Background: Genome-wide homology search is inconsistent with the emerging view of bacterial genome morphology.
Results: Stress-induced genome condensation proceeds through nonrandom convergence of sister chromosomes that culminates in spatial proximity of homologous sites.
Conclusion: Chromosome convergence enables repair of double strand DNA breaks.
Significance: Exposure to diverse stressful conditions primes bacteria to cope with detrimental DNA lesions.
Keywords: Biophysics, DNA Damage, DNA Physical Chemistry, DNA Structure, Escherichia coli, Fluorescence, Homologous Recombination, RecA, Replication Forks
Abstract
Genome condensation is increasingly recognized as a generic stress response in bacteria. To better understand the physiological implications of this response, we used fluorescent markers to locate specific sites on Escherichia coli chromosomes following exposure to cytotoxic stress. We find that stress-induced condensation proceeds through a nonrandom, zipper-like convergence of sister chromosomes, which is proposed to rely on the recently demonstrated intrinsic ability of identical double-stranded DNA molecules to specifically identify each other. We further show that this convergence culminates in spatial proximity of homologous sites throughout chromosome arms. We suggest that the resulting apposition of homologous sites can explain how repair of double strand DNA breaks might occur in a mechanism that is independent of the widely accepted yet physiologically improbable genome-wide search for homologous templates. We claim that by inducing genome condensation and orderly convergence of sister chromosomes, diverse stress conditions prime bacteria to effectively cope with severe DNA lesions such as double strand DNA breaks.
Introduction
A particularly deleterious type of DNA damage is double strand DNA breaks (DSBs)2 (1). An important repair mechanism of this lesion proceeds through homologous repair, which relies on the interaction of chromosomal ends generated by DSBs with homologous sequences that act as templates (2, 3). Accordingly, homologous repair strictly depends on the accessibility of such templates to severed DNA ends. In eukaryotes, this prerequisite is met by cohesin-mediated apposition of sister chromatids (4–6). In contrast, segregation of bacterial sister chromosomes shortly following replication (7–14) prevents on-site access of DSBs to their repair templates. It is thus generally assumed that for homologous repair to occur in bacteria, severed DNA ends need to conduct a genome-wide homology search (2, 3).
Genome-wide search is, however, inconsistent with the emerging view of genome morphology in bacteria. Specifically, recent studies demonstrated that chromosomal sites in bacteria are persistently localized at specific cytoplasmic addresses (9, 15–20) and that bacterial genomes consist of multiple domains that are spatially confined and mutually inaccessible (21, 22). Such a genome organization, which indicates a highly constrained motion of chromosomal sites, implies that a genome-wide search is unlikely. This claim is supported by the finding that diffusion of genomic sites is substantially slower than that revealed by proteins (23), indicating that homology search conducted by chromosomal sites for homologous templates cannot be considered in terms of a search performed by proteins for their cognate DNA sequences (24). Indeed, although significant insights into the process by which DSBs identify their homologous repair templates have recently been achieved (25, 26), the question how such lesions find these templates, which are embedded within a genomic-length DNA, remains unanswered (24).
It is becoming increasingly evident that exposure of bacteria to diverse detrimental conditions results in massive genome condensation (27–32). The physiological implications of this apparently generic stress response, which was recently demonstrated to occur also in archaea (33), remain unclear. By using fluorescent markers to locate specific Escherichia coli chromosomal sites, we show that stress-induced genome condensation proceeds through an orderly convergence of segregated sister chromosomes rather than through random DNA collapse. This convergence is suggested to initiate at the replication forks and to be mediated by the recently demonstrated ability of identical double-stranded DNA molecules to specifically identify each other and generate robust complexes (34, 35). We propose that the ensuing proximity of homologous chromosomal sites throughout chromosomal arms enables DSB repair in a mechanism that is independent of a genome-wide search yet consistent with the current physical understanding of the morphology of bacterial genomes. The results reported here imply that genome condensation, triggered even by relatively moderate stressful conditions and cellular damage, primes bacteria to rapidly and effectively cope with highly detrimental DNA lesions such as DSBs. Moreover, although our observations are consistent with the notion that RecA plays an essential role in the identification of accurate homologous template as well as in the formation of a stable complex between presynaptic filaments and their repair templates (2, 3, 36, 37), these findings imply that, in contrast to the widespread conviction, RecA-dependent genome-wide search is not required for DSB homologous repair.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions (Table 1)
TABLE 1.
Strains and plasmids
| Genotype | Source | |
|---|---|---|
| Strains | ||
| AB1157 | thr-1, araC14, leuB6(Am), Δ(gpt-proA)62, lacY1, tsx-33, qsr′-0, glnV44(AS), galK2(Oc), LAM-, Rac-0, hisG4(Oc), rfbC1, mgl-51, rpoS396(Am), rpsL31(strR), kdgK51, xylA5, mtl-1, argE3(Oc), thi-1 | |
| WBN2 | RecA− (AB1157 (ΔrecA306::Tn10)) | |
| IL05 | AB1157 (lacO240-Km]1801; tetO240-Gm]3908. | 7, 12 |
| WX51 | AB1157 (lacO240-Km at 90° and tetO240-Gm at 270°) | 7, 12 |
| IL05-RecA− | IL05 (ΔrecA306::Tn10) | This study |
| SMR8478 | MG1655 (Δattλ::PsulAΩgfp-mut2 attP21::pAH81-PBADΩI-SceI) | 1 |
| SMR8478-FseI | MG1655 (Δattλ::PsulAΩgfp-mut2 attP21::pAH81-PBADΩI-SceI::pZS*32-FseI) | This study |
| Plasmids | ||
| pLAU53 | pUC18 derivative containing genes for fusion proteins LacI-eCFP and TetR-eYFP under ara promoter | 7, 12 |
| pZS*32-MCS | pZ vector system | This study |
| pZS*32-FseI | FseI endonuclease under PLacO-1 promoter | This study |
E. coli wild-type strain used in this study was AB1157. WBN2 RecA− strain is an AB1157 derivative. IL05-RecA− was constructed by P1 transduction of the DrecA306::Tn10 locus from WBN2 into IL05 and selection for tetracycline, gentamicin, and kanamycin resistance. The recA deletion was confirmed by colony PCR and DNA sequencing. A strain carrying I-SceI endonuclease under PBAD arabinose-inducible promoter and a single I-SceI chromosomal cut site (SMR8478; gift from S. Rosenberg) (1) was used for DSB induction at a single chromosomal site (Table 1). To construct the SMR8478-FseI strain (Table 1) that enables generation of five DSBs, plasmid pZS*32-FseI that harbors FseI endonuclease under the control of the PLlacO-1 promoter was transformed into SMR8478 cells. Cells were regularly grown in LB at 37 °C. NA and chloramphenicol (Sigma) were added to mid-log cultures to concentrations of 50–500 or 100 μg/ml, respectively. For endonuclease induction, SMR8478 and SMR8478-FseI strains were grown in M9 minimal medium supplemented with 0.1% glucose to A600 = 0.3, centrifuged, resuspended in M9, including either 0.1% l-arabinose (for I-SceI induction) or 0.1% l-arabinose and 1 mm isopropyl 1-thio-β-d-galactopyranoside (for induction of both I-SceI and FseI endonucleases), and incubated at 37 °C for 1 h.
Fluorescent Repressor Operator Systems (FROS) Studies (7, 12)
E. coli strains IL05 (wild-type and RecA−) and WX51 were used for visualization of chromosomal sites. IL05 carries on its chromosome an array of 240 copies of tetO (tetO240-Gm) inserted close to the origin and an array of 240 copies of lacO (lacO240-Km) inserted close to the terminus site (Fig. 1A). A second strain for visualization of chromosomal sites, WX51, carries 240 copies of lacO and 240 copies of tetO inserted at 90° and 270° of the chromosome, respectively (Fig. 1B). A multicopy plasmid, pLAU53 (Table 1), expressing LacI-eCFP and TetR-eYFP (enhanced cyan and yellow fluorescent proteins, respectively) fusion repressors from PBAD arabinose promoter was used as a source of fluorescently labeled LacI and TetR. The FROS systems were kindly provided by D. Sherratt.
FIGURE 1.
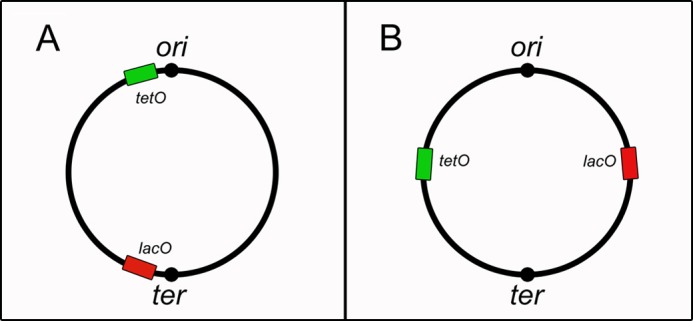
FROS (7, 12). A, FROS 1. tetO and lacO arrays were inserted close to the origin and terminal chromosomal sites, respectively (IL05 strain; Table 1). B, FROS 2. lacO and of tetO arrays were inserted at 90° and 270° of the chromosome, respectively (WX51 strain; Table 1).
Fluorescence Microscopy and Image Processing
Strains were grown to A600 ∼0.5, treated with antibiotics, and stained with the membrane stain FM4-64 (25 μg/ml; Invitrogen) and DAPI (0.5 ng/ml; Sigma) for 15 min. For FROS experiments, E. coli IL05 (wild-type and RecA−) or WX51 carrying pLAU53 was grown to A600 ∼0.3, induced with 0.01% l-arabinose for 30 min, and exposed to antibiotics as described above. Cells were then placed on 1% agarose pads prepared in a CoverWell Imaging Chamber (Sigma). Samples were visualized with ×100 oil immersion objective and photographed using a Deltavision microscope (Applied Precision). eCFP and eYFP were visualized by using ET470/24m and ET535/30m filters (Chroma), respectively. The signals of DAPI and FM4-64 were viewed by using ET457/40 and ET617/63 filters (Chroma), respectively. All images were identically de-convoluted and processed with the conservative SoftWorx package (Applied Precision).
Time-lapse studies were conducted on E. coli IL05 and WX51 cells carrying pLAU53. Cultures were grown in LB to A600 ∼0.3 and induced with 0.02% l-arabinose for 30 min. Cells were placed on 1% LB-agarose pads containing 500 μg/ml NA. Because of rapid bleaching of CFP, only YFP excitation was used in some time-lapse experiments, enabling localization of origin-proximal or 270° tetO foci. Images were de-convoluted with the conservative SoftWorx package and processed with ImageJ software.
RESULTS
Visualization of Chromosomal Sites
Chromosomal site localization was analyzed by using E. coli strains in which tetO and lacO arrays were inserted at various chromosomal sites (Fig. 1). These strains carry a plasmid encoding TetR-eYFP and LacI-eCFP repressors (FROS) (7, 12). Decoration of chromosomal sites following replication with the fluorescently tagged repressors provides a means for probing cellular addresses of homologous sites located on sister chromosomes.
Stressful Growth Conditions Result in a Nonrandom Genome Condensation and Convergence of Sister Chromosomes
In unstressed, exponentially growing E. coli cells (strains IL05 and AB1157; Table 1), chromatin is spread over the entire cytoplasm (Fig. 2A; Table 2), as is the case for other bacterial strains such as Bacillus subtilis (29) and in archaea (33). In contrast and as previously shown (28), E. coli cells exposed to NA, a pleiotropic drug that inflicts diverse DNA lesions (nicks, gaps, and DSBs) (38, 39), reveal condensed chromatin morphology (Fig. 2B; Table 2). In addition, exposure of IL05 cells to oxidative irradiation (γ, 137Cs source) that inflicts DNA lesions at doses of 100 and 200 gray resulted in genome condensation in 21 ± 2 and 34 ± 3% of the cells, respectively (data not shown). Notably, genotoxic stress was also shown to elicit genome condensation in B. subtilis (29) and archaea (33).
FIGURE 2.
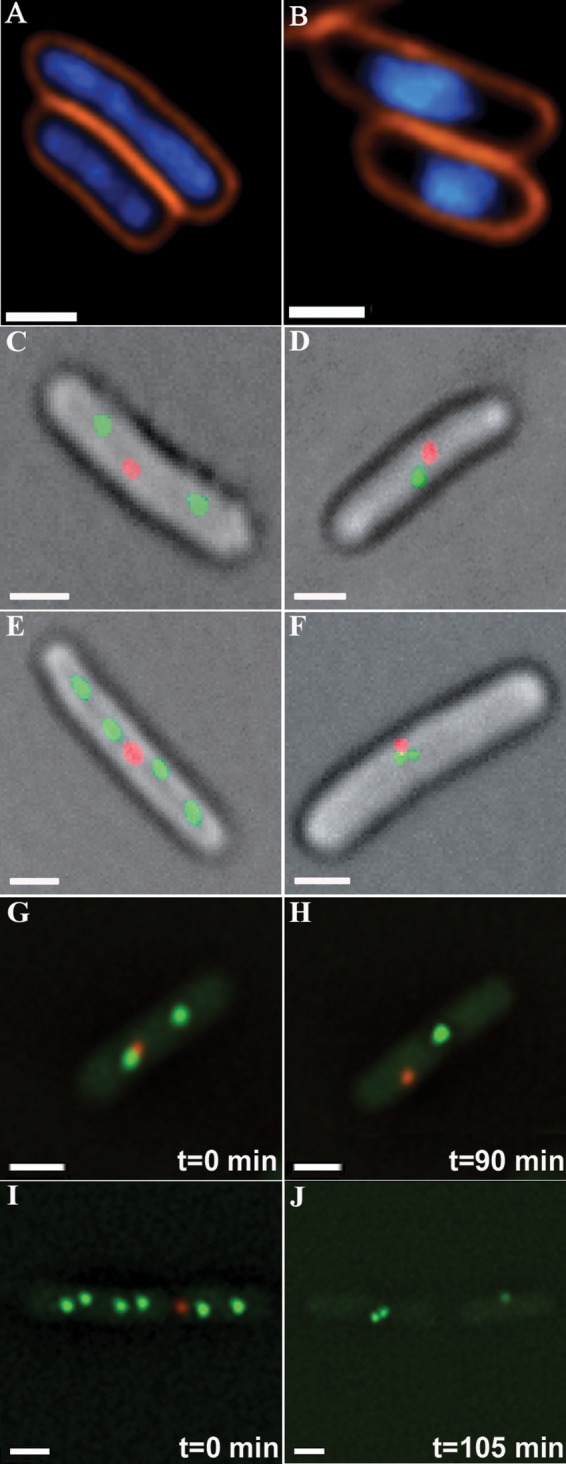
Chromosomal organization and site localization in E. coli (FROS 1) exposed to NA. A, expanded genomes in unstressed cells. B, genome condensation following exposure to NA (1 h; 350 μg/ml) (DNA stained blue with DAPI, and cell membranes stained orange with FM4-64). C, E, G, and I, unstressed cells carrying FROS 1 (Fig. 1A) reveal two or four origin-proximal foci (green) and a single terminus-proximal focus (red). D, F, H, and J, following NA treatment, two foci coalesce into a single focus, and four foci merge into two adjacent foci. G, I, H, and J are initial and final frames from time-lapse experiments (supplemental Movies S1 and S2, respectively). Notably, two cells are depicted in I and J. Scale bars, 0.5 μm.
TABLE 2.
Effects of NA on DNA condensation in WT E. coli (AB1157)
| NAa | Spread genome | Condensed genome | n/Nb |
|---|---|---|---|
| μg/ml | % of cells | % of cells | |
| 0 | 97 ± 2 | 3 ± 2 | 680/6 |
| 100 | 7 ± 2 | 93 ± 2 | 492/6 |
| 350 | 2 ± 2 | 98 ± 2 | 486/6 |
| 500 | 0 | 99 ± 2 | 703/6 |
a Exposure time was 60 min.
b n in all tables represents the total number of cells scored for a given set of conditions. N is the number of independent experiments conducted.
To locate specific chromosomal sites following stress-induced genome condensation, we used IL05 E. coli strain (Table 1), in which tetO and lacO arrays were inserted near the replication origin and chromosomal terminus, respectively (FROS 1; Fig. 1A) (7). Unstressed exponentially growing IL05 cells reveal two or four origin-proximal sites but only one terminus-proximal site (Fig. 2, C, E, G, and I), in keeping with multifork replication and rapid chromosome segregation (7, 12, 13, 40). In sharp contrast, NA-treated cells reveal only a single origin-proximal focus or two closely adjacent foci (Fig. 2, D, F, H, and J; Tables 3 and 4), which are always separated from the terminus-proximal site. These observations are supported by time-lapse experiments (supplemental Movies S1 and S2), which reveal progressive convergence of origin-proximal sites that culminates in their coalescence. Notably, whereas the inherently limited resolution of fluorescence microscopy does not allow unequivocal determination of the precise relative position of the origin-proximal foci, the progressive approach of these sites toward each other as well as the observation that once merged they remain co-localized support the notion that these foci do indeed coalesce. Collectively, these findings indicate co-localization of homologous sites (represented by origin-proximal foci) but not of nonhomologous sites (origin- and terminus-proximal sites).
TABLE 3.
Effects of NA (100 μg/ml) on the distribution of origin-proximal homologous sites in E. coli IL05
| NA incubation | 1 focus | 2 adjacent foci | ≥2 separated foci | n/N |
|---|---|---|---|---|
| min | % of cells | % of cells | % of cells | |
| 0 | 0 | 2 ± 1 | 98 ± 1 | 330/3 |
| 10 | 3 ± 1 | 4 ± 1 | 93 ± 1 | 178/3 |
| 20 | 14 ± 3 | 18 ± 3 | 68 ± 3 | 102/2 |
| 40 | 33 ± 3 | 37 ± 3 | 30 ± 3 | 304/3 |
| 60 | 43 ± 3 | 48 ± 3 | 9 ± 3 | 315/3 |
TABLE 4.
Effects of NA (350 μg/ml) on the distribution of origin-proximal homologous sites in E. coli IL05
| NA incubation | 1 focus | 2 adjacent foci | ≥2 separated foci | n/N |
|---|---|---|---|---|
| min | % of cells | % of cells | % of cells | |
| 10 | 5 ± 3 | 5 ± 3 | 90 ± 3 | 151/2 |
| 20 | 18 ± 2 | 20 ± 2 | 63 ± 2 | 176/2 |
| 40 | 38 ± 4 | 39 ± 4 | 23 ± 4 | 244/3 |
| 60 | 46 ± 2 | 51 ± 2 | 3 ± 2 | 375/3 |
We propose that this co-localization of chromosomal homologous sites results from the convergence of sister chromosomes. A single origin-proximal focus in NA-treated bacteria derives from cells that, prior to being exposed to stress, maintained a single pair of segregating sister chromosomes that originated from a single replication-firing event. Two contiguous origin-proximal foci are suggested to arise from the convergence of two pairs of chromosome arms that resulted from two successive replication-firing events (as indicated in the model depicted in Fig. 6). This conjecture, which is supported by our time-lapse movies (supplemental Movies S1–S6), is further substantiated by a statistical analysis conducted on a large number (>500) of unstressed and NA-exposed cells. This analysis demonstrated that in unstressed cells, the ratio between two and four origin-proximal foci is 0.41. This ratio is very similar to that of a single and two contiguous foci revealed in NA-treated cells (0.44), implying that chromosome convergence occurs between chromosome sisters that derive from an identical replication-firing event. Notably, NA removal results in a progressive increase in the number of fluorescent foci in a limited but significant cell population (Table 5), indicating reversibility of genome condensation and re-initiation of chromosome segregation.
FIGURE 6.
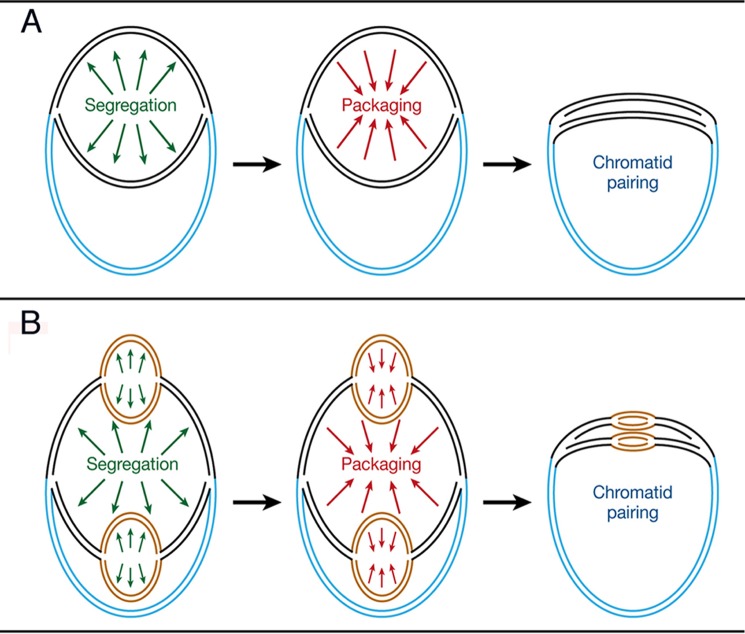
Schematic model of stress-induced convergence of chromosomal arms. Growing bacteria reveal either one (A) or two (B) pairs of newly replicated chromosome arms (black and brown ribbons, respectively). Adverse growth conditions promote genome condensation and sister chromosome convergence, which is initiated at the replication forks that act as nucleation sites and proceed in a zipper-like pathway. The resulting spatial proximity of homologous sites throughout chromosome arms enables interaction of DSBs with their homologous templates in a process that relies on random, short range, and diffusion-driven collisions rather than on a genome-wide homologous search.
TABLE 5.
Distribution of origin-proximal sites following NA (100 μg/ml) removal
| Time following NA removal | 1 focus | 2 foci | n/N |
|---|---|---|---|
| min | % of cells | % of cells | |
| 0 | 85 ± 5 | 15 ± 5 | 71/3 |
| 30 | 68 ± 5 | 32 ± 5 | 122/3 |
| 60 | 64 ± 6 | 36 ± 6 | 55/2 |
To corroborate the claim that stress-induced genome condensation promotes convergence of sister chromosomes and to eliminate the possibility that this convergence is limited to origin-proximal sites, we used an additional FROS strain (WX51; Table 1) in which lacO and tetO arrays were inserted at 90° and 270° sites of the chromosome, respectively (FROS 2; Fig. 1B) (12). Exponentially growing cells reveal two or four tetO and lacO foci (Fig. 3, A, C, and E; Table 6) that, following NA treatment, merge into a single tetO and a single lacO foci that are always spatially separated (Fig. 3, B, D, and F). Whereas only the tetO array (270° chromosomal site) could be probed in time-lapse experiments due to rapid eCFP bleaching, studies on cells fixed at different time points revealed that the nonhomologous tetO and lacO foci are consistently spatially separated, as shown in Fig. 3B. Notably, because FROS 2 probes the location of two distinct chromosomal sites, these observations, corroborated by time-lapse experiments (Fig. 3, C–F; supplemental Movie S3), further support the notion that genome condensation promotes convergence of chromosome arms throughout their entire length.
FIGURE 3.
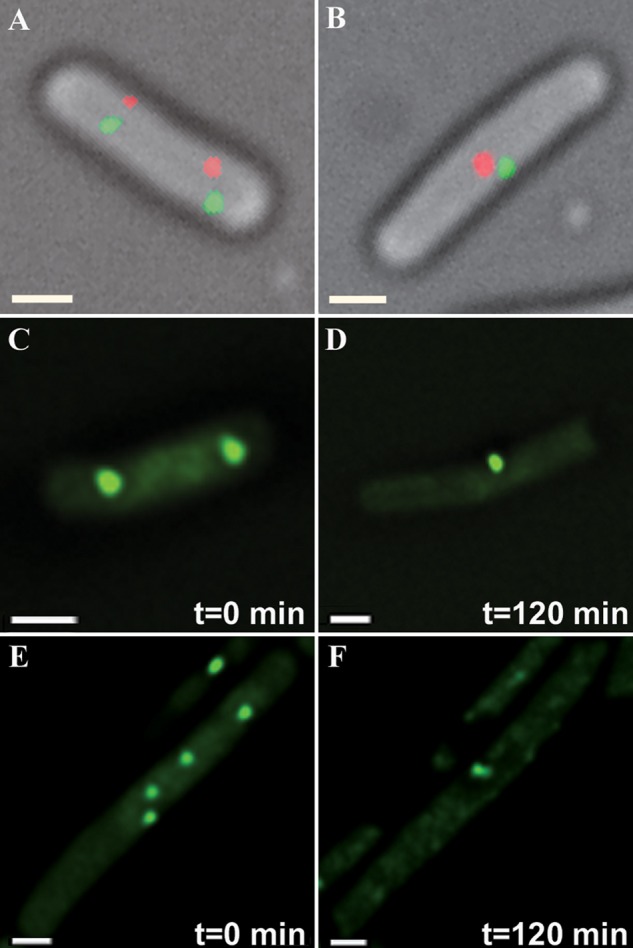
Chromosomal site localization in E. coli (FROS 2) exposed to NA. A, C, and E, distribution of chromosomal foci in nonstressed E. coli cells carrying FROS 2 (Fig. 1B). B, D, and F, cells exposed to NA, revealing co-localization of homologous but not of nonhomologous sequences (red and green foci). C–F (supplemental Movie S3) represent the initial and final frames from time-lapse experiments. In the time-lapse studies only the tetO array (270° chromosomal site) was probed due to rapid eCFP bleaching. Scale bars, 0.5 μm.
TABLE 6.
Effects of NA (350 μg/ml) on the distribution of site located at 270° of the chromosome in E. coli WX51
Similar results were obtained for the chromosomal site at 90°.
| NA incubation | 1 focus | 2 contiguous foci | ≥2 foci | n/N |
|---|---|---|---|---|
| min | % of cells | % of cells | % of cells | |
| 0 | 7 ± 0.5 | 0 | 93 ± 0.5 | 364/3 |
| 30 | 60 ± 2 | 12 ± 2 | 28 ± 2 | 514/3 |
| 60 | 81 ± 2 | 12 ± 2 | 7 ± 2 | 499/4 |
Insights from Induction of Double Strand DNA Breaks at Precise Number and Chromosomal Sites
The DNA-damaging agents described above (NA, oxidative irradiation) inflict lesions at random chromosomal sites and at a frequency that cannot be precisely controlled. We therefore sought to examine the correlation between genome condensation and generation of DNA lesions at a well defined number and at specific chromosomal sites.
Toward this aim, we used an E. coli strain (SMR8478) that carries the inducible double strand I-SceI endonuclease and a single I-SceI cut site, thus enabling the generation of a single DSB at a specific chromosomal site (1). Whereas induction of I-SceI was shown to generate a DSB in a large majority of the cells (1), such induction resulted in genome condensation in only a small percentage of the population (6 ± 1%; 88 cells probed in two experiments) (data not shown). In light of the fact that the I-SceI cut site is located near the origin of replication (355,395 bp from the origin in an ∼4,700-kbp chromosome site (1)), we suggest that the low extent of genome condensation reflects the fact that in most cells endonuclease induction occurred when replication forks have already proceeded through the I-SceI site and consequently is not affected by this lesion.
To verify this conjecture, we constructed a strain (SMR8478-FseI; Table 1) that enables infliction of four double strand DNA breaks at natively present FseI sites, in addition to the single DSB generated by the I-SceI endonuclease. Induction of both I-SceI and FseI, which generates five DSBs, resulted in genome condensation in 32 ± 2% of the cells (260 cells probed in three different experiments). We claim that this substantial increase in the population of cells that undergo genome condensation following infliction of five DSBs is consistent with the notion that generation of DNA lesions, the ensuing stalling of replication forks, and genome condensation are mechanistically related.
RecA and DSBs Are Required for Persistent Co-localization of Sister Chromosomes and Homologous Sites
To gain deeper insights into the mechanistic implications of stress-mediated genome condensation and chromosomal arm convergence, we studied these processes in two E. coli strains deficient in the recombination-repair protein RecA (WBN2 and IL05-RecA−; Table 1). Exposure of these strains to low NA doses resulted in genome condensation in a limited fraction of the bacterial population (Table 7), in keeping with previous observations demonstrating that RecA promotes genome condensation (28). Higher NA concentrations triggered condensation in a significantly larger population (Fig. 4, A and B; Table 7). Following exposure to such doses, convergence of origin-proximal sites into close spatial proximity was detected in 68 out of 80 cells (85%). However, and in clear contrast to wild-type cells, convergence did not result in persistent co-localization of homologous sites but rather in two adjacent foci or a single focus that, in most cases, subsequently segregated into two closely apposed foci (Fig. 4, C–G; supplemental Movies S4 and S5). These results indicate that, although genome condensation and sister chromosome convergence may occur in the absence of RecA, persistent sister co-localization is RecA-dependent. This observation is consistent with the fact that RecA is required for the formation of a lasting complex between presynaptic filaments generated at DSB sites and their homologous repair partners (2, 3).
TABLE 7.
Effects of NA on DNA packaging in RecA−E. coli (WBN2)
| NAa | Fully spread genome | Condensed genome | n/N |
|---|---|---|---|
| μg/ml | % of cells | % of cells | |
| 0 | 93 ± 2 | 7 ± 2 | 101/6 |
| 100 | 68 ± 5 | 32 ± 5 | 744/4 |
| 350 | 42 ± 3 | 58 ± 3 | 490/4 |
| 500 | 11 ± 0.5 | 89 ± 0.5 | 470/3 |
a Exposure time was 60 min.
FIGURE 4.
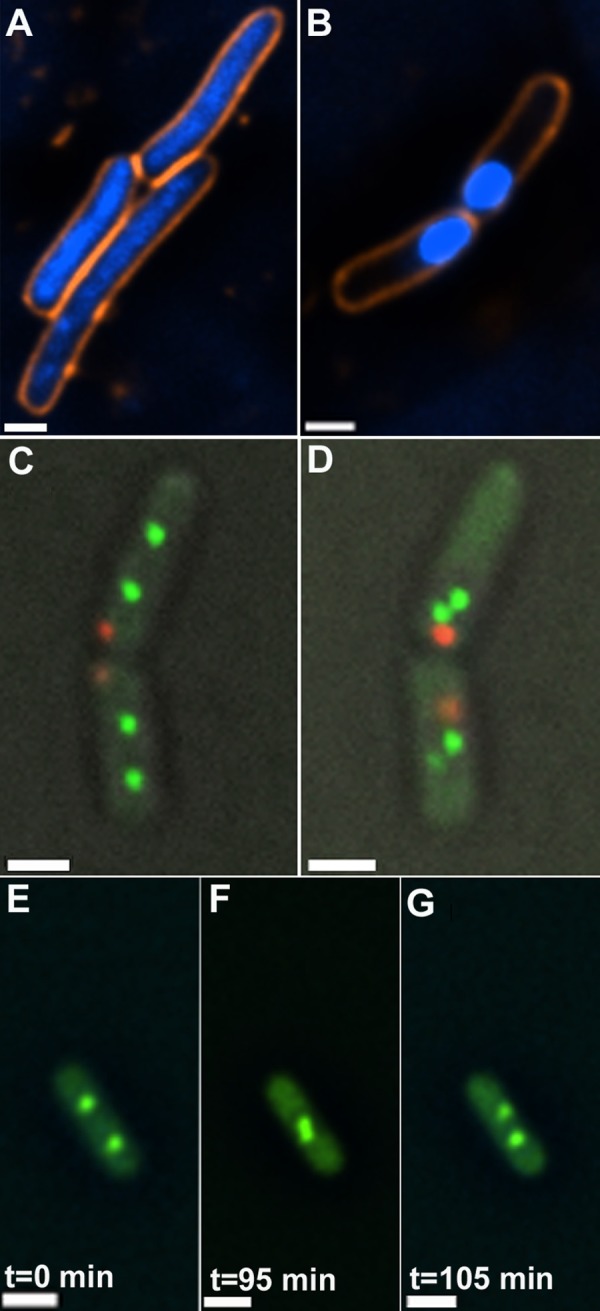
Genome organization and chromosomal foci in RecA− cells exposed to NA. A, genomes in untreated cells. B, genome condensation following NA treatment. C and D, cells at time 0 (C) and following 120 min of exposure to NA (D) reveal convergence, but not co-localization, of origin-proximal (green) sites. E–G, frames from supplemental Movie S4 showing co-localization and subsequent separation of origin-proximal foci. Scale bars, 0.5 μm.
Similar results were obtained when wild-type IL05 cells were exposed to chloramphenicol, an antibiotic that inhibits protein synthesis. As shown previously, chloramphenicol treatment results in chromatin condensation into toroidal structures (Fig. 5A) (41). Time-lapse studies in which 60 cells were probed following exposure to chloramphenicol revealed that in 52 cells (86.6%) homologous origin-proximal sites converged into close vicinity or merged into a single focus that then segregated into two closely apposed foci (Fig. 5, B–D; supplemental Movie S6). Because chloramphenicol does not inflict DNA lesions, these observations imply that DSBs are not required for sister chromosome convergence yet are essential for stabilizing their co-localization through the formation of robust and persistent presynaptic filaments-homologous template-RecA joint complexes, as is the case for RecA (2, 3).
FIGURE 5.
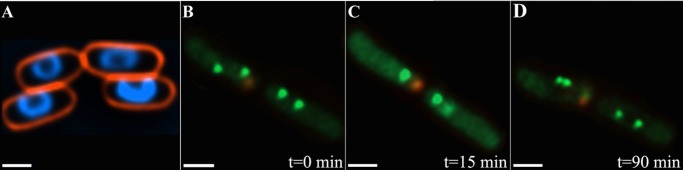
Effects of chloramphenicol on chromosomal foci localization. A, chromatin packaging into toroidal structures following exposure of IL05 to chloramphenicol. B–D, frames from a time-lapse experiment (supplemental Movie 6) in which IL05 cells were exposed to 150 μg/ml chloramphenicol, depicting merging followed by separation of homologous, origin-proximal foci in the right-side of the cell, and foci convergence followed by segregation in the left-side of the cell. Scale bars, 1.0 μm in A, and 0.5 μm in B–D.
DISCUSSION
This study was motivated by two considerations. The first is the increasing realization that exposure of bacteria to diverse stressful conditions, including starvation (27, 42), UV irradiation (28, 29), various antibiotics (31), and oxidative stress (32), results in massive genome condensation. Whereas this apparently generic stress response was previously proposed to promote DNA protection through physical sequestration (27, 43, 44) or to predispose bacteria to programmed cell death (31), the physiological implications of this process remain unclear. The second consideration was the unlikelihood of homologous repair pathways of double strand DNA breaks that rely on genome-wide search conducted by DNA ends for their repair templates. As indicated in the Introduction, such a search is inconsistent with the highly restricted motility of chromosomal sites, which is reflected by their precise and persistent localization at particular cytoplasmic addresses. The enigmatic nature of a genome-wide homology search has indeed been repeatedly highlighted (24). Intriguingly, studies conducted on yeast cells revealed an increased mobility of chromosomal sites following induction of DSBs and proposed that this enhanced motility promotes homology search (45, 46). A recent study (also conducted on yeast) demonstrated, however, that an a priori proximity between homologous sites, imposed by nuclear architectural features such as telomere and centromere clustering, plays a crucial role in enhancing the efficiency of DSB homologous repair (47).
Previous studies revealed that condensation of bacterial genome is mediated by specific stress-induced DNA-binding proteins such as Dps during starvation (27, 42) or RecA following infliction of DNA damage (28, 29). The observations reported here, according to which genome condensation occurs in RecA− cells as well as in wild-type bacteria in which protein synthesis is inhibited, imply that condensation can be induced through an alternative, generic, pathway. This notion is corroborated by recent observations according to which exposure of bacteria to oxidative stress results in genome packaging in a process that is independent of the main DNA-binding structural proteins such as Dps, H-NS, IHF, HU, and MukB (32).
Genome morphology in bacteria has been argued to represent a balance between the ever-present condensing factors that include the highly crowded cellular environment as well as attractive DNA-DNA interactions (48–51) and the expanding factors associated with metabolic activities (52, 53). These consist of DNA replication, transcription, chromosome segregation and, in particular, coupled transcription-translation and membrane insertion of membrane proteins that occur in bacteria (54). Indeed, a recent study demonstrated that expression of membrane proteins plays a crucial role in maintaining an expanded conformation of bacterial chromosomes (55). Collectively, these considerations imply that attenuated metabolic activity imposed by diverse stressful conditions such as starvation, drug-induced inhibition of protein synthesis, or exposure to DNA-damaging agents (previously demonstrated to induce replication arrest and cell cycle arrest by stalling replication forks (3, 56)) triggers DNA condensation. This condensation is presumably effected by tipping the balance between genome expansion and condensation toward the latter, as suggested previously (30, 57, 58).
On the basis of these considerations and the observations described above, we propose that stress-induced genome condensation maintains a crucial and heretofore unconsidered role in repair of bacterial DSBs. Specifically, we show that condensation does not proceed through random DNA collapse but rather through an orderly convergence and realignment of sister chromosomes throughout their length (Fig. 6). We suggest that this convergence is mediated by the long hypothesized (50, 59, 60) and recently in vitro demonstrated (34, 35, 61, 62) ability of identical (homologous) double-stranded DNA molecules to specifically identify each other and to generate relatively robust complexes. Significantly, this process was shown to be protein-independent and to be promoted by DNA condensation (35, 60, 63). We further propose that chromosome-realignment mediated by homologous dsDNA-dsDNA interactions is initiated at replication forks where ∼300,000 bp of the newly replicated sisters remain apposed before segregation is initiated (14). Such a priori paired regions may act as nucleation sites for convergence of the remaining sections of the sister chromosomes that then proceed sequentially in a zipper-like manner.
The conjecture that genome condensation and a nonrandom re-pairing of sister chromosomes are initiated at replication forks is supported by the results derived from the induction of DSBs by endonucleases, which imply that DNA lesions inflicted at sites that have already been replicated and as such do not encounter replication forks do not induce DNA packaging. This proposal is also corroborated by our time-lapse experiments, which reveal that two homologous sites merge into a single fluorescent focus whereas four sites, present on two pairs of homologous chromosomal arms, coalesce into two foci (as depicted in Fig. 6). It is further supported by our finding that the ratio between two and four fluorescent foci in unstressed cells is similar to the ratio between a single and two foci in NA-exposed cells, implying that a single pair of segregating chromosomes converges into a single focus (Fig. 6A), whereas two pairs of chromosome arms that result from two successive replication-firing events merge into two adjacent foci (Fig. 6B).
Following chromosome arm convergence, homologous sites on both arms are brought into close spatial proximity throughout the chromosome arms (Fig. 6). We suggest that this proximity enables DSBs to find their homologous repair templates through short range diffusion-driven random collisions that were shown to govern genomic DNA motion (23). Notably, although such a spatially constrained motion is incompatible with a genome-wide search, it is consistent with the notion that the main recombination protein RecA is not required for the homology search. RecA is, however, essential for accurate homology recognition and the subsequent formation of a stable recombination synapse (2, 3). This claim is supported by the finding that in RecA− cells exposed to NA, homologous sites converge but do not form lastingly co-localized foci. It is also consistent with the observation that in chloramphenicol-treated wild-type cells, chromosome arms converge but do not merge into lasting foci, as DSBs, along with RecA, are required for the formation of a robust joint recombination complex between presynaptic filaments and their homologous templates (2, 3).
It has been proposed that chromatid cohesion in eukaryotes evolved to enable DNA repair (4) and is, accordingly, enhanced following infliction of DSBs (5, 64). This notion, along with the bacterial DSB repair pathway proposed here, implies that the a priori juxtaposition of homologous partners is a critical prerequisite for homologous repair of DSBs, a prerequisite that is conserved in bacteria and eukaryotes. An intriguing implication is that genome condensation and chromosome re-pairing effected by diverse stressful conditions prime bacteria to rapidly and effectively cope with highly detrimental DNA lesions by facilitating homologous repair of DSBs. Moreover, by implying that stress-induced DNA condensation is initiated at replication forks, the observations presented here support the notion (65–71) that homologous recombination evolved mainly to repair stalled replication forks, rather than to generate genetic diversity. An additional corollary of this study is that the innate physicochemical properties of DNA molecules, including their tendency to condense under appropriate conditions and the ability of identical double-stranded DNA molecules to identify each other and generate a robust complex, are directly pertinent to their physiological activities (43, 49, 57, 72, 73).
Acknowledgments
We thank D. Sherratt for providing systems for chromosomal site localization and D. Ben-Halevy, G. Haran, Z. Livneh, and Y. Mutsafi for helpful comments. Fluorescence studies were conducted at the Moskowitz Center for Bio-Nano Imaging at the Weizmann Institute of Science.
This work was supported by the Israel Science Foundation funded by the Academy of Sciences and Humanities and by the Minerva Foundation, Germany.

This article contains supplemental Movies 1–6.
- DSB
- double strand DNA break
- NA
- nalidixic acid
- FROS
- Fluorescent Repressor Operator Systems
- eCFP
- enhanced cyan fluorescent protein
- eYFP
- enhanced yellow fluorescent protein.
REFERENCES
- 1. Pennington J. M., Rosenberg S. M. (2007) Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39, 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63, 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sjögren C., Nasmyth K. (2001) Sister chromatid cohesion is required for postreplicative double strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 [DOI] [PubMed] [Google Scholar]
- 5. Ström L., Karlsson C., Lindroos H. B., Wedahl S., Katou Y., Shirahige K., Sjögren C. (2007) Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317, 242–245 [DOI] [PubMed] [Google Scholar]
- 6. Unal E., Heidinger-Pauli J. M., Koshland D. (2007) DNA double strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317, 245–248 [DOI] [PubMed] [Google Scholar]
- 7. Lau I. F., Filipe S. R., Søballe B., Økstad O. A., Barre F. X., Sherratt D. J. (2003) Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49, 731–743 [DOI] [PubMed] [Google Scholar]
- 8. Sherratt D. J. (2003) Bacterial chromosome dynamics. Science 301, 780–785 [DOI] [PubMed] [Google Scholar]
- 9. Viollier P. H., Thanbichler M., McGrath P. T., West L., Meewan M., McAdams H. H., Shapiro L. (2004) Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U.S.A. 101, 9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nielsen H. J., Ottesen J. R., Youngren B., Austin S. J., Hansen F. G. (2006) The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62, 331–338 [DOI] [PubMed] [Google Scholar]
- 11. Nielsen H. J., Youngren B., Hansen F. G., Austin S. (2007) Dynamics of Escherichia coli chromosome segregation during multifork replication. J. Bacteriol. 189, 8660–8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X., Liu X., Possoz C., Sherratt D. J. (2006) The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20, 1727–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reyes-Lamothe R., Possoz C., Danilova O., Sherratt D. J. (2008) Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joshi M. C., Bourniquel A., Fisher J., Ho B. T., Magnan D., Kleckner N., Bates D. (2011) Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc. Natl. Acad. Sci. U.S.A. 108, 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Teleman A. A., Graumann P. L., Lin D. C., Grossman A. D., Losick R. (1998) Chromosome arrangement within a bacterium. Curr. Biol. 8, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 16. Niki H., Yamaichi Y., Hiraga S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 14, 212–223 [PMC free article] [PubMed] [Google Scholar]
- 17. Thanbichler M., Wang S. C., Shapiro L. (2005) The bacterial nucleoid: A highly organized and dynamic structure. J. Cell. Biochem. 96, 506–521 [DOI] [PubMed] [Google Scholar]
- 18. Thanbichler M., Shapiro L. (2008) Getting organized–how bacterial cells move proteins and DNA. Nat. Rev. Microbiol. 6, 28–40 [DOI] [PubMed] [Google Scholar]
- 19. White M. A., Eykelenboom J. K., Lopez-Vernaza M. A., Wilson E., Leach D. R. (2008) Nonrandom segregation of sister chromosomes in Escherichia coli. Nature 455, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 20. Montero Llopis P., Jackson A. F., Sliusarenko O., Surovtsev I., Heinritz J., Emonet T., Jacobs-Wagner C. (2010) Spatial organization of the flow of genetic information in bacteria. Nature 466, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Postow L., Hardy C. D., Arsuaga J., Cozzarelli N. R. (2004) Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18, 1766–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boccard F., Esnault E., Valens M. (2005) Spatial arrangement and macrodomain organization of bacterial chromosomes. Mol. Microbiol. 57, 9–16 [DOI] [PubMed] [Google Scholar]
- 23. Marshall W. F., Straight A., Marko J. F., Swedlow J., Dernburg A., Belmont A., Murray A. W., Agard D. A., Sedat J. W. (1997) Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7, 930–939 [DOI] [PubMed] [Google Scholar]
- 24. Weiner A., Zauberman N., Minsky A. (2009) Recombinational DNA repair in a cellular context: a search for the homology search. Nat. Rev. Microbiol. 7, 748–755 [DOI] [PubMed] [Google Scholar]
- 25. Chen Z., Yang H., Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 26. De Vlaminck I., van Loenhout M. T., Zweifel L., den Blanken J., Hooning K., Hage S., Kerssemakers J., Dekker C. (2012) Mechanism of homology recognition in DNA recombination from dual-molecule experiments. Mol. Cell 46, 616–624 [DOI] [PubMed] [Google Scholar]
- 27. Wolf S. G., Frenkiel D., Arad T., Finkel S. E., Kolter R., Minsky A. (1999) DNA protection by stress-induced biocrystallization. Nature 400, 83–85 [DOI] [PubMed] [Google Scholar]
- 28. Levin-Zaidman S., Frenkiel-Krispin D., Shimoni E., Sabanay I., Wolf S. G., Minsky A. (2000) Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc. Natl. Acad. Sci. U.S.A. 97, 6791–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith B. T., Grossman A. D., Walker G. C. (2002) Localization of UvrA and effect of DNA damage on the chromosome of Bacillus subtilis. J. Bacteriol. 184, 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cook P. R. (2002) Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 32, 347–352 [DOI] [PubMed] [Google Scholar]
- 31. Dwyer D. J., Camacho D. M., Kohanski M. A., Callura J. M., Collins J. J. (2012) Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 46, 561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ko K. C., Tai P. C., Derby C. D. (2012) Mechanisms of action of escapin, a bactericidal agent in the ink secretion of the sea hare Aplysia californica: Rapid and long-lasting DNA condensation and involvement of the OxyR-regulated oxidative stress pathway. Antimicrob. Agents Chemother. 56, 1725–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delmas S., Duggin I. G., Allers T. (2013) DNA damage induces nucleoid compaction via the Mre11-Rad50 complex in the archaeon Haloferax volcanii. Mol. Microbiol. 87, 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baldwin G. S., Brooks N. J., Robson R. E., Wynveen A., Goldar A., Leikin S., Seddon J. M., Kornyshev A. A. (2008) DNA double helices recognize mutual sequence homology in a protein free environment. J. Phys. Chem. B 112, 1060–1064 [DOI] [PubMed] [Google Scholar]
- 35. Danilowicz C., Lee C. H., Kim K., Hatch K., Coljee V. W., Kleckner N., Prentiss M. (2009) Single molecule detection of direct, homologous, DNA/DNA pairing. Proc. Natl. Acad. Sci. U.S.A. 106, 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. (1983) The double strand-break repair model for recombination. Cell 33, 25–35 [DOI] [PubMed] [Google Scholar]
- 37. Folta-Stogniew E., O'Malley S., Gupta R., Anderson K. S., Radding C. M. (2004) Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol. Cell 15, 965–975 [DOI] [PubMed] [Google Scholar]
- 38. Reece R. J., Maxwell A. (1991) DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26, 335–375 [DOI] [PubMed] [Google Scholar]
- 39. Bejerano-Sagie M., Oppenheimer-Shaanan Y., Berlatzky I., Rouvinski A., Meyerovich M., Ben-Yehuda S. (2006) A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125, 679–690 [DOI] [PubMed] [Google Scholar]
- 40. Nielsen H. J., Li Y., Youngren B., Hansen F. G., Austin S. (2006) Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 61, 383–393 [DOI] [PubMed] [Google Scholar]
- 41. Zimmerman S. B. (2002) Toroidal nucleoids in Escherichia coli exposed to chloramphenicol. J. Struct. Biol. 138, 199–206 [DOI] [PubMed] [Google Scholar]
- 42. Frenkiel-Krispin D., Levin-Zaidman S., Shimoni E., Wolf S. G., Wachtel E. J., Arad T., Finkel S. E., Kolter R., Minsky A. (2001) Regulated phase transitions of bacterial chromatin: a nonenzymatic pathway for generic DNA protection. EMBO J. 20, 1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Minsky A. (2004) Information content and complexity in the high-order organization of DNA. Annu. Rev. Biophys. Biomol. Struct. 33, 317–342 [DOI] [PubMed] [Google Scholar]
- 44. Frenkiel-Krispin D., Minsky A. (2006) Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis and D. radiodurans. J. Struct. Biol. 156, 311–319 [DOI] [PubMed] [Google Scholar]
- 45. Miné-Hattab J., Rothstein R. (2012) Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517 [DOI] [PubMed] [Google Scholar]
- 46. Dion V., Kalck V., Horigome C., Towbin B. D., Gasser S. M. (2012) Increased mobility of double strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14, 502–509 [DOI] [PubMed] [Google Scholar]
- 47. Renkawitz J., Lademann C. A., Kalocsay M., Jentsch S. (2013) Monitoring homology search during DNA double strand break repair in vivo. Mol. Cell 50, 261–272 [DOI] [PubMed] [Google Scholar]
- 48. Gronbech-Jensen N., Mashl R. J., Bruinsma R. F., Gelbart W. M. (1997) Counterion-induced attraction between rigid polyelectrolytes. Phys. Rev. Lett. 78, 2477–2480 [Google Scholar]
- 49. Strey H. H., Podgornik R., Rau D. C., Parsegian V. A. (1998) DNA-DNA interactions. Curr. Opin. Struct. Biol. 8, 309–313 [DOI] [PubMed] [Google Scholar]
- 50. Kornyshev A. A., Leikin S. (1999) Electrostatic zipper motif for DNA aggregation. Phys. Rev. Lett. 82, 4138–4141 [Google Scholar]
- 51. Todd B. A., Parsegian V. A., Shirahata A., Thomas T. J., Rau D. C. (2008) Attractive forces between cation condensed DNA double helices. Biophys. J. 94, 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woldringh C. L., Jensen P. R., Westerhoff H. V. (1995) Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol. Lett. 131, 235–242 [DOI] [PubMed] [Google Scholar]
- 53. Zimmerman S. B., Murphy L. D. (1996) Macromolecular crowding and the mandatory condensation of DNA in bacteria. FEBS Lett. 390, 245–248 [DOI] [PubMed] [Google Scholar]
- 54. Norris V. (1995) Hypothesis: chromosome separation in Escherichia coli involves autocatalytic gene expression, transertion and membrane-domain formation. Mol. Microbiol. 16, 1051–1057 [DOI] [PubMed] [Google Scholar]
- 55. Libby E. A., Roggiani M., Goulian M. (2012) Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 7445–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rudolph C. J., Upton A. L., Lloyd R. G. (2007) Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 21, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minsky A., Shimoni E., Frenkiel-Krispin D. (2002) Stress, order and survival. Nat. Rev. Mol. Cell Biol. 3, 50–60 [DOI] [PubMed] [Google Scholar]
- 58. Minsky A. (2003) Structural aspects of DNA repair: the role of restricted diffusion. Mol. Microbiol. 50, 367–376 [DOI] [PubMed] [Google Scholar]
- 59. Kornyshev A. A., Leikin S. (2001) Sequence recognition in the pairing of DNA duplexes. Phys. Rev. Lett. 86, 3666–3669 [DOI] [PubMed] [Google Scholar]
- 60. Kornyshev A. A., Wynveen A. (2009) The homology recognition well as an innate property of DNA structure. Proc. Natl. Acad. Sci. U.S.A. 106, 4683–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Inoue S., Sugiyama S., Travers A. A., Ohyama T. (2007) Self-assembly of double-stranded DNA molecules at nanomolar concentrations. Biochemistry 46, 164–171 [DOI] [PubMed] [Google Scholar]
- 62. Wang X., Zhang X., Mao C., Seeman N. C. (2010) Double-stranded DNA homology produces a physical signature. Proc. Natl. Acad. Sci. U.S.A. 107, 12547–12552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cherstvy A. G. (2011) DNA-DNA sequence homology recognition: physical mechanisms and open questions. J. Mol. Recognit. 24, 283–287 [DOI] [PubMed] [Google Scholar]
- 64. Ström L., Lindroos H. B., Shirahige K., Sjögren C. (2004) Postreplicative recruitment of cohesin to double strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 65. Cox M. M. (1998) A broadening view of recombinational DNA repair in bacteria. Genes Cells 3, 65–78 [DOI] [PubMed] [Google Scholar]
- 66. Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) The importance of repairing stalled replication forks. Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
- 67. Michel B., Flores M. J., Viguera E., Grompone G., Seigneur M., Bidnenko V. (2001) Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8181–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kuzminov A. (2001) DNA replication meets genetic exchange: Chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cox M. M. (2002) The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 510, 107–120 [DOI] [PubMed] [Google Scholar]
- 70. Lusetti S. L., Cox M. M. (2002) The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71, 71–100 [DOI] [PubMed] [Google Scholar]
- 71. McGlynn P. (2004) Links between DNA replication and recombination in prokaryotes. Curr. Opin. Gen. Dev. 14, 107–112 [DOI] [PubMed] [Google Scholar]
- 72. Goobes R., Minsky A. (2001) Thermodynamic aspects of triplex DNA formation in crowded environments. J. Am. Chem. Soc. 123, 12692–12693 [DOI] [PubMed] [Google Scholar]
- 73. Levin-Zaidman S., Englander J., Shimoni E., Sharma A. K., Minton K. W., Minsky A. (2003) Ringlike structure of the Deinococcus radiodurans genome: A key to radioresistance? Science 299, 254–256 [DOI] [PubMed] [Google Scholar]