Summary
In mitotic cells, RAD9A functions in repairing DNA double-strand breaks (DSBs) by homologous recombination and facilitates the process by cell cycle checkpoint control in response to DNA damage. DSBs occur naturally in the germline during meiosis but whether RAD9A participates in repairing such breaks is not known. In this study, we determined that RAD9A is indeed expressed in the male germ line with a peak of expression in late pachytene and diplotene stages, and the protein was found associated with the XY body. As complete loss of RAD9A is embryonic lethal, we constructed and characterized a mouse strain with Stra8-Cre driven germ cell-specific ablation of Rad9a beginning in undifferentiated spermatogonia in order to assess its role in spermatogenesis. Adult mutant male mice were infertile or sub-fertile due to massive loss of spermatogenic cells. The onset of this loss occurs during meiotic prophase, and there was an increase in the numbers of apoptotic spermatocytes as determined by TUNEL. Spermatocytes lacking RAD9A usually arrested in meiotic prophase, specifically in pachytene. The incidence of unrepaired DNA breaks increased, as detected by accumulation of γH2AX and DMC1 foci on the axes of autosomal chromosomes in pachytene spermatocytes. The DNA topoisomerase IIβ-binding protein 1 (TOPBP1) was still localized to the sex body, albeit with lower intensity, suggesting that RAD9A may be dispensable for sex body formation. We therefore show for the first time that RAD9A is essential for male fertility and for repair of DNA DSBs during meiotic prophase I.
Key words: RAD9A, Double-strand breaks, Meiosis, Spermatogenesis
Introduction
Mouse Rad9a is an evolutionarily conserved gene with a diverse set of functions important primarily for promoting genomic integrity (Lieberman, 2006). RAD9A protein is a member of the heterotrimeric 9-1-1 complex, which consists of RAD9A, HUS1 and RAD1, and carries out many of its activities as part of this complex. Deletion of Rad9a causes embryonic lethality (Hopkins et al., 2004) and mice with a conditional knock out in keratinocytes demonstrate enhanced susceptibility to the development of carcinogen-induced skin tumors (Hu et al., 2008). Mice heterozygous for Rad9a are prone to spontaneous as well as radiation-induced cataractogenesis (Kleiman et al., 2007). Conversely, aberrantly high levels of human RAD9A play a functional role in prostate carcinogenesis (Broustas and Lieberman, 2012; Zhu et al., 2008).
The RAD9A protein is intimately involved in the cellular response to DNA damage. It regulates cell cycle checkpoints in mitosis through participation in both the ATR as well as ATM signaling pathways (Furuya et al., 2004; Lee et al., 2007; Niida and Nakanishi, 2006). Human RAD9A colocalizes with the phosphorylated form of histone H2A variant X (γH2AX) after DNA damage, and this is independent of ATM function (Greer et al., 2003). Furthermore, RAD9A inactivation delays the appearance of ionizing radiation-induced γH2AX foci, which together argue that RAD9A may modulate chromatin structure in response to DNA damage (Pandita et al., 2006). RAD9A also influences DNA repair directly. It can physically interact with proteins involved in base excision repair, mismatch repair and homologous recombination repair, and consequently, modify their activity and modulate the respective DNA repair pathways (Chang and Lu, 2005; Gembka et al., 2007; Guan et al., 2007; He et al., 2008; Pandita et al., 2006; Park et al., 2009; Smirnova et al., 2005; Toueille et al., 2004; Wang et al., 2004). There is also evidence that RAD9A participates in nucleotide excision repair (Li et al., 2013).
DNA repair is an important part of the normal progression of meiosis and the function of the 9-1-1 complex for meiosis in vivo has been demonstrated in several non-mammalian systems. The S. cerevisiae homolog of RAD9A, Ddc1, is found at sites of double-strand breaks (DSBs) in meiosis and is required for the pachytene checkpoint (Hong and Roeder, 2002). It appears that in this model, the 9-1-1 complex is bound to the synaptonemal complex (SC) via Red1 and is essential for SC formation (Eichinger and Jentsch, 2010). A possible role in DNA repair for one member of the complex, HUS1, had been described during oogenesis in Drosophila melanogaster wherein females with a null hus1 mutation were sterile, mainly due to oocyte nuclear defects similar to those produced by mutations in the spindle class of DNA DSB repair enzymes (Abdu et al., 2007). Furthermore, the Drosophila hus1 gene is required for proper disassembly of the SC for efficient processing of DNA DSBs (Peretz et al., 2009).
The roles of mammalian RAD9A in cell cycle checkpoint control, apoptosis and DNA repair are well established in mitotically dividing cells. All of these activities are critical for gametogenesis, but no prior studies have documented involvement of the 9-1-1 complex and specifically RAD9A in mammalian meiosis. The early embryonic lethality of Rad9a null mice is accompanied by increased apoptotic activity and reduced cellular proliferation (Hopkins et al., 2004). While these findings clearly underscore that Rad9a is an essential gene in mammals, the embryonic lethality obviated understanding its function in the germ line by use of this null mutation. We initially determined RAD9A's pattern of expression in the male germ line and then examined the effect of loss of its function on the progression of cells through mitosis and meiosis. In order to investigate its function during spermatogenesis, we generated mice in which a conditional Rad9a allele was ablated specifically in undifferentiated spermatogonia by Cre recombinase expressed under the Stra8 promoter. While the mitotic divisions of spermatogonia appeared to be unaffected, RAD9A-deficiency had drastic effects on meiosis. We determined for the first time that RAD9A is essential in males for progression through meiosis and for the efficient repair of meiotic DNA DSBs.
Results
The distinct expression pattern of RAD9A in testicular cells suggests a role in meiosis
It is well established that in somatic cells RAD9A has multiple functions, including DNA repair and checkpoint control (Lieberman, 2006). To determine whether RAD9A has similar functions in male germ cells, we first characterized the pattern of expression of its protein and mRNA in wild type (WT) mouse testis. We examined the abundance of RAD9A protein by immunoblotting of total testis extracts from mice of ascending age (Fig. 1A). With the onset of meiosis at postnatal day (pnd) ∼10, the level of RAD9A increased and peaked coincident with the abundance of spermatocytes in late pachytene and diplotene stage at pnd18 (Bellvé et al., 1977). The appearance of haploid germ cells at ∼pnd20 and their relative prevalence in the testis thereafter was accompanied by a drop in RAD9A levels at pnd28 and in adult testis. This most likely reflected low or no expression in spermatids. To test this hypothesis and determine the cellular localization of the protein, we detected RAD9A in testis by immunohistochemistry and observed strong signal in the nuclei of prophase spermatocytes (Fig. 1B, red arrows). Lower levels of nonetheless consistent signal were also seen in Sertoli cells (Fig. 1B, green arrowheads). Spermatids contained a comparatively weak signal, if at all, and spermatogonia did not seem to express RAD9A. It is possible, however, that basal expression of RAD9A in spermatogonia is below the detection limits of the assay.
Fig. 1.
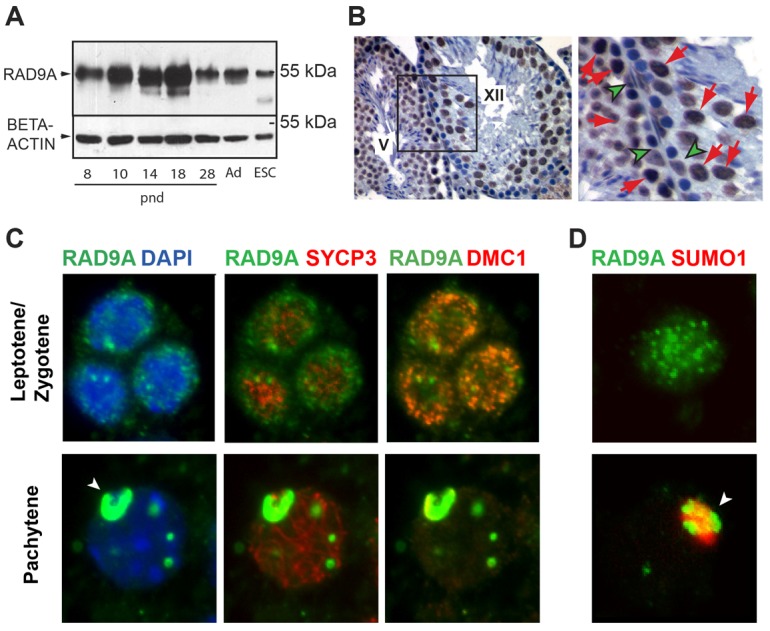
Levels of RAD9A protein in mouse testis. (A) RAD9A protein abundance detected by immunoblotting of total testis extracts (50 µg/lane) isolated from juvenile mice of ascending age, adult (Ad) wild-type testis and wild-type mouse ES cells. Beta-actin was a loading control. Mouse age (postnatal day, pnd) is indicated under each lane. (B) Localization of RAD9A protein in adult mouse testis determined by immunohistochemistry in sections from adult Rad9af/+ testis at 40× (left panel or at 100×, right panel). RAD9A signal (brown) was detected in the nucleus of pachytene spermatocytes (red arrows) and Sertoli cells (green arrowheads). Roman numerals indicate the stage of the mouse seminiferous epithelial cycle. Hematoxylin (blue) marks the nuclei. Spermatids were only weakly labeled. (C) Immunostaining for RAD9A, SYCP3 and DMC1 in squash preparations of adult seminiferous tubules. Left panels show RAD9A (green) and DAPI (blue; revealing nuclear localization); middle panels, SYCP3 (far red), RAD9A (green); right panels, DMC1 (red), RAD9A (green). Upper panels: leptotene/zygotene stage; lower panels: pachytene stage. In leptotene/zygotene spermatocytes, there are bright DMC1 foci at DSBs and the RAD9A signal is diffuse with some speckled aggregates. In pachytene, RAD9A localizes primarily to the XY body (arrowhead), with a few additional regions staining positive, in distinct foci not associated with chromatin. (D) Colocalization of RAD9A (green) with SUMO1 (red). SUMO1 was not detected in zygotene spermatocytes (upper panel). In pachytene spermatocytes (lower panel) RAD9A localized primarily to the XY body (arrowhead) with a few additional foci as in C. Colocalization of RAD9A and SUMO1 in a pachytene spermatocyte confirms localization of RAD9A to the XY body (arrowhead).
These data are consistent with the profile of Rad9a RNA expression measured by qRT-PCR in total RNA isolated from testes of similarly aged juvenile mice (supplementary material Fig. S1A). RNA levels reflected RAD9A protein expression, with an increase at the entry of germ cells into meiosis and peaking toward the end of the first meiotic prophase at pnd17. This trend was supported by the higher levels of Rad9a RNA in isolated primary spermatocytes as compared to round spermatids (supplementary material Fig. S1A). The cellular distribution of Rad9a RNA was also confirmed by in situ hybridization analysis on adult testis, where Rad9a signal was detected in meiotic prophase spermatocytes and also in Sertoli cells, but not in round spermatids nor in spermatogonia (supplementary material Fig. S1B).
To determine the distribution and precise sub-cellular localization of RAD9A in spermatocytes, we carried out a detailed immunofluorescence study on squash preparations of testicular tubules. Staging of meiotic prophase I nuclei was based on morphology of the SC (detected with mouse anti-SYCP3 antibody), as well as the presence of foci of DNA DSBs (visualized with rabbit anti-DMC1 antibody). Leptotene-to-zygotene spermatocytes are characterized by fragmentary SYCP3 signal on homologous chromosomes and the presence of multiple DMC1 foci (Fig. 1C, top row) (Moens et al., 2002). At this stage, we found RAD9A clustered in multiple foci as well as diffusely distributed throughout the nucleus. In contrast, by mid-pachytene, when DSBs on autosomes are repaired and chromosome synapsis is complete, RAD9A localizes primarily to the XY body (Fig. 1C, bottom row). There were a few remaining strong RAD9A foci throughout the nucleus, which apparently were not colocalized with SYCP3 at axial elements (AE) on chromosomes. In pachytene spermatocytes, SUMO1 appears exclusively at the X and Y bivalent (Rogers et al., 2004). Colocalization of RAD9A with SUMO1 confirmed its association with the XY body at this stage (Fig. 1D). This distinct nuclear distribution of RAD9A in spermatocytes suggests at least two possible functions for RAD9A during male meiosis – participation in the repair of DSBs and/or a role in the formation and maintenance of the XY body.
Mice with conditional deletion of Rad9a in the testis are infertile
Whole body Rad9a deletion in mice is embryonic lethal (Hopkins et al., 2004), thus precluding analysis of RAD9A-deficient germ cells under these conditions. Therefore, to study the role of the protein in spermatogenesis it was necessary to generate animals with a male germ cell-specific deletion of a floxed Rad9a allele. As we reported previously (Hu et al., 2008), Rad9af/f mice are viable and normal, consistent with our expectation that the ‘floxed’ Rad9a allele is functionally WT. We crossed homozygous Rad9af/f mice with a ‘deleter’ strain, in which Cre expression is driven by the Stra8 promoter, shown previously to drive Cre expression in undifferentiated type A spermatogonia (Sadate-Ngatchou et al., 2008). Excised floxed alleles are designated as Rad9adel to distinguish them from the originally obtained Rad9a− alleles (Hopkins et al., 2004). We compared expression of Rad9a in Rad9af/+, heterozygous-like Rad9af/del, and mutant Rad9af/delCre+ testes, and found about half the levels of Rad9af/+ expression in Rad9af/del testes and over a 3.5-fold reduction of RNA levels in the mutant testes (Fig. 2A).
Fig. 2.
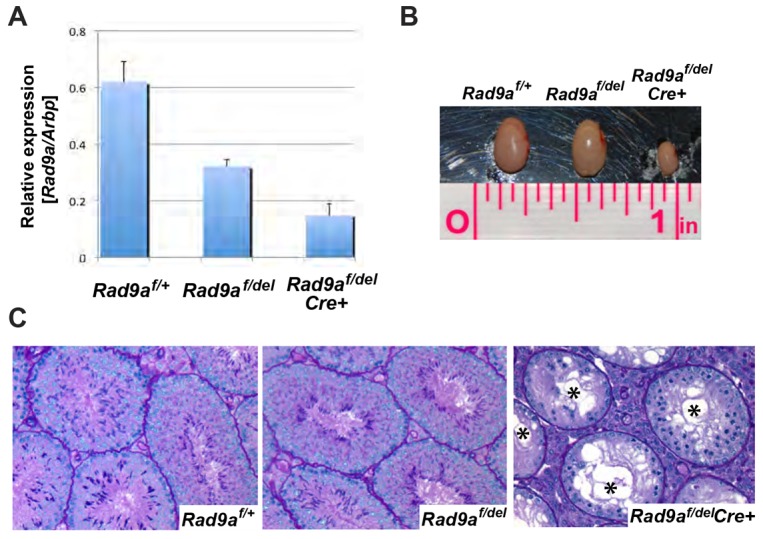
Testis-specific deletion of Rad9a leads to reduced testis size and loss of germ cells. (A) Quantitative RT-PCR for Rad9a mRNA in whole testis from Rad9af/+, Rad9af/del and Rad9af/delCre+ mice. Arbp is used as an internal control. The twofold reduction in Rad9a mRNA level in the heterozygous-like Rad9af/del mouse testis is similar to that in Rad9a+/− ES cells (Hopkins et al., 2004). In the Rad9af/delCre+ mutant, Rad9a message was reduced to less than a third of that in the Rad9af/+ control. Expression of Rad9a in Sertoli cells probably accounts for the levels detected. (B) Testes from Rad9af/+, Rad9af/del and Rad9af/delCre+ mice. Rad9af/+ and Rad9af/del mouse testes were equivalent in size. However, the Rad9af/delCre+ mouse testis was one-third the size of the other two. (C) Histological examination of testicular sections from adult Rad9af/+ (left), Rad9af/del (middle) and Rad9af/delCre+ (right) littermates stained with periodic acid-Schiff (PAS). The testicular section from a sterile Rad9af/delCre+ male shows depletion of germ cell populations in most tubules, vacuolization, agametic tubules (asterisks) and lack of round or elongating spermatids. Tubules with one layer of cells were found at stages where there should normally be three to four layers.
Next, we determined if loss of RAD9A function in the germ line affects the reproductive potential of the mutant males. Three rounds of mating of an initial cohort of Rad9af/delCre+ males (n = 7) each with two wild-type CD1 females resulted in complete infertility (Table 1; supplementary material Fig. S2A–C, group Rad9af/delCre+-1). A second cohort of Rad9af/delCre+ males (n = 7) was produced by further matings of the Rad9af/+Cre+ progeny, which was maintained on a mixed 129SvEv, C57Bl/6 and FVB background to Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J originating from a cross between 129X1/SvJ and C57Bl/6J. Most mutant mice from this cohort were initially sub-fertile rather than completely infertile (Table 1; supplementary material Fig. S2A–C, group Rad9af/delCre+-2). Overall, nine out of fourteen (64.3%) males did not produce any pups over the course of three months. The five remaining males exhibited declining fertility as they only fathered pups during the first or the first and second rounds of mating. Statistical analysis of the three rounds of matings was performed and mouse cohorts with respective genotypes were grouped into classes (‘a’ and ‘b’) according to similarities in statistically significant differences in litter size. These analyses demonstrated that after the first round, the fertility of the first Rad9af/delCre+ cohort (Table 1, mice 1–7; supplementary material Fig. S2A, Rad9af/delCre+-1) formed a class of its own (supplementary material Fig. S2A, class b) and was significantly different from the three other groups (supplementary material Fig. S2A, class a), while that of the second Rad9af/delCre+ cohort (Table 1, mice 8–14; supplementary material Fig. S2A, Rad9af/delCre+-2) was not significantly different from that of Rad9af/+ and Rad9af/del littermates (Table 1; supplementary material Fig. S2A, class a). However, after the second and third matings (supplementary material Fig. S2B,C), the fertility of Rad9af/+ and the second Rad9af/delCre+ cohort was found to be significantly different and they fell into two different classes, a and b, respectively.
Table 1. Summary of Rad9af/+, Rad9af/del and Rad9af/delCre+ mouse fertility study.
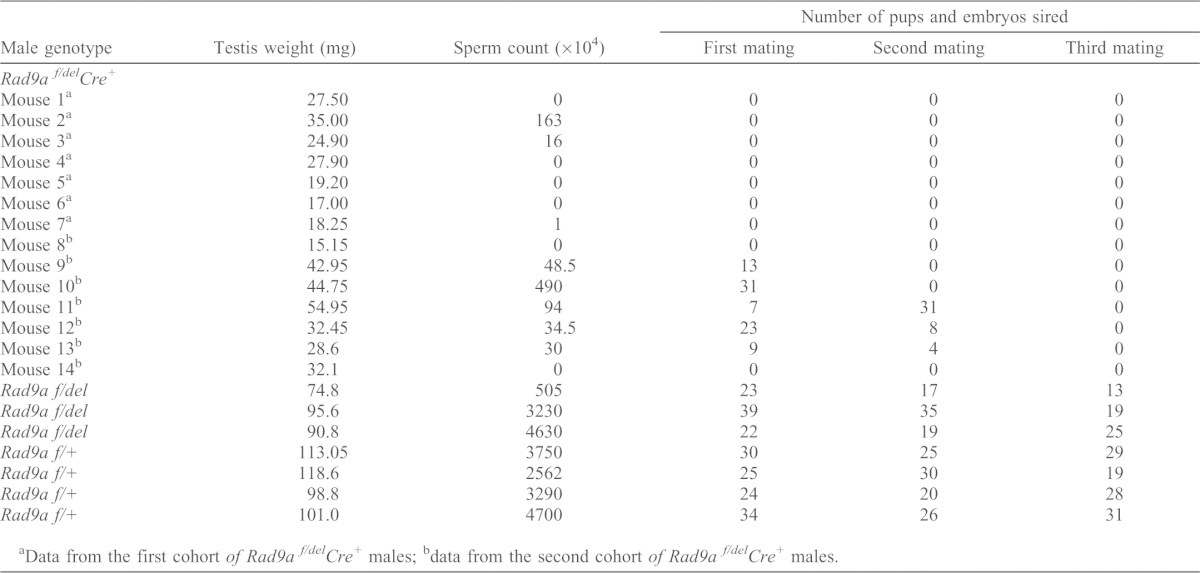
Data from the first cohort of Rad9a f/delCre+ males;
data from the second cohort of Rad9a f/delCre+ males.
After completion of the study at 5.5 months of age, the reproductive tracts of the mated males were excised and examined at the gross morphological and histological levels. Testes of the completely infertile Rad9af/delCre+ males were about a third of the size of Rad9af/+ or Rad9af/delCre− testes (Fig. 2B), and testicular sections showed severe loss of germ cells (Fig. 2C). The most severely affected tubules had gross vacuolization (Fig. 2C, asterisks) and appeared almost empty, containing mainly Sertoli cells, spermatogonia and an occasional spermatocyte.
We next compared the morphology and the degree of disruption of spermatogenesis in the sub-fertile cohort with that of the completely infertile mutant males (supplementary material Fig. S3). Histological analysis of epididymal sections showed that at the end of the study, both the initially sub-fertile (supplementary material Fig. S3B) and the completely infertile (supplementary material Fig. S3C) mutants were virtually devoid of sperm, consistent with the low sperm counts and lack of progeny at the third mating (Table 1). Although many testicular tubules from sub-fertile males contained elongating spermatids and even sperm (supplementary material Fig. S3E), they also displayed a range of abnormalities characteristic of the completely infertile cohort (supplementary material Fig. S3F) but not seen in controls (supplementary material Fig. S3D). In testes of infertile mice, between 91.2 and 97.8% of tubules exhibited gross vacuolization with a single exception of a sterile male having only 12.5% of tubules containing more than two large vacuoles. In contrast, only 2–10% of testicular tubules in sub-fertile males were highly vacuolized. Note that such vacuolization was not observed in the Rad9afl/+ testes. Multinucleated giant cells were present in ∼7% of testicular tubules from infertile males (supplementary material Fig. S3F, red arrows) and in ∼2% of tubules in sub-fertile males (supplementary material Fig. S4A,B, red arrows). Among the multiple abnormalities found in both cohorts were condensed, apoptotic nuclei of early prophase I spermatocytes (supplementary material Fig. S3E,F,and Fig. S4, black arrows) and abnormal metaphase figures (supplementary material Fig. S4F, arrowheads). Partial germ cell loss in sub-fertile testes (supplementary material Fig. S3E) was typically characterized by lack of second and/or third germ cell layers on half or more of the tubule perimeter (supplementary material Fig. S3E, asterisk and Fig. S4C, asterisks). Interestingly, spermatogonia appeared normal (supplementary material Fig. S4, marked with ‘s’).
We next asked whether incomplete excision of the floxed allele could contribute to the heterogeneity of the mutant phenotypes. We first examined the efficiency of the Rad9a ‘flox’ allele excision by immunostaining for RAD9A protein in Rad9af/+ and Rad9af/delCre+ testicular sections. Rad9af/+ pachytene spermatocytes stain positive for RAD9A at the XY body and form a continuous layer in the tubule (Fig. 3A). However, there were RAD9A-positive spermatocytes along part of the perimeter of the tubules in mutant testes (Fig. 3B, arrows), suggesting that Cre excision had not occurred in these cells. Immunohistochemical staining of testicular sections with our anti-RAD9A antibody revealed that RAD9A was still expressed in ∼37% of the remaining spermatocytes in testes of sub-fertile (supplementary material Fig. S5B) and infertile (supplementary material Fig. S5C,D) Rad9af/delCre+ males. This was not surprising in view of the previously reported lack of expression of the Stra8-Cre transgene (Sadate-Ngatchou et al., 2008) in some PLZF-positive spermatogonia, resulting in a failure to excise a floxed allele (Hobbs et al., 2012). The RAD9A-deficient spermatocytes detected appeared abnormal, as indicated by a weak and fragmentary SYCP3 signal (Fig. 3B, right panel).
Fig. 3.
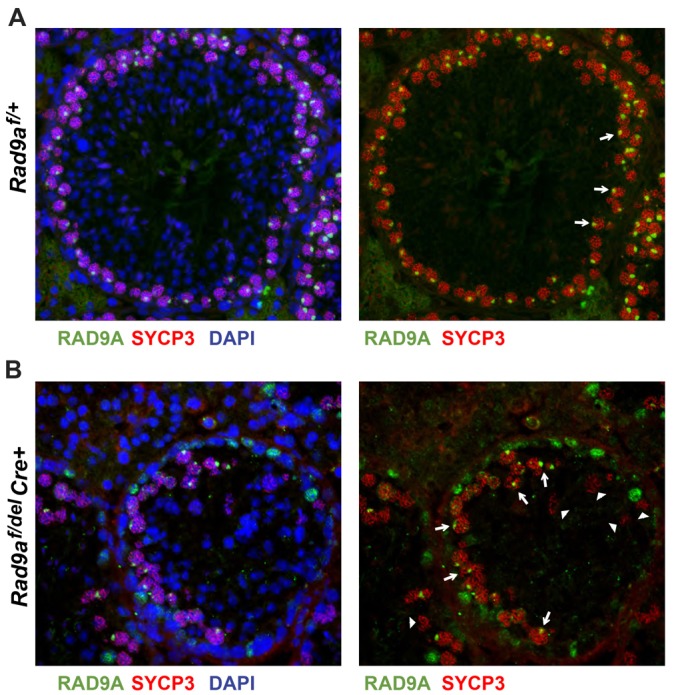
Spermatocytes in Rad9af/delCre+ testis can escape floxed allele excision. (A,B) Immunofluorescence was performed on testicular sections from adult Rad9af/+ (A) and Rad9af/delCre+ (B) mice. Left panels: detection of RAD9A (green), SYCP3 (red) and nuclei (by DAPI staining; blue). Right panels: for clarity, only RAD9A and SYCP3 are shown. Pachytene spermatocytes (red SYCP3 on chromosomes) in Rad9af/+ testis formed an uninterrupted concentric circle with bright RAD9A signal (green) on the XY body (A, right panel, white arrows). RAD9A-positive spermatocytes were readily detected in Rad9af/delCre+ tubules indicating lack of ‘flox’ allele excision (B, right panel, white arrows). A few spermatocytes with interrupted chromosome cores and lower intensity of SYCP3 appear RAD9A-negative (white arrowheads). Rad9af/+ tubules were filled with structured layers including round and elongated haploid spermatids, and fewer ‘DAPI only’ haploid nuclei were detected in the lumen of Rad9af/delCre+ tubules, confirming that these cells have escaped excision of the conditional Rad9a allele.
We therefore asked whether this inefficiency of excision was due to low Cre activity or was dependent on locus context. To assess Cre activity, we crossed Rad9af/delCre+ mice with a second mouse line, Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG), designed as a reporter for efficiency of excision (Muzumdar et al., 2007). The efficiency of excision of the tdTomato transgene was assessed by scoring the fraction of cells expressing EGFP, i.e. fluorescence turned from red to green, in single-cell suspensions from Rad9af/+Cre+Tom+ and Rad9af/delCre+Tom+ testes using a previously reported flow cytometric assay (Bastos et al., 2005; Vasileva et al., 2009). In the Rad9af/+Cre+Tom+ adult testes, 91.4% of the prophase spermatocytes were EGFP positive, while in the Rad9af/delCre+Tom+ male there were 96.5% EGFP-positive spermatocytes (these were the mice used in cohort 2 of the fertility studies), which corroborates previously published >95% efficiency of excision for the Stra8-Cre transgene (Sadate-Ngatchou et al., 2008; Yang et al., 2012). Therefore, incomplete excision of the Rad9a allele is not likely a consequence of low Cre activity or frequent occurrence of cells lacking Cre expression. Incomplete excision, as demonstrated by an unrecombined floxed allele, was also observed in offspring born to wild-type females mated to Rad9af/delCre+ males from the fertility study. Taken together, these data indicate that Rad9a deletion in the Stra8-Cre model was incomplete and that the heterogeneity of the Rad9a mutant phenotype may be due to inefficient excision precluded by locus-specific genomic context of the Rad9a conditional allele within a subset of spermatogonia.
RAD9A-deficient germ cells are eliminated by apoptosis
It is well established in cell lines that RAD9A has an anti-apoptotic function (Kobayashi et al., 2004), and Rad9a−/− mouse ES cells demonstrate high spontaneous levels of apoptosis (Zhu et al., 2005). We therefore assessed the number of apoptotic cells by TUNEL staining in histological testicular sections obtained from the infertile mice used in the fertility study (Fig. 4A–C). As is standard in such analysis of apoptosis in testicular tubules, quantification of apoptosis is presented as the number of apoptotic cells counted per 100 tubules and by calculating the apoptotic index (by multiplying the percentage of tubules containing apoptotic germ cells by the number of apoptotic germ cells per tubule; Fig. 4D) (Salazar et al., 2005). While the Rad9af/+ testes had the expected occasional apoptotic spermatocyte (6–9 TUNEL-positive cells/100 tubules), in the mutant there were numerous positive spermatocytes (Fig. 4B,C). We determined the approximate stage of the seminiferous cycle of the mutant tubules and observed apoptotic spermatocytes in tubules corresponding to stages beyond mid-pachytene (Fig. 4B,C, red arrowheads). Mutant males 1, 2, 3, 4 and 7 had intermediate loss of germ cells (Fig. 4B) and showed an ∼3-fold increase in the number of apoptotic cells/100 tubules, and more than 20-fold increase in apoptotic index, relative to controls. As expected, values for both the total apoptotic cells and the apoptotic index were downwardly skewed in mutants containing mainly agametic tubules (males 5 and 6), due to massive overall germ cell loss (Fig. 4C).
Fig. 4.

Elevated levels of apoptosis in adult testes of infertile Rad9af/delCre+ mice. (A–C) Testes from adult Rad9af/+ (A) and Rad9af/delCre+ (B,C) littermates were stained using the TUNEL technique, and sections were counterstained with Hematoxylin. Higher numbers of apoptotic cells appear in Rad9af/delCre+ testes with intermediate (B) as opposed to nearly complete germ cell loss (C). Tubules were staged according to Russell et al. (Russell et al., 1990) and stages indicated with Roman numerals. Asterisks after Roman numerals indicate that mutant tubules were staged with the closest possible approximation to a normal seminiferous tubule cycle. In the Rad9af/delCre+ testes prophase I spermatocytes were observed in tubules of all stages (red arrowheads). (D) Quantification of TUNEL-stained apoptotic cells was performed in round cross sections of tubules. Data were collected from at least 100 tubules per section from three different sections and average total apoptotic cells/100 tubules (gray columns) or apoptotic index (black columns) are shown. Numbers for the Rad9af/+ genotype were from two randomly picked males in this group (see Table 1; results from Rad9af/delCre+ males are listed individually).
Spermatocytes lacking RAD9A arrest at the late zygotene or early pachytene stage of meiotic prophase I
To identify the specific stage at which apoptosis was occurring, we quantified the proportion of each germ cell stage with specific attention to the number of spermatocytes at different stages of prophase I using flow cytometry as previously described (Bastos et al., 2005; Vasileva et al., 2009). In most mouse strains, the first meiotic division in the first spermatogenic wave occurs around pnd18–19 and round spermatids appear at ∼pnd20 (Bellvé et al., 1977). We chose this age for our analysis as differences in cellularity between mutant and Rad9af/+ testes would likely be the direct result of loss of RAD9A function rather than a secondary effect of massive germ cell loss due to improper cell–cell contact and general degeneration of seminiferous tubules, which is observed in adult mutant testes (Chung et al., 2004). As assays performed on different days tend to have variable intensity of the Hoechst staining, we adjusted the parameters in each assay by matching a control Rad9af/+ littermate for every mutant littermate. Testes of Rad9af/+ and of Rad9af/delCre+ littermates contained similar numbers of spermatogonia and preleptotene spermatocytes (Table 2; supplementary material Fig. S6). Strikingly, a notable reduction in the number of late prophase spermatocytes was detected in Rad9af/delCre+ as compared to Rad9af/+ littermates. This was occasionally accompanied by an increase in the numbers of early prophase I leptotene/zygotene spermatocytes. We conclude that the majority of RAD9A-deficient spermatocytes fail to progress through pachytene or they arrest early in this stage. Spermatids were readily detected in Rad9af/+ pnd20 littermates (∼7.07%) but not in mutant testes cell suspensions.
Table 2. Percentage of each germ cell population in testes of Rad9af/+ or Rad9af/delCre+ littermates at pnd20.
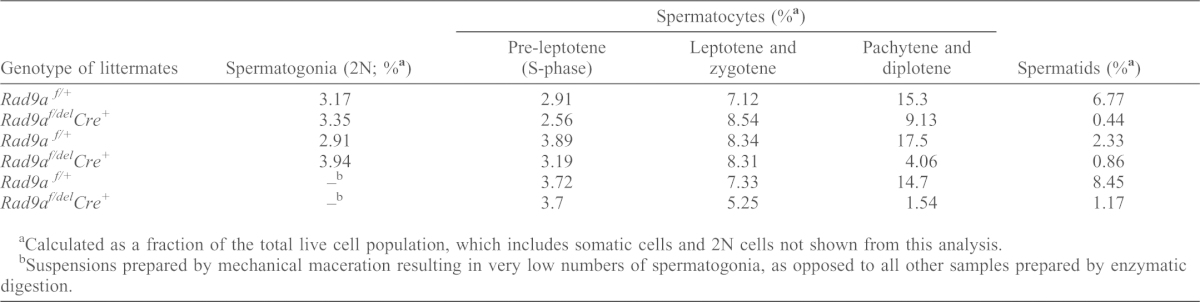
Calculated as a fraction of the total live cell population, which includes somatic cells and 2N cells not shown from this analysis.
Suspensions prepared by mechanical maceration resulting in very low numbers of spermatogonia, as opposed to all other samples prepared by enzymatic digestion.
RAD9A is essential for efficient repair of DNA DSBs on autosomes during prophase I
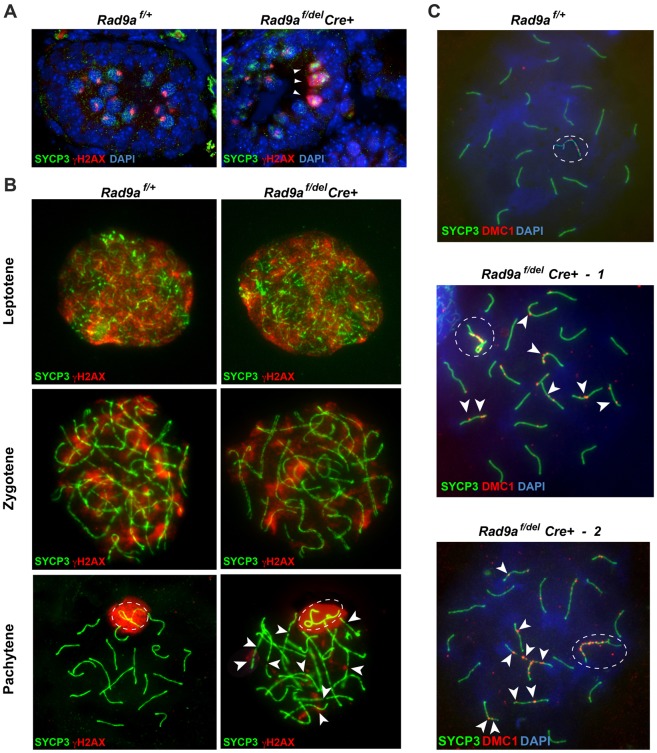
We next asked what might trigger arrest in early pachytene and ensuing apoptosis of RAD9A-deficient spermatocytes. We speculated that, given the role of RAD9A in DNA repair, spermatocytes lacking RAD9A may have defects in repairing the DNA DSBs necessary for homologous recombination. The pattern of distribution of γH2AX, a marker for DSBs, during meiosis is well documented (Bellani et al., 2005; Mahadevaiah et al., 2001). In leptotene and zygotene spermatocytes, it is usually interspersed throughout the nucleus and disappears by late zygotene, whereas in pachytene, strong γH2AX signal is solely restricted to the sex body (Fig. 5A, left panel). However, in some RAD9A-deficient pachytene spermatocytes, γH2AX was still seen throughout the nuclei (Fig. 5A, right panel). To investigate this characteristic at higher resolution, we assessed γH2AX in spermatocyte spreads from Rad9af/+ (Fig. 5B, left panels) and Rad9af/delCre+ testes (Fig. 5B, right panels). Both Rad9af/+ and Rad9af/delCre+ leptotene and zygotene spermatocytes displayed γH2AX signal (red) associated with SC at DSBs formed on autosomes (Fig. 5B, top and middle, left and right panels, respectively). While pachytene spermatocytes in the Rad9af/+ exhibited normal localization of γH2AX to the sex body (Fig. 5B, lower left panel), mutant spermatocytes exhibited various patterns of γH2AX distribution. Approximately 29% of RAD9A-deficient mid-pachytene spermatocytes and 21% of those late spermatocytes detected displayed normal localization of γH2AX to the sex body (Fig. 5B, lower right panel; supplementary material Fig. S7Ai and Bi, green arrows and Table S1). However, we also detected γH2AX associated with autosomes indicating unrepaired DNA DSBs. The γH2AX distribution in the remaining RAD9A-deficient spermatocytes appeared multilobular (supplementary material Fig. S7Bi, asterisks; Table S1).
Fig. 5.
Rad9af/delCre+ spermatocytes contain residual, unrepaired DNA DSBs. (A) Co-immunofluorescence on paraffin-embedded testis tissue from pnd13 Rad9af/+ (left panel) and Rad9af/delCre+ (right panel) littermates. Pachytene spermatocytes have fully synapsed chromosomes marked by SYCP3 (green) and correctly formed XY bivalents stained with γH2AX (red). Rad9af/delCre+ spermatocytes often have excessive γH2AX staining resembling an apoptotic response due to DNA damage (arrowheads). DAPI was used to stain nuclei. (B) Examples of typical Rad9af/+ and Rad9af/delCre+ leptotene (top panels) and zygotene (middle panels) spermatocytes, with partially synapsed chromosomes as indicated by interrupted SYCP3 signal and ‘clouds’ of γH2AX localized to sites of obligatory DNA DSBs. Lower panels: normal localization of γH2AX at the XY body of a Rad9af/+ (left panel) and Rad9af/delCre+ (right panel) pachytene spermatocytes. Autosomes in RAD9A-deficient pachytene spermatocytes retained γH2AX indicating persistent DNA damage (right panel). (C) Double immunolabeling of DMC1 (red) and SYCP3 (green) was performed on spermatocyte spreads from Rad9af/+ (top panel) and two different Rad9af/delCre+ (middle and bottom panel) testes. DAPI was used to stain nuclei. Mid-pachytene stage was determined by the extent of axial element formation and chromosome compaction and lack of thickening at the ends. In Rad9af/+ pachytene spermatocytes, DMC1 is cleared from autosomes, indicating complete repair of DNA DSBs. Multiple DMC1 foci were present on at least three autosomes in Rad9af/delCre+ spermatocytes at mid-pachytene (arrowheads). Dashed ovals indicate the location of the sex body.
To further evaluate the efficiency of DNA DSB repair in the Rad9a mutant and obtain data independent of γH2AX, which also localizes to broken DNA in cells undergoing apoptosis, we stained spread spermatocytes with antibodies against DMC1, which specifically mark meiotic DSBs. Rad9af/+ mid-pachytene spermatocytes had cleared DMC1 from autosomes completely (Fig. 5C, top panel), while same stage spermatocytes from mutant testes had retained numerous DMC1 foci on AEs (Fig. 5, white arrowheads). As expected DMC1 foci were found on the X chromosome (Moens et al., 2002), but appeared more prominent compared to foci in Rad9af/+ spermatocytes (Fig. 5C, encircled XY).
TOPBP1 localization in RAD9A-deficient spermatocytes
Persistence of γH2AX and DMC1 foci in RAD9A-deficient pachytene spermatocytes indicates a requirement for RAD9A protein in the repair of DNA DSBs. In S. pombe the Rad9 interacting protein TOPBP1 functions together with ATR in the activation of the recombination checkpoint during late prophase (Perera et al., 2004). A study in mitotic mammalian cells further demonstrated that RAD9A has a role in homologous recombination repair (Pandita et al., 2006). TOPBP1 is found on asynapsed, but not on synapsed chromosome axes in zygotene and also in the sex body in pachytene spermatocytes (Perera et al., 2004).
We therefore examined the localization of TOPBP1 in Rad9a conditional knockout spermatocytes. The distribution of TOPBP1 was similar in leptotene and early zygotene Rad9af/+ and mutant spermatocytes (Fig. 6A,Bi for Rad9af/+ and Fig. 6E,Fi for Rad9af/delCre+, respectively). In Rad9af/+ control late zygotene nuclei, we observed multiple TOPBP1 foci aligned with the SC only on asynapsed autosomes (Fig. 6Bii, blue arrowheads). This is in contrast to the previously reported localization of TOPBP1 on multiple asynapsed autosomes in zygotene (Perera et al., 2004). However, in RAD9A-deficient late zygotene spermatocytes strong TOPBP1 signal was present along the axes of multiple synapsed autosomes (Fig. 6Fii, blue arrowheads).
Fig. 6.
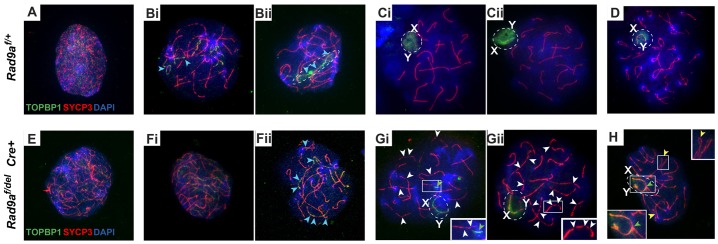
Localization of TOPBP1 in Rad9af/+ and Rad9af/delCre+ spermatocytes. Rad9af/+ (top row) and confirmed RAD9-deficient Rad9af/delCre+ (bottom row) nuclei labeled with anti-TOPBP1 (green), anti-SYCP3 (red), anti-RAD9A (far red, not shown) and DAPI (blue) are shown. Both Rad9af/+ (A) and Rad9af/delCre+ (E) leptotene spermatocytes contain multiple TOPBP1 foci throughout the nucleus. In Rad9af/+ (Bi,Bii) and Rad9af/delCre+ (Fi,Fii) zygotene spermatocytes, TOPBP1 is found along the unsynapsed regions of chromosome axes (blue arrowheads in Bi, Bii and Fii) and presumably on the X and Y (dashed oval in Bii). TOPBP1 signal in Rad9af/+ (Ci,Cii) and RAD9A-deficient (Gi,Gii) pachytene nuclei was observed at the unsynapsed regions of the X and Y (dashed ovals, X and Y label); however, it was quite weak in Rad9af/delCre+ pachytene-like spermatocytes (Gi,Gii,H) as opposed to a strong signal encompassing the entire sex body in Rad9af/+ spermatocytes (Ci,Cii). RAD9A-deficient nuclei retain diffuse TOPBP1 staining along the SC of autosomes; white arrowheads indicate interruptions in the SC in RAD9A-deficient pachytene spermatocytes (also insets in Gi,Gii); yellow arrowheads in H indicate paired homologs with partial asynapsis (top inset); green arrowhead in Gi shows TOPBP1 retained on autosomes in pachytene stage; green arrowhead in H indicates a mixed XY+autosomal domain (also H, bottom inset). Diplotene spermatocytes appeared normal in spreads from Rad9af/+ testis (D) and were not found in RAD9A-deficient spermatocytes from Rad9af/delCre+ testes.
Rad9af/+ pachytene and diplotene spermatocytes exhibited the characteristic intense cloud-like TOPBP1 signal encompassing the sex body (Fig. 6Ci, Cii and D). Although other characteristics such as γH2AX and SUMO1 localization in Rad9a mutant pachytene spermatocytes appear normal, TOPBP1 signal had lower intensity and was confined to the chromosome axes of the XY chromosomes in the sex body. In addition, RAD9A-deficient pachytene spermatocytes retained diffuse TOPBP1 signal on autosomes (Fig. 6Gi and Gii). We did not observe TOPBP1 at sites of fragmentary SC (Fig. 6Gi and Gii, white arrowheads, magnified in insets), and aligned, but not fully synapsed bivalents were typically observed in mid-pachytene nuclei (Fig. 6H, yellow arrowheads, upper inset). Along with fragmentary SC, RAD9A-deficient spermatocytes occasionally contained an extended XY body domain encompassing autosomes (Fig. 6Gi and H, green arrowheads, insets).
Discussion
Studies of meiosis in yeasts, fly and nematode indicate that the 9-1-1 complex plays critical roles in several activities related to this process, including DNA repair, partner choice for homologous recombination and induction of the meiotic checkpoint (Grushcow et al., 1999; Hofmann et al., 2002; Lieberman, 2006; Peretz et al., 2009). However, a meiosis-specific function for mammalian 9-1-1, and in particular the RAD9A component, has not yet been demonstrated, complicated in part by the fact that the Rad9a null is embryonic lethal (Hopkins et al., 2004). Therefore, we constructed and characterized a mouse strain bearing a conditional Rad9a knockout in undifferentiated spermatogonia to assess its role in spermatogenesis.
RAD9A is expressed in a distinct pattern in meiotic prophase spermatocytes
We demonstrated that RAD9A was readily detected in spermatocytes but surprisingly not in spermatogonia, implying a specific requirement during the meiotic phase of male germ cell differentiation. Early in meiosis, during the leptotene stage, RAD9A protein was present throughout the nucleus. Subsequently, in zygotene, this diffuse nuclear staining was accompanied by foci of RAD9A presumably associated with the chromosome axes, similar to the distribution of DMC1 foci. This observation, together with the previously reported physical association of RAD9A with RAD51 in mitotic cells (Pandita et al., 2006), suggested a similar interaction between RAD9A and the recombination related protein DMC1 during homology-directed repair of DSBs in meiosis. However, RAD9A foci did not fully overlap with DMC1 foci (Fig. 1C), which suggests that rather than a direct role in DNA end binding and homology search, RAD9A might be affecting DNA DSB repair indirectly, via its association with the checkpoint apparatus. Similar observations were made for RAD1, which did not colocalize completely with DMC1 in early meiotic prophase (Freire et al., 1998). The precise spatial and functional relationship between early recombination nodules containing DMC1 and mouse RAD9A foci needs further resolution.
It was previously shown that TOPBP1, a well-known binding partner of RAD9A (Mäkiniemi et al., 2001), shares a similar localization pattern at DNA repair foci together with ATR (Perera et al., 2004). Human CHK1, which requires phosphorylation by TOPBP1-bound ATR for full activation, was also found on meiotic chromosomes and has been proposed to function in DNA damage-induced checkpoint responses (Flaggs et al., 1997). The association of RAD9A with the sex body during pachytene and diplotene is similar to the pattern reported for RAD1, TOPBP1 and ATR, all strongly localizing to the XY pair (Freire et al., 1998; Perera et al., 2004). This overlapping localization pattern indicates that, although the sex body is formed, TOPBP1-related checkpoint signaling during late prophase I may be RAD9A-dependent. Therefore, it is possible that RAD9A may play multiple functions in meiosis, during both early and later stages.
Rad9af/delCre+ male mice display variable fertility
Male mice deficient in RAD9A function from the undifferentiated spermatogonial stage developed normally and showed no unusual reproductive behavior. However, they exhibited reduced testis size and low sperm counts that led to infertility or sub-fertility, underscoring the essential role of RAD9A in male germ cell differentiation. While there was heterogeneity in the severity of the infertility, all males examined eventually became sterile. A failure to fully excise the floxed Rad9a allele in some spermatogonia due to the lack of expression of Stra8-Cre in some spermatogonia (Hobbs et al., 2012; Sadate-Ngatchou et al., 2008) may have contributed in part to this heterogeneity. Nonetheless, the loss of RAD9A results in a striking and surprising meiotic phenotype.
Histological analysis of the testis of Rad9af/delCre+ mutant males indicated the presence of the various types of spermatogonia and the cells appear to enter into meiotic prophase normally. However, few of the spermatocytes progressed beyond zygotene into pachytene and virtually no RAD9A-deficient diplotene spermatocytes are seen. Our TUNEL assay revealed that RAD9A-deficient testis demonstrated higher levels of apoptosis, predominantly in spermatocytes as judged by positive signal in the first and second layers of cells from the basal membrane of the seminiferous tubules.
A prominent meiotic defect in RAD9A-deficient spermatocytes is unrepaired DNA DSBs
In mammals, spermatocyte apoptosis can be activated in mutants that fail to complete meiotic DNA repair and recombination (e.g. Dmc1−/−, Msh4−/− or Msh5−/−), and both male and female mice with such mutations are sterile (Edelmann et al., 1999; Pittman et al., 1998). Also, spermatocytes with defective SC formation or DSB repair are eliminated at mid-pachytene, demonstrating that coordination between SC dynamics and DSB repair is necessary to permit meiotic progression (Bolcun-Filas et al., 2009; Hamer et al., 2008a; Hamer et al., 2008b). Synapsis and DNA repair are interrelated and interdependent events, making it difficult to discriminate which process, either one or the other or both, is affected by RAD9A deficiency. Human RAD9A is retained at sites of damaged DNA and colocalizes with γH2AX. Most of the γH2AX foci on autosomes of RAD9A-deficient spermatocytes were presumably at sites of DNA DSBs generated in leptotene and not at sites of asynapsed chromatin. Unlike in SPO11-deficient spermatocytes (Barchi et al., 2005), in those cells lacking RAD9A, DNA DSBs were normally produced and sex bodies seem to form. We found RAD9A-deficient spermatocytes with significant retention of γH2AX throughout the nucleus up to early pachytene stage, which pointed to unrepaired DNA damage. This was accompanied by a significant increase in the number of leptotene/zygotene spermatocytes in the mutant concomitant with a decrease of cells in pachytene. Persistent DMC1 foci on autosomes in mid-pachytene further support this idea and indicate that the abnormal γH2AX presence in pachytene was a direct result of compromised DNA DSB repair in the absence of RAD9A. Together, these data strongly indicate that aberrantly persistent γH2AX foci in mutant spermatocytes were at sites of unrepaired DNA damage and that early apoptosis has been triggered either due to a failure of RAD9A to aid homologous repair directly or because of failed activation of the DNA DSB checkpoint, normally controlled by the 9-1-1 clamp.
These data demonstrate a function for RAD9A during early prophase I. However, the distinct pattern of RAD9A localization to the sex body during late prophase I may indicate an additional role at this stage. Spermatocyte apoptosis at mid-pachytene can also result from abnormal sex body formation and defective meiotic sex chromosome inactivation (MSCI) (Mahadevaiah et al., 2008; Royo et al., 2010). Definitive analysis of such a role in meiosis was precluded in RAD9A-deficient spermatocytes due to induction of apoptosis in early pachytene spermatocytes, and late-pachytene spermatocytes were rare. However, sex bodies formed and key events of MSCI such as localization of γH2AX and SUMO1 at the XY pair appeared normal in RAD9A-deficient spermatocytes that did reach mid-pachytene. Therefore, while RAD9A is not likely required for the formation of the sex body, a weak TOPBP1 signal and its restriction to the axes of the X and Y suggest that RAD9A may be important for the functioning of the sex body and could play a role in MSCI.
There is also the interesting possibility that, at different stages, RAD9A physically interacts with different protein complexes to mediate distinct activities. For example, it is known that in somatic cells RAD51 binds to RAD9A, which in our system could be involved in the repair of obligatory DSBs. Studies in S. pombe have also shown that the Rad9 interacting protein TopBP1 functions in the meiotic recombination checkpoint (Perera et al., 2004). Since RAD9A and TOPBP1 are found to interact in mitotic mammalian cells, it is likely that RAD9A and TOPBP1 are in complexes functioning in mammalian meiotic recombination as well as in late prophase I.
In summary, our findings demonstrate that RAD9A is required for progression through prophase I of mammalian male meiosis. RAD9A deficiency causes distinct abnormalities in prophase I spermatocytes, mainly the accumulation of DNA DSBs and increased apoptosis. This in turn results in infertility or sub-fertilility. RAD9A is a multifunctional protein with activities in DNA repair, cell cycle checkpoint control and apoptosis, studied primarily in the context of the mitotic cell cycle. Investigations reported herein reveal a role for RAD9A in events in the meiotic cell cycle and thus importantly provide an avenue to evaluate the significance of specific RAD9A-related functions in mitotic versus meiotic processes, as well as impact on male fertility.
Materials and Methods
mRNA expression analyses
Q-RT-PCR
Total testis RNA was isolated from mice of the indicated genotype and age using Trizol reagent (Invitrogen) according to the manufacturer's recommendations, treated with DNase I (Invitrogen) and reverse-transcribed using random primers (Invitrogen). In each real-time PCR 100 ng cDNA was combined with qPCR Mastermix Plus for SYBR Green I (Applied Biosystems) containing 300 nM of specific primers: Rad9a-q forward: 5′-GGCTGTCCATTCGCTATCCC-3′ Rad9a-q reverse: 5′-GTGGGGCAAAAAGGAAGCAG-3′ designed to anneal to exon two and three. Intervening intron precludes the amplification of a larger fragment under these conditions.
Arbp was amplified with primers: Arbp-q forward: 5′-CAAAGCTGAAGCAAAGGAAGAG-3′ Arbp-q reverse: 5′-AATTAAGCAGGCTGACTTGGTTG-3′ as previously reported (Yabuta et al., 2011). Reactions were performed in triplicate in an ABI 7300 Real-time PCR System (Applied Biosystems, Foster City, CA) and RNA load normalized with Arbp.
In situ hybridization
Rad9a mRNA was detected in situ in 5 µm paraffin sections of adult wild-type mice using digoxigenin-labeled riboprobes as described previously (Batourina, et al., 2001; Chung, et al., 1998b). A cDNA fragment spanning nucleotides 1–180 of the Rad9a mRNA was cut with NdeI (Invitrogen, Carlsbad, CA) from full ORF cloned in pGEMT and T7 polymerase used to generate the sense probe. Antisense probe was prepared by AatII (Invitrogen, Carlsbad, CA) digest followed by SP6 polymerase conversion.
Protein extracts and immunoblotting
Cytoplasmic and nuclear extracts from fresh testis were prepared as described previously (Andrews and Faller, 1991; Revenkova et al., 2001). Immunoblot analysis was performed according to standard protocols with the following antibodies: mouse monoclonal against RAD9A (cat. no. 200784, Zen Biosciences, CA) and goat polyclonal against beta-actin (cat. no. sc-163637, Santa Cruz Biotechnology, CA).
Generation of Rad9af/f or Rad9af/del and Stra8-Cre Rad9a male mice
All mice were housed in a pathogen-free facility, and manipulations performed in strict adherence to IACUC guidelines. The generation of the conditional Rad9a flox allele has been described previously (Hu et al., 2008). Rad9af/f mice were interbred with Stra8-Cre mice (Sadate-Ngatchou et al., 2008) obtained from The Jackson Labs in order to generate Rad9af/+Cre+ mice. In a second round of mating, Rad9af/+Cre+ males were crossed with Rad9af/f females to generate Rad9af/delCre+ males. For assessment of male fertility, female mice of the inbred CD1 line purchased from Charles River Labs (Wilmington, MA) were mated with adult Rad9af/delCre+ males.
Rad9af/delCre+ mice were crossed with Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) (Muzumdar et al., 2007) to assess efficiency of excision by the Stra8-Cre driver. The mouse colony was maintained in a mixed genetic background of 129 SvEv, C57BL/6 and FVB/NJ.
Fertility studies
The fertility of males with Rad9af/delCre+ and Rad9af/fCre+ genotypes was assessed, beginning at 3 months of age, by mating to two, 8-week-old CD1 females. At day 30 females were sacrificed and the number of live pups and developing embryos recorded. At the end of each mating period two new 8-week-old CD1 females were added for another month, for a total of three rounds. Statistical analysis was performed for litter size using a negative binomial model with correction for multiple comparisons. Data were plotted at the 95% confidence limit. At the completion of the three mating periods, males were sacrificed and their testis weight and sperm counts from cauda epididymes determined as described (Wang, 2003). Progeny of mutant males were genotyped to determine the efficiency of conditional allele excision.
Tissue preparation and histology
Testes of mice at indicated ages were dissected from anesthetized animals, weighed, then fixed in either Bouin's solution (Sigma) or 3.7% paraformaldehyde overnight at 4°C. Fixed tissues were embedded in paraffin, sectioned at 5-µm thick, and mounted on slides. Periodic acid-Schiff (PAS) staining was used for histological analysis, according to standard procedures (Sotomayor and Handel, 1986). Staging of spermatogenesis was performed according to a published method (Russell et al., 1990).
TUNEL staining of apoptotic cells and calculation of the AI
TUNEL staining was performed on tissue sections using the in situ cell death detection kit (Roche Diagnostics, Indianapolis, IN), according to the manufacturer's instructions. Briefly, histological sections were treated as for immunohistochemistry without subsequent antigen retrieval. The tailing labeling reaction was carried out in the terminal deoxynucleotidyl transferase (TdT) buffer with HRP-labeled UTP. TUNEL-positive cells were visualized with DAB/H2O2 and sections counterstained with Hematoxylin. Only clearly stained cells were scored as apoptotic, and only tubules cut perpendicular to the length of the tubule (yielding round tubules in section) were evaluated. At least 100 tubules per section from six different testicular sections (more than 60 µm apart) of the same animal were counted. Apoptosis level quantified by the apoptotic index (AI), was determined as described (Woolveridge et al., 1999; Yu et al., 2001). Tubules containing three or more TUNEL-positive cells were considered apoptotic. The AI was assessed by multiplying the percentage of apoptotic tubules by the mean number of TUNEL-positive cells per tubule. Significant differences (P<0.01) between groups were assessed by statistical analysis.
Preparation of chromosome spreads and tubule squashes
Testis cell preparations were generated from juvenile or adult (2- to 4-months old) mice. Usually, one testis was freshly frozen in OCT or fixed in 4% PFA to be later embedded in paraffin and used for immunohistochemistry, while the other was processed for surface spreading according to established techniques (Peters et al., 1997). For long-term storage, slides were kept at −80°C.
For squashes, tubules were dispersed and fixed for 10 minutes in 2% formaldehyde in PBS with 0.05% Triton X-100, placed on slides, pressed with a coverslip, frozen in liquid nitrogen and kept at −80°C prior to processing. Washes were carried out in 0.1% Triton X-100 and 10% antibody dilution buffer (ADB: 10% donkey serum, 3% BSA, 0.05% Triton X-100) in PBS. Then primary antibodies at dilutions of 1∶50–1∶100 in ADB in PBS were incubated overnight at 4°C. Secondary antibodies diluted 1∶300–1∶500 were added to the tubule preparations and incubated at room temperature. After washing, slides were mounted in ProLong Gold antifade reagent (Molecular Probes, Eugene, OR, USA), with 5 µg/ml DAPI.
Antibody generation and immunohistochemistry
The full-length Rad9a cDNA fragment (encoding amino acid residues 1–390) was subcloned into the Escherichia coli expression vector pGEX-4T-3 (Invitrogen), expressed and purified on glutathione agarose (Sigma). A mixture of native and denatured protein was used for immunization of rabbits and polyclonal antibodies were generated (Cocalico Biologicals, Reamstown, PA). Specific antibodies were purified on Protein A–Sepharose Fast-flow (Invitrogen), followed by passage through GST–Sepharose to reduce background. The resulting antibodies were designated as α-RAD9A. The specificity of the antibodies was confirmed by immunoblotting and used to detect RAD9A protein in paraffin embedded testicular tissue sections by immunohistochemistry.
Immunohistochemical analyses were performed using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA), as previously described (Liu et al., 1998). Antigen unmasking was performed by boiling slides in a microwave twice for 5 minutes in 0.01 M citrate buffer, pH 6.0 (Shi et al., 1991). Endogenous peroxidase activity was saturated in 0.3% H2O2 for 30 minutes. Antibody staining with RAD9A primary antibodies at 1∶100 dilution was performed according to the Vectastain ABC kit manufacturer's instructions. RAD9A protein was visualized with DAB and nuclei counterstained with Hematoxylin. Slides were viewed on a Nikon photomicroscope under brightfield optics and images were captured with a Spot digital camera using SPOT software.
Immunofluorescence
Labeling of paraffin and frozen sections of mouse testis was performed as described previously (Revenkova et al., 2004). The following antibodies were used: goat polyconal anti-RAD9A (sc-10465, Santa Cruz Biotechnology), mouse monoclonal anti-SYCP3 (ab-97672, Abcam), rabbit polyclonal anti-SYCP3 (ab-15093, Abcam), rabbit polyclonal anti-DMC1 (sc-22768, Santa Cruz Biotechnology), rabbit polyclonal anti-TOPBP1 (ab-105109, Abcam), mouse monoclonal anti-γH2AX Ser-139 (cat. no. 05-636, Millipore) and anti-SUMO1 (sc-5508, Santa Cruz Biotechnology). For double or triple immunostaining with primary antibodies from the same species, appropriate monovalent Fab fragments were employed. Secondary antibodies used were DyLight-647-labeled donkey anti-mouse IgG (1∶300; Jackson Immunoresearch), DyLight-488-labeled donkey anti-goat (Jackson Immunoresearch, PA), DyLight-594-labeled donkey anti-rabbit IgG (Jackson Immunoresearch, PA) and Alexa-Fluor-647-labeled donkey anti-goat IgG (Invitrogen). Nuclei of testis sections were visualized by staining with DAPI. In addition to the antibodies listed above, on squash preparations the following pairs of secondary antibodies were applied at 1∶500 dilution: anti-rabbit-549 (cat. no. 111-605-144, Jackson Immunoresearch, PA) and anti-mouse Alexa-647 (SUMO1 and SYCP3; cat no. A-31571, Molecular Probes, Life Technologies, CA) and anti-goat Alexa-488 (RAD9A; cat. no. A-11055, Molecular Probes, Life Technologies, CA). Both PBT and ADB buffers were made using immunoglobulin G-free protease-free BSA (cat. no. 001-000-161, Jackson ImmunoResearch, PA).
Analysis of excision efficiency by Stra8-Cre and quantification of germ cell populations by flow cytometry
For staining of individual germ cell types, single-cell suspensions were prepared from testes as described previously (Bastos et al., 2005; Vasileva et al., 2009) with minor modifications. When the same testes were used for spermatocyte spreads, suspensions were prepared by mechanical maceration as opposed to enzymatic digestion, a modification, which resulted in very low numbers of spermatogonia (Table 2). To discriminate between cell types based on their DNA content and the progressive loss of activity of a nuclear ATP pump, which excludes dye from the nucleus as cells differentiate, we stained cells with Hoechst 33342 and performed flow cytometry. Single-cell suspensions from Rad9af/+Cre+Tom+ and Rad9af/delCre+Tom+ testes were used to determine the efficiency of excision of the tdTomato allele as the percentage of EGFP-positive germ cells from the total germ cell pool. Suspensions were analyzed on BD LSRII (BD Biosciences, Seattle, WA), with UV laser excitation wavelength 355 nm and emission filters UV-Blue 460/50 and UV-Red 692/40. Data were acquired using FACSDiva software (BD Biosciences, CA) and analyzed with FlowJo 8.7 (Tree Star, Inc., OR) as described (Bastos et al., 2005; Vasileva et al., 2009). Cells with 2N DNA content and inactive ATP pump include somatic cells and germ cells (secondary spermatocytes) are not shown. The subpopulation of 4N primary spermatocytes was subdivided into three regions, Ho-redlow, Ho-redmed and Ho-redhi, which contained, respectively, leptotene and zygotene, pachytene and diplotene spermatocytes. The identity of each population was confirmed by assessing the type of SCP3 immunofluorescence on chromosomes of sorted cells from each subset. To confirm that cell populations, which by Hoechst staining appeared as 4N are indeed spermatocytes, we crossed the Rad9af/delCre+ line to transgenic mice expressing EGFP under the meiosis-specific Smc1β promoter described previously (Adelfalk et al., 2009). In this mouse line EGFP expression starts at the onset of prophase I.
Supplementary Material
Acknowledgments
We thank Alastair Tulloch for help with quantification of the various testicular abnormalities, Dr Marcia Manterola for expert advice on pachytene stage matching and Dr Sanny S.W. Chung for help with staging of mutant tubules, and comments on the manuscript.
Footnotes
Author contributions
A.V., D.J.W. and H.B.L. contributed equally to interpretation of data and writing of the manuscript. A.V. helped formulate the study design and performed all experiments apart from setting up fertility studies. K.M.H. and X.W. provided technical assistance with genotyping and histology. M.M.W. organized and conducted the fertility experiments. R.A.F. organized the fertility data and performed statistical analyses. D.J.W. and H.B.L. initially conceived and designed the study and provided oversight.
Funding
This work was supported by the National Institutes of Health [grant numbers R01CA130536 to H.B.L., R01GM079107 to H.B.L. and D.J.W.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.126763/-/DC1
References
- Abdu U., Klovstad M., Butin-Israeli V., Bakhrat A., Schüpbach T. (2007). An essential role for Drosophila hus1 in somatic and meiotic DNA damage responses. J. Cell Sci. 120, 1042–1049 10.1242/jcs.03414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelfalk C., Janschek J., Revenkova E., Blei C., Liebe B., Göb E., Alsheimer M., Benavente R., de Boer E., Novak I. et al. (2009). Cohesin SMC1beta protects telomeres in meiocytes. J. Cell Biol. 187, 185–199 10.1083/jcb.200808016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N. C., Faller D. V. (1991). A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499 10.1093/nar/19.9.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi M., Mahadevaiah S., Di Giacomo M., Baudat F., de Rooij D. G., Burgoyne P. S., Jasin M., Keeney S. (2005). Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell. Biol. 25, 7203–7215 10.1128/MCB.25.16.7203-7215.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos H., Lassalle B., Chicheportiche A., Riou L., Testart J., Allemand I., Fouchet P. (2005). Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry 65A, 40–49 10.1002/cyto.a.20129 [DOI] [PubMed] [Google Scholar]
- Batourina E., Gim S., Bello N., Shy M., Clagett-Dame M., Srinivas S., Costantini F., Mendelsohn C. (2001). Vitamin A controls epithelial/mesenchymal interactions through Ret expression. Nat. Genet. 27, 74–78 10.1038/83792 [DOI] [PubMed] [Google Scholar]
- Bellani M. A., Romanienko P. J., Cairatti D. A., Camerini-Otero R. D. (2005). SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J. Cell Sci. 118, 3233–3245 10.1242/jcs.02466 [DOI] [PubMed] [Google Scholar]
- Bellvé A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M., Dym M. (1977). Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 74, 68–85 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Hall E., Speed R., Taggart M., Grey C., de Massy B., Benavente R., Cooke H. J. (2009). Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 5, e1000393 10.1371/journal.pgen.1000393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas C. G., Lieberman H. B. (2012). Contributions of Rad9 to tumorigenesis. J. Cell. Biochem. 113, 742–751 10.1002/jcb.23424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. Y., Lu A. L. (2005). Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 280, 408–417 [DOI] [PubMed] [Google Scholar]
- Chung S. S., Sung W., Wang X., Wolgemuth D. J. (2004). Retinoic acid receptor alpha is required for synchronization of spermatogenic cycles and its absence results in progressive breakdown of the spermatogenic process. Dev. Dyn. 230, 754–766 10.1002/dvdy.20083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. S., Zhu L. J., Mo M. Y., Silvestrini B., Lee W. M., Cheng C. Y. (1998). Evidence for cross-talk between Sertoli and germ cells using selected cathepsins as markers. J. Androl. 19, 686–703 10.1002/j.1939-4640.1998.tb02078.x [DOI] [PubMed] [Google Scholar]
- Edelmann W., Cohen P. E., Kneitz B., Winand N., Lia M., Heyer J., Kolodner R., Pollard J. W., Kucherlapati R. (1999). Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127 10.1038/5075 [DOI] [PubMed] [Google Scholar]
- Eichinger C. S., Jentsch S. (2010). Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc. Natl. Acad. Sci. USA 107, 11370–11375 10.1073/pnas.1004248107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaggs G., Plug A. W., Dunks K. M., Mundt K. E., Ford J. C., Quiggle M. R., Taylor E. M., Westphal C. H., Ashley T., Hoekstra M. F. et al. (1997). Atm-dependent interactions of a mammalian chk1 homolog with meiotic chromosomes. Curr. Biol. 7, 977–986 10.1016/S0960-9822(06)00417-9 [DOI] [PubMed] [Google Scholar]
- Freire R., Murguía J. R., Tarsounas M., Lowndes N. F., Moens P. B., Jackson S. P. (1998). Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev. 12, 2560–2573 10.1101/gad.12.16.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Poitelea M., Guo L., Caspari T., Carr A. M. (2004). Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 18, 1154–1164 10.1101/gad.291104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gembka A., Toueille M., Smirnova E., Poltz R., Ferrari E., Villani G., Hübscher U. (2007). The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 35, 2596–2608 10.1093/nar/gkl1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer D. A., Besley B. D., Kennedy K. B., Davey S. (2003). hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 63, 4829–4835 [PubMed] [Google Scholar]
- Grushcow J. M., Holzen T. M., Park K. J., Weinert T., Lichten M., Bishop D. K. (1999). Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Madabushi A., Chang D. Y., Fitzgerald M. E., Shi G., Drohat A. C., Lu A. L. (2007). The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 35, 6207–6218 10.1093/nar/gkm678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G., Novak I., Kouznetsova A., Höög C. (2008a). Disruption of pairing and synapsis of chromosomes causes stage-specific apoptosis of male meiotic cells. Theriogenology 69, 333–339 10.1016/j.theriogenology.2007.09.029 [DOI] [PubMed] [Google Scholar]
- Hamer G., Wang H., Bolcun-Filas E., Cooke H. J., Benavente R., Höög C. (2008b). Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J. Cell Sci. 121, 2445–2451 10.1242/jcs.033233 [DOI] [PubMed] [Google Scholar]
- He W., Zhao Y., Zhang C., An L., Hu Z., Liu Y., Han L., Bi L., Xie Z., Xue P. et al. (2008). Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res. 36, 6406–6417 10.1093/nar/gkn686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R. M., Fagoonee S., Papa A., Webster K., Altruda F., Nishinakamura R., Chai L., Pandolfi P. P. (2012). Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 10, 284–298 10.1016/j.stem.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann E. R., Milstein S., Boulton S. J., Ye M., Hofmann J. J., Stergiou L., Gartner A., Vidal M., Hengartner M. O. (2002). Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12, 1908–1918 10.1016/S0960-9822(02)01262-9 [DOI] [PubMed] [Google Scholar]
- Hong E. J., Roeder G. S. (2002). A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev. 16, 363–376 10.1101/gad.938102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K. M., Auerbach W., Wang X. Y., Hande M. P., Hang H., Wolgemuth D. J., Joyner A. L., Lieberman H. B. (2004). Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 24, 7235–7248 10.1128/MCB.24.16.7235-7248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Liu Y., Zhang C., Zhao Y., He W., Han L., Yang L., Hopkins K. M., Yang X., Lieberman H. B. et al. (2008). Targeted deletion of Rad9 in mouse skin keratinocytes enhances genotoxin-induced tumor development. Cancer Res. 68, 5552–5561 10.1158/0008-5472.CAN-07-5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman N. J., David J., Elliston C. D., Hopkins K. M., Smilenov L. B., Brenner D. J., Worgul B. V., Hall E. J., Lieberman H. B. (2007). Mrad9 and atm haploinsufficiency enhance spontaneous and X-ray-induced cataractogenesis in mice. Radiat. Res. 168, 567–573 10.1667/rr1122.1 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Hirano A., Kumano T., Xiang S. L., Mihara K., Haseda Y., Matsui O., Shimizu H., Yamamoto K. (2004). Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells 9, 291–303 10.1111/j.1356-9597.2004.00728.x [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. (2007). The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 282, 28036–28044 10.1074/jbc.M704635200 [DOI] [PubMed] [Google Scholar]
- Li T., Wang Z., Zhao Y., He W., An L., Liu S., Liu Y., Wang H., Hang H. (2013). Checkpoint protein Rad9 plays an important role in nucleotide excision repair. DNA Repair (Amst.) 12, 284–292 10.1016/j.dnarep.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Lieberman H. B. (2006). Rad9, an evolutionarily conserved gene with multiple functions for preserving genomic integrity. J. Cell. Biochem. 97, 690–697 10.1002/jcb.20759 [DOI] [PubMed] [Google Scholar]
- Liu D., Matzuk M. M., Sung W. K., Guo Q., Wang P., Wolgemuth D. J. (1998). Cyclin A1 is required for meiosis in the male mouse. Nat. Genet. 20, 377–380 10.1038/3855 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Turner J. M., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W. M., Burgoyne P. S. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276 10.1038/85830 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Bourc'his D., de Rooij D. G., Bestor T. H., Turner J. M., Burgoyne P. S. (2008). Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J. Cell Biol. 182, 263–276 10.1083/jcb.200710195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkiniemi M., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Mäkelä T. P. et al. (2001). BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276, 30399–30406 10.1074/jbc.M102245200 [DOI] [PubMed] [Google Scholar]
- Moens P. B., Kolas N. K., Tarsounas M., Marcon E., Cohen P. E., Spyropoulos B. (2002). The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622 [DOI] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Niida H., Nakanishi M. (2006). DNA damage checkpoints in mammals. Mutagenesis 21, 3–9 10.1093/mutage/gei063 [DOI] [PubMed] [Google Scholar]
- Pandita R. K., Sharma G. G., Laszlo A., Hopkins K. M., Davey S., Chakhparonian M., Gupta A., Wellinger R. J., Zhang J., Powell S. N. et al. (2006). Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 26, 1850–1864 10.1128/MCB.26.5.1850-1864.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. J., Park J. H., Hahm S. H., Ko S. I., Lee Y. R., Chung J. H., Sohn S. Y., Cho Y., Kang L. W., Han Y. S. (2009). Repair activities of human 8-oxoguanine DNA glycosylase are stimulated by the interaction with human checkpoint sensor Rad9-Rad1-Hus1 complex. DNA Repair (Amst.) 8, 1190–1200 10.1016/j.dnarep.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Perera D., Perez-Hidalgo L., Moens P. B., Reini K., Lakin N., Syväoja J. E., San-Segundo P. A., Freire R. (2004). TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol. Biol. Cell 15, 1568–1579 10.1091/mbc.E03-06-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz G., Arie L. G., Bakhrat A., Abdu U. (2009). The Drosophila hus1 gene is required for homologous recombination repair during meiosis. Mech. Dev. 126, 677–686 10.1016/j.mod.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Peters A. H., Plug A. W., van Vugt M. J., de Boer P. (1997). A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 10.1023/A:1018445520117 [DOI] [PubMed] [Google Scholar]
- Pittman D. L., Cobb J., Schimenti K. J., Wilson L. A., Cooper D. M., Brignull E., Handel M. A., Schimenti J. C. (1998). Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697–705 10.1016/S1097-2765(00)80069-6 [DOI] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Gross B., Jessberger R. (2001). Novel meiosis-specific isoform of mammalian SMC1. Mol. Cell. Biol. 21, 6984–6998 10.1128/MCB.21.20.6984-6998.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Hodges C. A., Hunt P. A., Liebe B., Scherthan H., Jessberger R. (2004). Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6, 555–562 10.1038/ncb1135 [DOI] [PubMed] [Google Scholar]
- Rogers R. S., Inselman A., Handel M. A., Matunis M. J. (2004). SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma 113, 233–243 10.1007/s00412-004-0311-7 [DOI] [PubMed] [Google Scholar]
- Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M., Bradley A., de Rooij D. G., Burgoyne P. S., Turner J. M. (2010). Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20, 2117–2123 10.1016/j.cub.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Russell L. D., Ettlin R. A., Sinha Hikim A. P., Clegg E. D. (1990). Histological and Histopathological Evaluation of the Testis Clearwater, FL: Cache River Press [Google Scholar]
- Sadate-Ngatchou P. I., Payne C. J., Dearth A. T., Braun R. E. (2008). Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738–742 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G., Joshi A., Liu D., Wei H., Persson J. L., Wolgemuth D. J. (2005). Induction of apoptosis involving multiple pathways is a primary response to cyclin A1-deficiency in male meiosis. Dev. Dyn. 234, 114–123 10.1002/dvdy.20533 [DOI] [PubMed] [Google Scholar]
- Shi S. R., Key M. E., Kalra K. L. (1991). Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 39, 741–748 10.1177/39.6.1709656 [DOI] [PubMed] [Google Scholar]
- Smirnova E., Toueille M., Markkanen E., Hübscher U. (2005). The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 389, 13–17 10.1042/BJ20050211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor R. E., Handel M. A. (1986). Failure of acrosome assembly in a male sterile mouse mutant. Biol. Reprod. 34, 171–182 10.1095/biolreprod34.1.171 [DOI] [PubMed] [Google Scholar]
- Toueille M., El-Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M. O., Hübscher U. (2004). The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 32, 3316–3324 10.1093/nar/gkh652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A., Tiedau D., Firooznia A., Müller-Reichert T., Jessberger R. (2009). Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 19, 630–639 10.1016/j.cub.2009.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. (2003). Epididymal sperm count. Curr. Protoc. Toxicol. 14, 16.6.1–16.6.5 [DOI] [PubMed] [Google Scholar]
- Wang W., Brandt P., Rossi M. L., Lindsey-Boltz L., Podust V., Fanning E., Sancar A., Bambara R. A. (2004). The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA 101, 16762–16767 10.1073/pnas.0407686101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolveridge I., de Boer-Brouwer M., Taylor M. F., Teerds K. J., Wu F. C., Morris I. D. (1999). Apoptosis in the rat spermatogenic epithelium following androgen withdrawal: changes in apoptosis-related genes. Biol. Reprod. 60, 461–470 10.1095/biolreprod60.2.461 [DOI] [PubMed] [Google Scholar]
- Yabuta Y., Ohta H., Abe T., Kurimoto K., Chuma S., Saitou M. (2011). TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 192, 781–795 10.1083/jcb.201009043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wei Q., Adelstein R. S., Wang P. J. (2012). Non-muscle myosin IIB is essential for cytokinesis during male meiotic cell divisions. Dev. Biol. 369, 356–361 10.1016/j.ydbio.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Brain J., Laneuville P., Osmond D. G. (2001). Suppressed apoptosis of pre-B cells in bone marrow of pre-leukemic p190bcr/abl transgenic mice. Leukemia 15, 819–827 10.1038/sj.leu.2402079 [DOI] [PubMed] [Google Scholar]
- Zhu A., Zhou H., Leloup C., Marino S. A., Geard C. R., Hei T. K., Lieberman H. B. (2005). Differential impact of mouse Rad9 deletion on ionizing radiation-induced bystander effects. Radiat. Res. 164, 655–661 10.1667/RR3458.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Zhang C. X., Lieberman H. B. (2008). Rad9 has a functional role in human prostate carcinogenesis. Cancer Res. 68, 1267–1274 10.1158/0008-5472.CAN-07-2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.