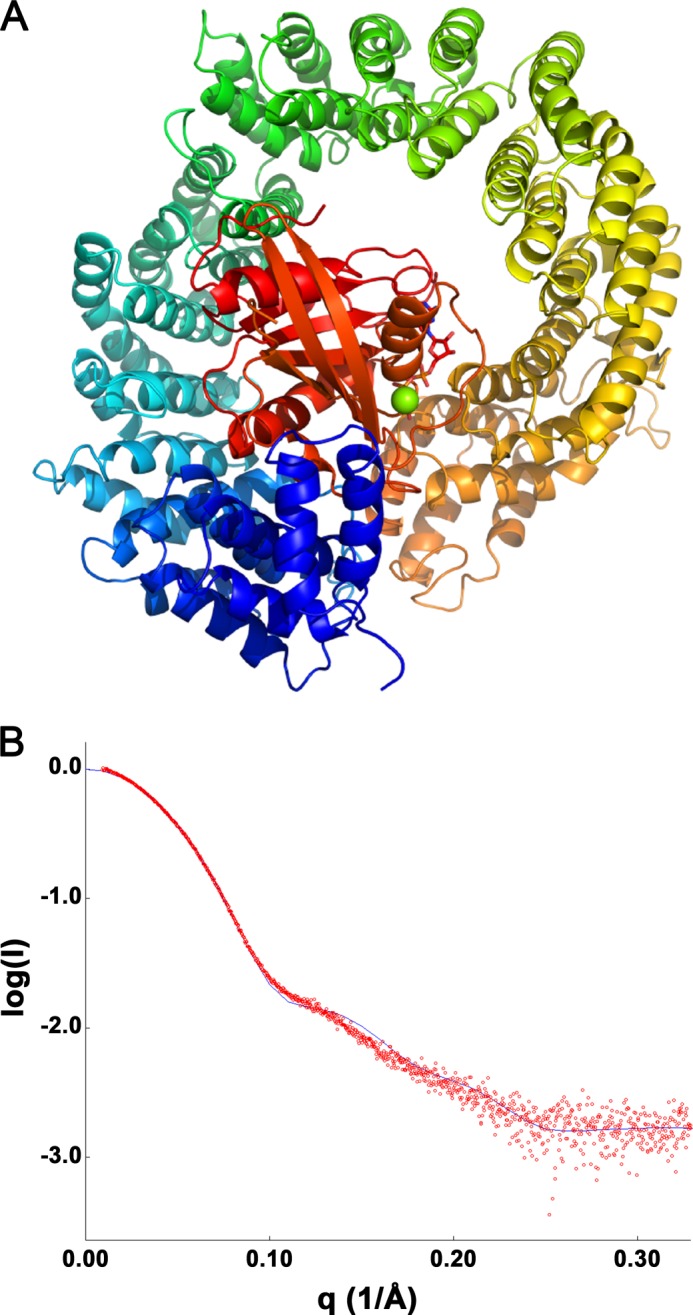
Homology model for TRN-SR2-RanGTP corroborated by experimental SAXS data. A, shown is a homology model of the TRN-SR2-RanGTP complex. B, shown is the experimental SAXS curve for the complex at 8.1 mg/ml (red circles) overlaid with the theoretical scattering (blue line) calculated from the homology model. The goodness of fit is χ = 1.846.
Importantly, we have calculated a theoretical SAXS curve from the obtained homology model of the TRN-SR2-RanGTP complex and compared it to the experimental data. A good match was obtained (goodness of fit χ = 1.846, Fig. 6B) which confirms that in solution TRN-SR2 and RanGTP indeed form a 1:1 complex. For further validation, we have introduced E145Q,V149A,E152Q,E153Q mutations into TRN-SR2 (referred to as TRN-SR2EVEE), which the modeling predicts to be at the interaction interface with RanGTP (supplemental Fig. S4A). Indeed, although DSF showed that the stability of free TRN-SR2 was unaffected by the substitutions (Tm = 45.7 ± 0.5 °C and 47.1 ± 0.9 °C for TRN-SR2WT and TRN-SR2EVEE, respectively), the temperature shift upon the addition of 3 μm RanGTP was significantly smaller for TRN-SR2EVEE compared with TRN-SR2WT (ΔT = 5.8 ± 0.9 °C versus 11.8 ± 0.3 °C, respectively, supplemental Fig. S4B). The smaller shift indicates that a complex can still be formed between RanGTP and TRN-SR2EVEE, likely due to the multiple interfaces between RanGTP and TRN-SR2 (see below), but that the affinity is significantly reduced. Together, these results support the accuracy of the proposed model despite the relatively low sequence identity of the template used (below 30%, the so-called “twilight zone”).
Detailed Interactions between TRN-SR2 and RanGTP in the Complex
TRN-SR2 is an all-helical protein (Fig. 6A and supplemental Fig. S3), as can be expected for a member of the karyopherin-β family. In the case of TRN-SR2 these helices are arranged into 19 typical HEAT repeats with A (inside) and B (outside) helices and one C-terminal capping 3-helix HEAT repeat (A, B, and C helices). On a larger scale, the HEAT repeats stack on top of one another into a flexible toroid shape. When binding to RanGTP, the TRN-SR2 toroid wraps around the smaller Ran protein, most likely compressing its toroid shape and bringing the N and C terminus closer together (Fig. 6A). Despite contacts with the TRN-SR2 middle and C-terminal parts (see below), RanGTP mainly occupies the inside of the N-terminal half of the toroid. This N-terminal RanGTP binding is a general observation for members of the karyopherin family, which is reflected in a higher degree of sequence conservation in this region (supplemental Fig. S3).
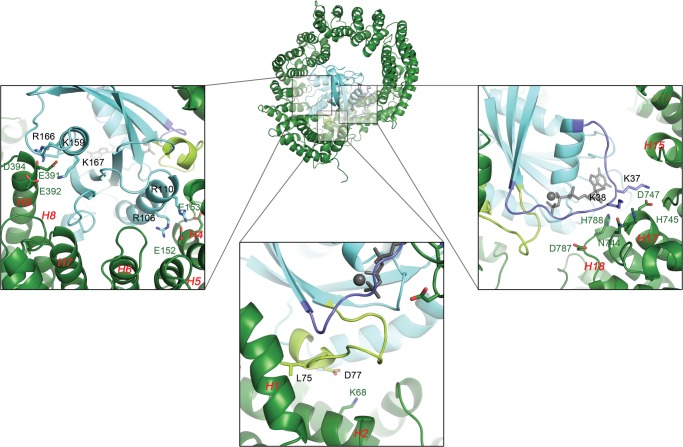
The small GTPase Ran has been extensively studied and characterized (57–59). Upon exchanging GDP for GTP or vice versa, two stretches of this protein undergo a conformational change, adequately called the switch I and II regions (amino acids Thr-32–Val-45 and Thr-66–Tyr-80 respectively) (60). These switches are responsible for specific recognition of Ran in its GTP-bound state by importins and hence for import cargo displacement in the nucleus (59, 61). In our model, like with Imp13 and exportin Crm1, switch I establishes interactions with HEAT repeats 16 to 19 (Fig. 7, right zoom). Notably, two lysines on RanGTP (Lys-37 and Lys-38) are predicted to engage in electrostatic interactions with residues Asn-744, His-745, and Asp-747 from HEAT repeat 17 and Asp-787 and His-788 from repeat 18. As evident from the alignment (supplemental Fig. S3), these interactions are largely conserved between TRN-SR2 and Imp13 (TRN-SR2 Asp-787 can potentially assume the role of Imp13 Glu-830 here). The Ran switch II region also shows electrostatic complementarity (Fig. 7, bottom zoom), as a salt bridge is formed between Lys-68 on the TRN-SR2 HEAT 2 B-helix and Asp-77 on Ran. Meanwhile, the hydrophobic Leu-74 is buried in a superficial groove lined by conserved Leu-18, Tyr-19, Leu-34, Gln-38, and Phe-62 side chains and located between HEAT 1 and 2 B-helices (Fig. 7, bottom, supplemental Fig. S3). The area next to switch II on RanGTP is also in close contact and further contributes to electrostatic interactions with the TRN-SR2 N terminus; two Ran arginine residues (Arg-106, Arg-110) balance the charge of two conserved glutamic acids (Glu-152, Glu-153) located on the TRN-SR2 HEAT 4 B-helix (Fig. 7, left zoom, supplemental Fig. S3). A final interaction site was formed in the middle of TRN-SR2 involving an acidic stretch encompassing Glu-391, Glu-392, and Asp-394 on HEAT 9 and its basic counterpart on Ran involving residues Lys-159, Arg-166, and Lys-167 (Fig. 7 left, supplemental Fig. S3). This central interaction is found in other karyopherins as well, albeit with small differences concerning the exact position of the acidic stretch (56). Overall, mainly electrostatic interactions seem to steer complex formation between TRN-SR2 and RanGTP.
FIGURE 7.
Interactions between RanGTP and TRN-SR2 predicted by homology modeling. The central image shows a graphic representation of the TRN-SR2-RanGTP complex. The lower panels zoom in on the three main interaction sites between both proteins. Main interacting amino acids are rendered as sticks and are labeled in the color of the corresponding protein/region. HEAT repeats are labeled in red, and the Ran switch I and II regions (amino acids 32–45 and 66–80) are colored purple and pale green, respectively. On the left the interaction interface in the middle of TRN-SR2 is displayed. An acidic stretch in the TRN-SR2 HEAT 9 repeat containing Glu-391, Glu-392, and Asp-394 allows formation of salt bridges with RanGTP residues Lys-159, Arg-166, and Lys-167. Similarly, two arginines from RanGTP (Arg-106, Arg-110) balance the charges of two glutamic acid residues (Glu-152, Glu-153) in the HEAT 4 B-helix. These last two residues lie next to the interface TRN-SR2 makes with the Ran switch II region, depicted in the middle panel. Here another salt bridge is established between the HEAT 2 B-helix Lys-68 and a Asp-77 counterpart in the switch. Additionally, Leu-74 from switch II inserts its hydrophobic side chain into a shallow pocket formed between the HEAT 1 and 2 B-helices. On the right is the interface of the RanGTP switch I region with HEAT repeats 16–19. Main interactions are again polar in nature as residues Lys-37 and Lys-38 from the switch reach out to a patch containing TRN-SR2 residues Asn-744, His-745, and Asp-747 on HEAT 17 and Asp-787 and His-788 on HEAT 18.
DISCUSSION
Although TRN-SR2 is generally accepted to act as a cofactor for HIV replication, its exact role remains unclear. According to one of the (non-exclusive) hypotheses, HIV nuclear import is mediated by the direct interaction between TRN-SR2 and HIV integrase present in the PIC (14). All karyopherin-mediated nuclear import is directed by a gradient of RanGTP/GDP (41). As a step in the elucidation of the nuclear import mechanism of HIV, here we studied the biochemical and structural aspects of the interaction between TRN-SR2 and RanGTP.
First we demonstrated in AlphaScreen that Ran binds TRN-SR2 in its GTP-loaded form only (Fig. 1). This result was confirmed with DSF, which showed increased thermostability upon binding to RanGTP, also pointing to stable complex formation (supplemental Fig. S1). RanGTP is further able to displace recombinant HIV integrase from TRN-SR2, much alike its well known cellular cargo ASF-SF2 (Fig. 2). This result is consistent with the model in which nuclear import is mediated by the direct interaction between TRN-SR2 and HIV IN. According to this model RanGTP will dissociate the complex after its arrival in the nucleus.
Next, we performed SEC (Figs. 3 and 4), DLS (supplemental Fig. S1, Table 1), SAXS (Fig. 5, Table 2), and mutagenesis and homology modeling (Fig. 6) to study the structure of TRN-SR2 and its complex with RanGTP in solution (Fig. 7). Importins in general show high conformational flexibility and variability (62, 63) allowing them to adapt their conformations in response to their cargo and the highly dynamic environment of the NPC. The original view of the accommodation of different binding partners via induced-fit mechanisms was replaced by a concept of population shift between preexisting alternative conformations (64). This makes studies of conformational states of these flexible helicoids very demanding, especially at the level of interpretation of the obtained data. The elastic behavior of solenoid proteins like TRN-SR2 is dominated by non-polar interactions between HEAT repeats (65). In particular, the hypothesis of a soft nanospring has arisen with a molten globule-like hydrophobic core (65), where small changes between HEAT repeats accumulate across the molecule to result in a relatively large change in the overall conformation.
In line with a recent report, our SAXS data for TRN-SR2 alone indicate that the protein predominantly exists as a dimer in solution (30). For TRN-SR2-RanGTP, SEC, SAXS, and homology modeling data strongly support a 1:1 stoichiometry of the complex. The remarkable increase in the SEC elution volume (Ve) as well as the decreased values of Rg (SAXS) and Rh (DLS) may hence result from dissociation of TRN-SR2 dimers as well as the conformational changes in monomeric TRN-SR2 while wrapping around RanGTP. As we are likely observing two processes at the same time, dissociation of TRN-SR2 dimers and formation of the 1:1 TRN-SR2-RanGTP complex, it is difficult to make conclusions on conformational changes within monomeric TRN-SR2 itself upon RanGTP binding. For importin β, which stayed monomeric in solution, both Rgand Dmax decreased upon binding of RanGTP. In contrast, for TNPO1 the Rg value increased and Dmax remained the same upon binding of Ran (62). The fact that importin β and TNPO1 are more related in the sequence-based phylogenetic tree and distant from TRN-SR2 (66) suggests that the structural changes may be dictated rather by the higher order structural elements (HEAT repeats) than by the primary sequence. Potentially extensive conformational changes in TRN-SR2 versus TRN-SR2-RanGTP are in accordance with SAXS measurements available for importin β and TNPO1 (62). A more compact and rigid conformation of the complex with RanGTP in comparison with the free karyopherin was also demonstrated by (non)equilibrium molecular dynamics simulations for importin β (66). However, to fully understand the role of preexisting alternative conformations (such as TRN-SR2 dimers), additional studies have to be conducted. Due to the intrinsic flexibility of TRN-SR2, it will also be of interest to perform molecular dynamics simulation of the contacts involved in the interaction of the proteins.
Last, because of the good fit between experimental and model back-calculated SAXS curves for the TRN-SR2-RanGTP complex and the additional support from mutagenesis and DSF, we can consider the molar ratio of 1:1 between both protein partners as corroborated. Furthermore, this validated model provides detailed insights into the complex (Fig. 7). RanGTP establishes elaborate contacts with TRN-SR2; while mainly occupying the N-terminal part of the TRN-SR2 toroid, Ran also contacts several central and C-terminal HEAT repeats. The interface between both proteins is characterized by extensive charge complementarity, notably involving the switch I and II regions of Ran, which determine nucleotide-dependent recognition of protein partners. Similar to other importins, mainly electrostatic interactions seem to steer complex formation between TRN-SR2 and RanGTP. These details on the TRN-SR2-RanGTP interaction and on TRN-SR2 itself may provide important insights into its function as a nuclear import factor for HIV-1. Additionally, these structural insights provide a first stepping stone toward modulation of TRN-SR2 function or inhibition of its interaction with HIV-1 IN for therapeutic purposes.
Acknowledgments
We thank Nam Joo Van der Veken and Linda Desender for excellent technical assistance.
This work was supported by CellCoVir SBO Grant 60813 of the Flemish IWT (Innovation by Science and Technology), FWO Grant G.0530.08, and FP7 Grants THINC and CHAARM, the Research Fund and the IOF (Industrial Research Fund) Program of the KU Leuven (FU-0T).

This article contains supplemental Figs. S1–S4.
- PIC
- preintegration complex
- CA
- capsid
- NPC
- nuclear pore complex
- TRN-SR2
- Transportin-SR2 (Transportin-3, TNPO3)
- IN
- integrase
- SR proteins
- serine/arginine-rich proteins
- CPSF
- cleavage and polyadenylation specificity factor subunit 6
- SEC
- size-exclusion chromatography
- SAXS
- small angle x-ray scattering
- DSF
- differential scanning fluorometry
- Imp13
- Importin-13.
REFERENCES
- 1. Suzuki Y., Craigie R. (2007) The road to chromatin - nuclear entry of retroviruses. Nat. Rev. Microbiol. 5, 187–196 [DOI] [PubMed] [Google Scholar]
- 2. De Rijck J., Vandekerckhove L., Christ F., Debyser Z. (2007) Lentiviral nuclear import. A complex interplay between virus and host. Bioessays 29, 441–451 [DOI] [PubMed] [Google Scholar]
- 3. Arhel N. (2010) Revisiting HIV-1 uncoating. Retrovirology 7, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gallay P., Stitt V., Mundy C., Oettinger M., Trono D. (1996) Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70, 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gallay P., Hope T., Chin D., Trono D. (1997) HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U.S.A. 94, 9825–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fassati A. (2003) Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22, 3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zielske S. P., Stevenson M. (2005) Importin 7 may be dispensable for human immunodeficiency virus type 1 and simian immunodeficiency virus infection of primary macrophages. J. Virol. 79, 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ao Z., Huang G., Yao H., Xu Z., Labine M., Cochrane A. W., Yao X. (2007) Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 282, 13456–13467 [DOI] [PubMed] [Google Scholar]
- 9. Ao Z., Danappa Jayappa K., Wang B., Zheng Y., Kung S., Rassart E., Depping R., Kohler M., Cohen E. A., Yao X. (2010) Importin α3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 84, 8650–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monette A., Panté N., Mouland A. J. (2011) HIV-1 remodels the nuclear pore complex. J. Cell Biol. 193, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matreyek K. A., Engelman A. (2011) The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 85, 7818–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woodward C. L., Prakobwanakit S., Mosessian S., Chow S. A. (2009) Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J. Virol. 83, 6522–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zaitseva L., Myers R., Fassati A. (2006) tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 4, e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christ F., Thys W., De Rijck J., Gijsbers R., Albanese A., Arosio D., Emiliani S., Rain J.-C., Benarous R., Cereseto A., Debyser Z. (2008) Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18, 1192–1202 [DOI] [PubMed] [Google Scholar]
- 15. Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 16. König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C.-Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008) Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cribier A., Ségéral E., Delelis O., Parissi V., Simon A., Ruff M., Benarous R., Emiliani S. (2011) Mutations affecting interaction of integrase with TNPO3 do not prevent HIV-1 cDNA nuclear import. Retrovirology 8, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krishnan L., Matreyek K. A., Oztop I., Lee K., Tipper C. H., Li X., Dar M. J., Kewalramani V. N., Engelman A. (2010) The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 84, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai M. C., Lin R. I., Huang S. Y., Tsai C. W., Tarn W. Y. (2000) A human importin-β family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275, 7950–7957 [DOI] [PubMed] [Google Scholar]
- 20. Lai M. C., Lin R. I., Tarn W. Y. (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kataoka N., Bachorik J. L., Dreyfuss G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145, 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Logue E. C., Taylor K. T., Goff P. H., Landau N. R. (2011) The cargo-binding domain of transportin 3 is required for lentivirus nuclear import. J. Virol. 85, 12950–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schaller T., Ocwieja K. E., Rasaiyaah J., Price A. J., Brady T. L., Roth S. L., Hué S., Fletcher A. J., Lee K., KewalRamani V. N., Noursadeghi M., Jenner R. G., James L. C., Bushman F. D., Towers G. J. (2011) HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7, e1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Iaco A., Santoni F., Vannier A., Guipponi M., Antonarakis S., Luban J. (2013) TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valle-Casuso J. C., Di Nunzio F., Yang Y., Reszka N., Lienlaf M., Arhel N., Perez P., Brass A. L., Diaz-Griffero F. (2012) TNPO3 is required for HIV-1 replication after nuclear import but prior to integration and binds the HIV-1 core. J. Virol. 86, 5931–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Iaco A., Luban J. (2011) Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus. Retrovirology 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou L., Sokolskaja E., Jolly C., James W., Cowley S. A., Fassati A. (2011) Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 7, e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ocwieja K. E., Brady T. L., Ronen K., Huegel A., Roth S. L., Schaller T., James L. C., Towers G. J., Young J. A., Chanda S. K., König R., Malani N., Berry C. C., Bushman F. D. (2011) HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 7, e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee K., Ambrose Z., Martin T. D., Oztop I., Mulky A., Julias J. G., Vandegraaff N., Baumann J. G., Wang R., Yuen W., Takemura T., Shelton K., Taniuchi I., Li Y., Sodroski J., Littman D. R., Coffin J. M., Hughes S. H., Unutmaz D., Engelman A., KewalRamani V. N. (2010) Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larue R., Gupta K., Wuensch C., Shkriabai N., Kessl J. J., Danhart E., Feng L., Taltynov O., Christ F., Van Duyne G. D., Debyser Z., Foster M. P., Kvaratskhelia M. (2012) Interaction of the HIV-1 intasome with Transportin 3 protein (TNPO3 or TRN-SR2). J. Biol. Chem. 287, 34044–34058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shah V. B., Shi J., Hout D. R., Oztop I., Krishnan L., Ahn J., Shotwell M. S., Engelman A., Aiken C. (2013) The host proteins Transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 87, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koh Y., Wu X., Ferris A. L., Matreyek K. A., Smith S. J., Lee K., KewalRamani V. N., Hughes S. H., Engelman A. (2013) Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J. Virol. 87, 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thys W., De Houwer S., Demeulemeester J., Taltynov O., Vancraenenbroeck R., Gérard M., De Rijck J., Gijsbers R., Christ F., Debyser Z. (2011) Interplay between HIV entry and transportin-SR2 dependency. Retrovirology 8, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Price A. J., Fletcher A. J., Schaller T., Elliott T., Lee K., KewalRamani V. N., Chin J. W., Towers G. J., James L. C. (2012) CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 8, e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fricke T., Valle-Casuso J. C., White T. E., Brandariz-Nuñez A., Bosche W. J., Reszka N., Gorelick R., Diaz-Griffero F. (2013) The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology 10, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De Houwer S., Demeulemeester J., Thys W., Taltynov O., Zmajkovicova K., Christ F., Debyser Z. (2012) Identification of residues in the C-terminal domain of HIV-1 integrase that mediate binding to the transportin-SR2 protein. J. Biol. Chem. 287, 34059–34068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andrade M. A., Perez-Iratxeta C., Ponting C. P. (2001) Protein repeats. Structures, functions, and evolution. J. Struct. Biol. 134, 117–131 [DOI] [PubMed] [Google Scholar]
- 38. Cook A., Bono F., Jinek M., Conti E. (2007) Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 39. Sorokin A. V., Kim E. R., Ovchinnikov L. P. (2007) Nucleocytoplasmic transport of proteins. Biochemistry Mosc. 72, 1439–1457 [DOI] [PubMed] [Google Scholar]
- 40. D'Angelo M. A., Hetzer M. W. (2008) Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee S. J., Matsuura Y., Liu S. M., Stewart M. (2005) Structural basis for nuclear import complex dissociation by RanGTP. Nature 435, 693–696 [DOI] [PubMed] [Google Scholar]
- 42. Quimby B. B., Lamitina T., L'Hernault S. W., Corbett A. H. (2000) The mechanism of ran import into the nucleus by nuclear transport factor 2. J. Biol. Chem. 275, 28575–28582 [DOI] [PubMed] [Google Scholar]
- 43. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 44. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) ATSAS2.1. Toward automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40, s223-s228 [Google Scholar]
- 45. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., Svergun D. I. (2012) New developments in the ATSASprogram package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 47. Fischer H., Oliveira Neto M., de, Napolitano H. B., Polikarpov I., Craievich A. F. (2009) Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43, 101–109 [Google Scholar]
- 48. Svergun D., Barberato C., Koch M. H. J. (1995) CRYSOL. A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 49. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 50. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 51. Forwood J. K., Lonhienne T. G., Marfori M., Robin G., Meng W., Guncar G., Liu S. M., Stewart M., Carroll B. J., Kobe B. (2008) Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation. Implications for nuclear import complex assembly dynamics. J. Mol. Biol. 383, 772–782 [DOI] [PubMed] [Google Scholar]
- 52. Stewart M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 53. Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. (1994) RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. U.S.A. 91, 2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Svergun D. I., Richard S., Koch M. H., Sayers Z., Kuprin S., Zaccai G. (1998) Protein hydration in solution. Experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. U.S.A. 95, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bono F., Cook A. G., Grünwald M., Ebert J., Conti E. (2010) Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of importin 13. Mol. Cell 37, 211–222 [DOI] [PubMed] [Google Scholar]
- 56. Grünwald M., Bono F. (2011) Structure of Importin13-Ubc9 complex. Nuclear import and release of a key regulator of sumoylation. EMBO J. 30, 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chook Y. M., Blobel G. (1999) Structure of the nuclear transport complex karyopherin-β2-Ran × GppNHp. Nature 399, 230–237 [DOI] [PubMed] [Google Scholar]
- 58. Vetter I. R., Arndt A., Kutay U., Görlich D., Wittinghofer A. (1999) Structural view of the Ran-importin β interaction at 2.3 A resolution. Cell 97, 635–646 [DOI] [PubMed] [Google Scholar]
- 59. Vetter I. R., Nowak C., Nishimoto T., Kuhlmann J., Wittinghofer A. (1999) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue. Implications for nuclear transport. Nature 398, 39–46 [DOI] [PubMed] [Google Scholar]
- 60. Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. (1990) Molecular switch for signal transduction. Structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945 [DOI] [PubMed] [Google Scholar]
- 61. Scheffzek K., Klebe C., Fritz-Wolf K., Kabsch W., Wittinghofer A. (1995) Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature 374, 378–381 [DOI] [PubMed] [Google Scholar]
- 62. Fukuhara N., Fernandez E., Ebert J., Conti E., Svergun D. (2004) Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 279, 2176–2181 [DOI] [PubMed] [Google Scholar]
- 63. Conti E., Müller C. W., Stewart M. (2006) Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 16, 237–244 [DOI] [PubMed] [Google Scholar]
- 64. Nevo R., Stroh C., Kienberger F., Kaftan D., Brumfeld V., Elbaum M., Reich Z., Hinterdorfer P. (2003) A molecular switch between alternative conformational states in the complex of Ran and importin β1. Nat. Struct. Biol. 10, 553–557 [DOI] [PubMed] [Google Scholar]
- 65. Kappel C., Zachariae U., Dölker N., Grubmüller H. (2010) An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys. J. 99, 1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zachariae U., Grubmüller H. (2008) Importin-β. Structural and dynamic determinants of a molecular spring. Structure 16, 906–915 [DOI] [PubMed] [Google Scholar]