Abstract
Objective
To investigate the antibiotic resistance genes inserted into class 1 and class 2 integrons in Acinetobacter baumannii (A. baumannii) isolates obtained from nine different cities in Turkey.
Methods
A collection of 281 A. baumannii clinical isolates were collected from nine diferent state hospitals in Turkey and were confirmed as A. baumannii by conventional biochemical, API testing and bla-OXA-51 specific PCR. The isolates were examined by PCR for existence of class 1 and 2 integron gene cassettes.
Results
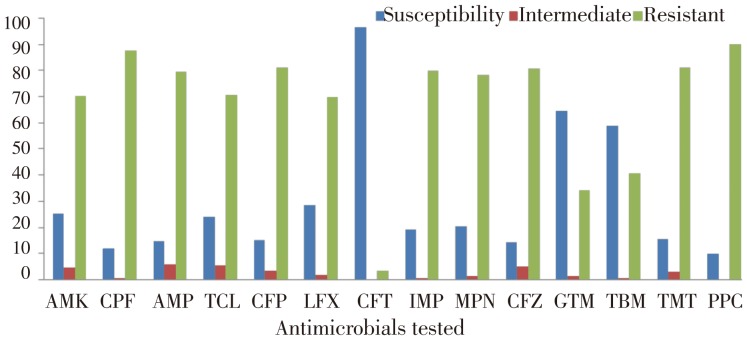
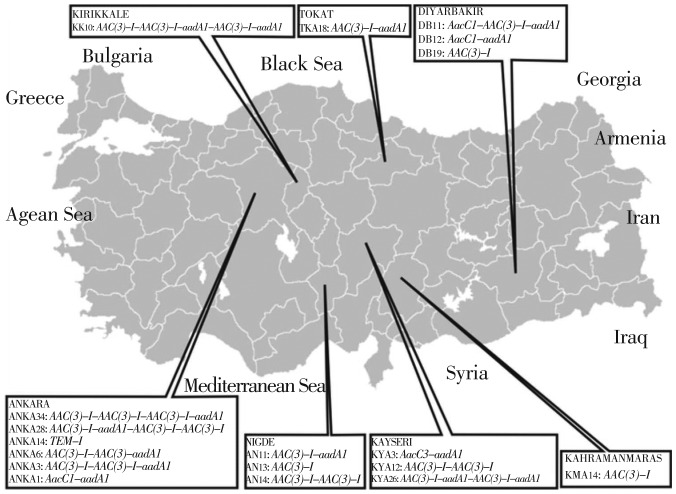
They were characterized by antimicrobial susceptibility testing and the highest resistance rates were determined for piperacillin (90.03%), ciprofloxacin (87.54%), cefepime and trimethoprim/sulfamethoxazole (81.13%). The lowest resistance rates was for cefotaxime (3.55%). class I integrons were detected in 6.4% (18/281) of A. baumannii strains and no class 2 integron was detected. The gene cassettes of class 1 integrons AacC1-AAC(3)I-aadA1, AacC1-aadA1, AAC(3)-I, AAC(3)-I -AAC(3)-I -aadA1, TEM-1, AAC(3)-I-aadA1 - AAC(3)-I -AAC(3)-I, AAC(3)-I -AAC(3)-I -AAC(3)-I -aadA1, AAC(3)-I - aadA1, AAC(3)-I-AAC(3)-I, AAC(3)-I-aadA1- AAC(3)-I-aadA1, AAC(3)-I- AAC(3)-I- aadA1-AAC(3)-I-aadA1 were detected in eighteen strains. The aac genes family were most frequently found integrated into the class 1 integrons and it was followed by aadA genes and TEM-1 genes.
Conclusions
This is an extensive study on the distribution of class 1 integron among A. baumannii in Turkey. In addition to these, two new alleles were observed. Their percentage rates of similarity to other cassettes are 95% aadA1 ( TKA18) and 89% aadA1 (ANKA3).
Keywords: Acinetobacter baumannii, Class 1 integron, Gene cassette, Resistance
1. Introduction
Acinetobacter baumannii (A. baumannii) is an important opportunistic pathogen responsible for a nosocomial infections, such as pneumonia, urinary tract infections, bacteraemia and meningitis[1]–[3]. Its clinical significance is due to its rapid acquisition of a wide variety of antibiotic resistance genes so it has caused difficulties in the control[4]. This extremely rapid development of resistance has caused serious therapeutic problems worldwide[5],[6]. A major acquisition and dissemination of resistance determinants is mobile elements, including plasmids, transposons and integrons[7]–[9]. Integrons are one of the main types of mobile elements currently known to be the natural gene capture systems in bacteria. Class 1 integrons are consisted of a 5′-conserved segment (5′CS) carrying a gene named intI encoding a site-specific integrase protein, and a variable region containing one or more antibiotic resistance genes[10],[11]. Class 2 integrons are related with a mobile genetic element, Tn7 transposon, and their structures resemble to class 1 integrons[10],[12]. The goals of this study were to screen the antibiotic resistance genes inserted into class 1 and class 2 integrons in A. baumannii isolates obtained from nine different cities in Turkey. This is also first nationwide investigation of the class 1 and 2 integrons among isolates of A. baumannii in Turkey.
2. Materials and methods
2.1. Bacterial strain and antibiotics susceptibilities
A total of 281 isolates of A. baumannii were investigated in this study. All strains were isolated from different hospitals in Turkey between 2011 and 2012. All clinical isolates were identified using the API 32GN system (bioMerieux, Marcy-l'Etoile, France). The isolates were also identified using blaOXA-51 specific PCR[13]. VITEK cards for antibiotic susceptibility testing (GNS-424 cards) were inoculated and incubated according to their commendations of the manufacturer (BioMerieux, Hazelwood, Mo.). The following antibiotics were tested: ampicillin-sulbactam, piperacillin, ceftazidime, cefepime, amikacin, gentamicin, tobramycin, ciprofloxacin, cefotaxime, levofloxacin, tetracycline, trimethoprim/sulfamethoxazole, imipenem meropenem according to Clinical and Laboratory Standards Institute[14].
2.2. DNA extraction
Genomic DNA used as a template for PCR assays was obtained from bacterial suspension grown overnight in Luria Broth with shaking incubator at 37 °C. Bacterial suspension was centrifuged at 13 000 r/min for 5 min. Pellet was suspended in 500 µL distilled water and subsequently boiled in a water bath for 10 min. Debris was centrifuged at 13 000 r/min for 5 min. Five microliters of supernatant was used as a template for PCR assays.
2.3. PCR assays for integrons
All isolates were tested for the presence of class 1 and class 2 integrases conserved region. Primers used for detection for intI1: intI1F (5′-ACATGTGATGGCGACGCACGA-3′) and intI1R (5′-ATTTCTGTCCTGGCTGGCGA-3′); for intI2: intI2F (5′-CACGGATATGCGACAAAAAGGT-3′) and intI2R (5′-GTAGCAAACGAGTGACGAAATG-3′); for class 1 integron gene cassette: 5′CS (5′-GGCATCCAAGCAGCAAG-3′) and for 3′CS (5′-AAGCAGACTTACCTGA-3′). PCRs were performed in a final volume of 50 µL. PCR mix component was as follows; 5 µL of 10X PCR buffer [0.1 mol/L Tris-HCl (pH 8.8), 0.5 mol/L KCl, 1% Triton X-100], 3 µL of 0.025 mol/L MgCl2, 5 µL of 10X dNTP (0.002 mol/L dATP, dCTP, dGTP and dTTP), 2 µL each of primer (25 pmol/µL), 34 µL deionised sterile water, 1 U of Taq DNA polymerase and 5 µL of template DNA. PCR amplification condition was as follows: initial denaturation at 94 °C for 3 min for initial denaturation, 94 °C for 45 seconds, 55 °C for 1 min, 72 °C for 3 min followed by 34 cycles and 5 min at 72 °C with a final extension[15],[16].
2.4. DNA sequencing and analysis
Electrophoresis of PCR products was performed with 1% agarose containing 0.5 µg/mL ethidium bromide, and subsequently visualized under UV light. The PCR products were ligated into pGEM-T easy vector (Promega) at 16 °C for 16 h by T4 DNA ligase. Ligation mixture were then transformed into Escherichia coli JM101 strain prepared according to Sambrook et al. and spread in ampicilline plates (50 µg/mL) containing X-gal (40 µg/mL)[17]. We collected the white colonies and plasmids were purified from these colonies by using Promega Plasmid Purification Kits. Recombinant plasmids carrying amplicons of class 1 integrons were sent to Macrogen Inc., Seoul, Korea for sequencing by using the universal oligonucleotide primers, T7 and SP6. Sequencing results were analysed using alignment search tool, BLAST (http://www.ncbi.nlm.nih.gov/BLAST)[18] and the multiple sequence alignment program, CLUSTAL W(http://www.ebi.ac.uk/clustalw).
3. Results
All isolates were tested for susceptibility to 14 antimicrobials. The frequency of resistance of the two hundred and eighty-one clinical isolates to each of the antibiotics tested can be observed in Figure 1. Resistance to tobramycin (40.56%), to piperacillin (90.03%), to ceftazidime (80.78%), to amikacin (70.01%), to gentamicin (34.16%), to imipenem (80.07%), to meropenem (78.29%), to ciprofloxacin (87.54%), to levofloxacin (69.75%), to cefepime (81.13%), to ampicillin-sulbactam (79.35%), to cefotaxime (3.55%), to tetracycline (70.46%) and to trimethoprim/sulfamethoxazole (81.13%) were observed. The lowest resistance rates was for cefotaxime (3.55%). The highest resistance rates were for piperacillin (90.03%), ciprofloxacin (87.54%), cefepime and trimethoprim/sulfamethoxazole (81.13%).
Figure 1. Antibiotics susceptibility of A. baumannii strains.
AMK: Amikacin; CPF: Ciprofloxacin; AMP: Ampicillin-sulbactam; TCL: Tetracycline; CFP: Cefepime; LFX: Levofloxacin; CFT: Cefotaxime; IMP: Imipenem; MPN: Meropenem; CFZ: Ceftazidime; GTM: Gentamicin; TBM: Tobramycin; TMT: Trimethoprim/sulfamethoxazole; PPC: Piperacillin.
Among the 281 isolates analysed by PCR, eighteen A. baumannii isolates were found to have the class 1 integron and no class 2 integron was detected. Integrase gene (Int1) were detected in 38.7% (109/281) of the total isolates. Of the 109 Int1 positive isolates which 18 of them were integron positive. A total of 12 different gene cassettes were observed in 18 isolates.The integrons were observed to contain 1-4 gene cassettes and location of the cassettes differ (Figure 2).
Figure 2. The geographical distribution of class 1 integrons found in A. baumannii strains in Turkey.
The gene cassettes of class 1 integrons AacC1-AAC(3) I-aadA1, AacC1-aadA1, AAC(3)-I, AAC(3)-I-AAC(3)-I-aadA1, TEM-1, AAC(3)-I-aadA1-AAC(3)-I-AAC(3)-I, AAC(3)-I-AAC(3)-I-AAC(3)-I-aadA1, AAC(3)-I-aadA1, AAC(3)-I-AAC(3)-I, AAC(3)-I-aadA1-AAC(3)-I-aadA1, AAC(3)-I-AAC(3)-I-aadA1-AAC(3)-I-aadA1. Among these profiles, aac genes family were found most frequently in class 1 integrons gene cassettes.
In addition to these, two new gene cassettes array were observed. Their percentage rates of similarity to other cassettes are 95% aadA1 (TKA18) and 89% aadA1 (ANKA3).
4. Discussion
A. baumannii is resistant to various antimicrobial agents such as penicillins, cephalosporines, aminoglycosides, tetracyclines[19],[20]. Because of the multidrug resistance quickly spread in hospital population, A. baumannii has a clinical significance, requiring epidemiologic monitoringas a measure for control of nosocomial infection[20].
Integrons play a significant role in the dissemination of antimicrobial resistance through horizontal transmission amoung Gram-negative bacteria[21],[22]. In the present study, we screened and characterised integrons in A. baumannii isolates from nine center in Turkey. 109 of 281 isolates contained integrase gene and 18 of IntI positive strains included gene cassetes. According to these results, 18 isolates contain gene cassettes and 91 strains do not include. The first reasons for this may be mutations or deformition at the 3′CS. The second reasons may be gene cassette array in novel, complex or unusual class 1 integrons[23],[24]. And finally perhaps variable region was too long to be amplified. The gene cassettes included those encoding resistance to aminoglycosides and the β-lactamase. Among 18 gene cassettes, there were 12 different gene arrays in the integron gene cassette systems and two new alleles were observed (95% aadA1 and 89% aadA1) in A. baumannii.
The production of aminoglycoside modifying enzymes (AMEs) confer to bacteria the ability to modify antibiotic's amino or hydroxyl functions and it's the main mechanism for the bacteria that provide resistance to aminoglycosides. Aminoglycoside modifying enzymes include the aminoglycoside acetyltransferase (AAC), aminoglycoside adenyltransferase (AAD), aminoglycoside nucleoside transferase (ANT), aminoglycoside phosphotransferase (APH) and their isozyme[25]. AAC (3)-I and AAC (6′)-I were found frequently in integron gene cassettes[26]. In our study, in A. baumannii, aminoglycoside resistance determinants, including aac(3)-I, aadA1 were predominantly. The high prevalence of aminoglycoside resistance genes in the A. baumannii integrons described here was comparable to that observed in other studies[27]–[30]. However our study is the first to report the presence of AacC1-AAC(3) I-aadA1, AacC1-aadA1, AAC(3)-I, AAC(3)-I-AAC(3)-I-aadA1, TEM-1, AAC(3)-I-aadA1-AAC(3)-I-AAC(3)-I, AAC(3)-I-AAC(3)-I-AAC(3)-I-aadA1, AAC(3)-I-aadA1, AAC(3)-I-AAC(3)-I, AAC(3)-I-aadA1-AAC(3)-I-aadA1, AAC(3)-I-AAC(3)-I-aadA1-AAC(3)-I-aadA1 in A. baumannii from clinical isolates in Turkey. Aminoglycoside resistance genes containing isolates were found in seven of nine different cities in Turkey. This finding indicates that the aminoglycoside resistance genes are predominant in A. baumannii in Turkey, to our knowledge.
The main mechanism of resistance to β-lactams in A. baumannii is enzymatic degradation by β-lactamases[31]. AmpC cephalosporinase is chromosomally encoded by A. baumannii, other β-lactamases TEM-92, TEM-116, SHV-12, CTX-M-2, TEM-1 and TEM-2 have been described, and we have identified TEM-1 type beta-lactamases in one strain (ANKA14). In conclusion, class 1 integron appear to be widely disseminated amongst clinical isolate of A. baumannii from Turkey. These data show that, 18 resistance gene cassettes were detected and 12 different gene cassette arrays were identified from A. baumannii. The wide distribution of integron and A. baumanni may pose a serious threat to the development of future effective antimicrobial therapies. This is the first extensive study on the distribution of class 1 integron among A. baumannii in Turkey.
Acknowledgments
This work was supported by Recep Tayyip Erdogan University, Research Fund Grants BAP-2012.102.03.4. and BAP-2013.102.03.4.
Comments
Background
A. baumannii is an opportunistic pathogen that is mostly cause of septicaemia, pneumonia and urinary tract infection. Recently, antibiotic resistance has increased in A. baumannii. The reason for this is gene acquisition. The gene acquisition is mediated by mobile genetic elements such as plasmids, transposons and integrons.
Research frontiers
A. baumannii is the important nosocomial infection agent in Turkey. It has acquired quickly wide variety of antibiotic resistance genes. Therefore, this is important to investigate the antibiotic resistance genes in these bacteria in Turkey.
Related reports
Mirnejad et al. (2013) have studied prevalence of integrons class 1 and class 2 in A. baumannii in Tehran, Iran. They showed that class 1 and class 2 integrons are prevalent in clinical isolates of A. baumannii in Iran.
Innovations and breakthroughs
This is first nationwide study of class 1 integron amoung clinical isolates of A. baumannii in Turkey. This study has found two novel alleles and 12 different gene cassettes have determined. Data showed that aminoglycoside modifying enzymes are widely found in Turkey.
Applications
It may be important to know the existence of class 1 integron in A. baumannii and distribution of A. baumannii in hospitalized patients. The results of this study suggest that class 1 integron is an important role in the spread of antibiotic resistance among clinical isolates in Turkey. Therefore, it is significant to monitor antibiotic resistance gene by PCR and sequence analysis.
Peer review
This is a good study in which authors evaluated the antibiotic resistance and carriage class 1 integron in clinical isolates of A. baumannii. The results are very valuable and suggested that class 1 integron is common in A. baumannii isolated from different hospitals in Turkey.
Footnotes
Foundation Project: Supported by Recep Tayyip Erdogan University (Grant No. BAP-2012.102.03.4. and BAP-2013.102.03.4).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. 2007;20:79–114. doi: 10.1128/CMR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontana C, Favaro M, Minelli S, Bossa MC, Testore GP, Leonardis F, et al. et al. Acinetobacter baumannii in intensive care unit: a novel system to study clonal relationshipamong the isolates. BMC Infect Dis. 2008;8:79–88. doi: 10.1186/1471-2334-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42:692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 5.Michalopoulos A, Falagas ME. Treatment of Acinetobacter infections. Expert Opin Pharmacother. 2010;11:779–788. doi: 10.1517/14656561003596350. [DOI] [PubMed] [Google Scholar]
- 6.Neonakis IK, Spandidos DA, Petinaki E. Confronting multidrugresistant Acinetobacter baumannii: a review. Int J Antimicrob Agents. 2011;37:102–109. doi: 10.1016/j.ijantimicag.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti-Bragadin C, et al. et al. Molecular characterizationof integrons in epidemiologically unrelated clinical isolatesof Acinetobacter baumannii from Italian hospitals reveals a limiteddiversity of gene cassette arrays. Antimicrob Agents Chemother. 2002;46:3665–3668. doi: 10.1128/AAC.46.11.3665-3668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 9.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 10.Labbate M, Case RJ, Stokes HW. The integron/gene cassette system: an active player in bacterial adaptation. Methods Mol Biol. 2009;532:103–125. doi: 10.1007/978-1-60327-853-9_6. [DOI] [PubMed] [Google Scholar]
- 11.Gillings M, Holley M, Stokes H, Holmes A. Integrons in Xanthomonas: a source of species genome diversity. Proc Natl Acad Sci U S A. 2005;102:4419–4424. doi: 10.1073/pnas.0406620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur A, Prakash P, Anupurba S, Mohapatra TM. Possible role of integrase gene polymerase chain reaction as an epidemiological marker: study of multidrug-resistant Acinetobacter baumannii isolated from nosocomial infections. Int J Antimicrob Agents. 2007;29:446–450. doi: 10.1016/j.ijantimicag.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012;35:317–325. [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing (M100-S23) Wayne: CLSI; 2013. [Google Scholar]
- 15.Rosser SJ, Young HK. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44:11–18. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 16.Lévesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. Cold Spring Harbor, New York: Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahbar M, Mehrgan H, Aliakbari NH. Prevalence of antibioticresistant Acinetobacter baumannii in a 1000-bed tertiary care hospital in Tehran, Iran. Indian J Pathol Microbiol. 2010;53:290–293. doi: 10.4103/0377-4929.64333. [DOI] [PubMed] [Google Scholar]
- 20.Taherikalani M, Maleki A, Sadeghifard N, Mohammadzadeh D, Soroush S, Asadollahi P, et al. et al. Dissemination of class 1, 2 and 3 integrons among different multidrug resistant isolates of Acinetobacter baumannii in Tehran hospitals, Iran. Pol J Microbiol. 2011;60:169–174. [PubMed] [Google Scholar]
- 21.Roy CP, Ingold A, Vanegas N, Martínez E, Merlino J, Merkier AK, et al. et al. Disseminationof multiple drug resistance genes by class 1 integrons in Klebsiella pneumoniae isolates from four countries: a comparative study. Antimicrob Agents Chemother. 2011;55:3140–3149. doi: 10.1128/AAC.01529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramírez MS, Stietz MS, Vilacoba E, Jeric P, Limansky AS, Catalano M, et al. et al. Increasing frequency of class 1 and 2 integrons in multidrug-resistant clones of Acinetobacter baumannii reveals the need for continuous molecular surveillance. Int J Antimicrob Agents. 2011;37:175–177. doi: 10.1016/j.ijantimicag.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Lee MF, Peng CF, Hsu HJ, Toh HS. Use of inverse PCR for analysis of class 1 integronscarrying an unusual 3′ conserved segment structure. Antimicrob Agents Chemother. 2011;55:943–945. doi: 10.1128/AAC.00988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu K, Wang F, Sun J, Wang Q, Chen Q, Yu S, et al. et al. Class 1 integron gene cassettes in multidrug-resistant Gram-negative bacteria insouthern China. Int J Antimicrob Agents. 2012;40:264–267. doi: 10.1016/j.ijantimicag.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Update. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Xu B, Yang Y, Liu D, Yang M, Wang J, et al. et al. A high throughput multiplex PCR assay for simultaneous detection of seven aminoglycoside-resistance genes in Enterobacteriaceae. BMC Microbiol. 2013;14:13–58. doi: 10.1186/1471-2180-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kansakar P, Dorji D, Chongtrakool P, Mingmongkolchai S, Mokmake B, Dubbs P. Local dissemination of multidrug-resistant Acinetobacter baumannii clones in a Thai hospital. Microb Drug Resist. 2011;17:109–119. doi: 10.1089/mdr.2010.0062. [DOI] [PubMed] [Google Scholar]
- 28.D'Arezzo S, Capone A, Petrosillo N, Visca P, Ballardini M, Bartolini S, et al. et al. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy) Clin Microbiol Infect. 2009;15:347–357. doi: 10.1111/j.1469-0691.2009.02668.x. [DOI] [PubMed] [Google Scholar]
- 29.Koo SH, Kwon KC, Cho HH, Sung JY. Genetic basis of multidrugresistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong Province, Korea. Korean J Lab Med. 2010;30:498–506. doi: 10.3343/kjlm.2010.30.5.498. [DOI] [PubMed] [Google Scholar]
- 30.Huang LY, Chen TY, Lu PL, Tsai CA, Cho WL, Chang FY, et al. et al. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin Microbiol Infect. 2008;14:1010–1019. doi: 10.1111/j.1469-0691.2008.02077.x. [DOI] [PubMed] [Google Scholar]
- 31.Zarrilli R, Giannouli M, Tomasone F, Triassi M, Tsakris A. Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J Infect Dev Ctries. 2009;5:335–341. doi: 10.3855/jidc.240. [DOI] [PubMed] [Google Scholar]