Summary
The majority of cancer cells rely on elevated telomerase expression and activity for rapid growth and proliferation. Telomerase-negative cancer cells, by contrast, often employ the alternative lengthening of telomeres (ALT) pathway to maintain telomeres. ALT cells are characterized by long and dynamic telomeres and the presence of ALT-associated promyelocytic leukemia (PML) bodies (APBs). Previous work has shown the importance of APBs to the ALT pathway, but their formation and precise role remain unclear. Here, we demonstrate that a homeobox-containing protein known as HMBOX1 can directly bind telomeric double-stranded DNA and associate with PML nuclear bodies. Hence, we renamed this protein TAH1 for telomere-associated homeobox-containing protein 1. TAH1 knockdown significantly reduced the number of APBs and led to an increase in DNA damage response signals at telomeres. Importantly, TAH1 inhibition also notably reduced the presence of telomere C-circles, indicating altered ALT activity. Our findings point to TAH1 as a novel link between pathways that regulate DNA damage responses, PML nuclear bodies, and telomere homeostasis in ALT cells, and provide insight into how ALT cells may achieve sustained growth and proliferation independent of the telomerase.
Key words: Telomere, HMBOX1, TAH1, Homeobox, DNA damage response, Alternative lengthening of telomeres, ALT pathway
Introduction
Telomeres have evolved to overcome the end replication problem and maintain genome stability in organisms with linear chromosomes (Blackburn, 2001; Feldser et al., 2003). In humans, for example, telomerase and a network of telomere-associated proteins are responsible for maintaining telomere length and integrity (Blackburn et al., 2006; Cong et al., 2002; de Lange, 2005; Venteicher et al., 2009; Venteicher et al., 2008; Xin et al., 2008). The expression and activity of telomerase are highly regulated. For instance, human telomerase reverse transcriptase expression and activity are very low or non-detectable in normal somatic cells (Kim et al., 1994). In contrast, 85–90% of cancer cells have elevated telomerase expression and activity (Shay and Bacchetti, 1997). Such mechanisms appear critical for the relentless growth of cancer cells. The remaining 10–15% of cancer cells, predominantly mesenchymal or neuroepithelial precursors, appear to extend their telomeres through alternative lengthening of telomeres (ALT), a mechanism that depends on homologous recombination (HR) rather than the telomerase (Bryan et al., 1997; Cesare and Reddel, 2010; Heaphy et al., 2011). ALT cells often contain heterogeneous, unstable, and excessively long telomeres (Bryan and Reddel, 1997; Dunham et al., 2000; Scheel et al., 2001) as well as abundant extra-chromosomal telomeric DNA (e.g. double-stranded and single-stranded circles) that has been implicated in telomere HR and the ALT pathway (Cesare and Griffith, 2004; Henson et al., 2009; Nabetani and Ishikawa, 2009; Wang et al., 2004).
Another characteristic of the ALT cells is the presence of ALT-associated PML (promyelocytic leukemia) bodies or APBs (Yeager et al., 1999), which contain both telomeric DNA and telomere-binding proteins. APBs are likely sites of ALT activity (Cesare and Reddel, 2010). Proteins that are involved in HR (e.g. RAD51, RAD52, RPA, HP1, MUS81, and SMC5/6) (Jiang et al., 2009; Potts and Yu, 2007; Yeager et al., 1999; Zeng et al., 2009) as well as DNA repair (e.g. RAD50, MRE11, NBS1, and Sp100) (Jiang et al., 2005; Wu et al., 2000; Zhong et al., 2007) have also been shown to localize to PML bodies and participate in telomere maintenance in ALT cells. A number of these proteins (e.g. PML, HP1, RAD50, MRE11, and NBS1) play important roles in the formation of APBs (Jiang et al., 2007; Wu et al., 2000). In ALT cells, members of the telosome/shelterin complex have been shown to recruit proteins that participate in other pathways to the telomeres. For example, the RAP1–TRF2 complex likely targets the MRN complex to telomeres through their interaction (Jiang et al., 2007). And TRF1 and TRF2 were shown to interact with PML, suggesting that they may bring PML bodies and telomeres together (Brouwer et al., 2009). TRF1, TRF2, and RAP1, which localize to the telomeres, are important for APB formation (Wu et al., 2000; Yeager et al., 1999). However, the precise roles of telomere-targeted proteins in the ALT pathway remain poorly understood.
Through a screen using the technique called proteomics of isolated chromatin segments (PICh), the homeobox containing protein 1 (HMBOX1) was recently found to associate with telomeres, and could co-localize with RAP1 in the ALT cell line WI38-VA13 as well as the telomerase-positive HeLa cells when exogenously expressed (Déjardin and Kingston, 2009). It can also function as a transcription factor and its homeodomain was predicted to bind double-stranded DNA (Berger et al., 2008; Chen et al., 2006). However, the molecular basis and functional significance for HMBOX1 telomeric localization is unclear. Here, we refer to the protein as telomere-associated homeodomain-containing protein 1 (TAH1), which better represents its function. We report that the homeodomain of TAH1 can directly bind to telomeric DNA and is required for TAH1 targeting to telomeres. In the ALT cell line we examined, inhibition of TAH1 impacted the number of APBs and induced DNA damage responses at telomeres. RNAi knockdown of TAH1 also led to a reduction in extra-chromosomal telomere DNA. These results indicate that TAH1 may help to bridge PML bodies to telomeres and contribute to telomerase-independent maintenance of telomeres in ALT cancer cells.
Results
TAH1 localizes more frequently in ALT cells than in telomerase positive cells
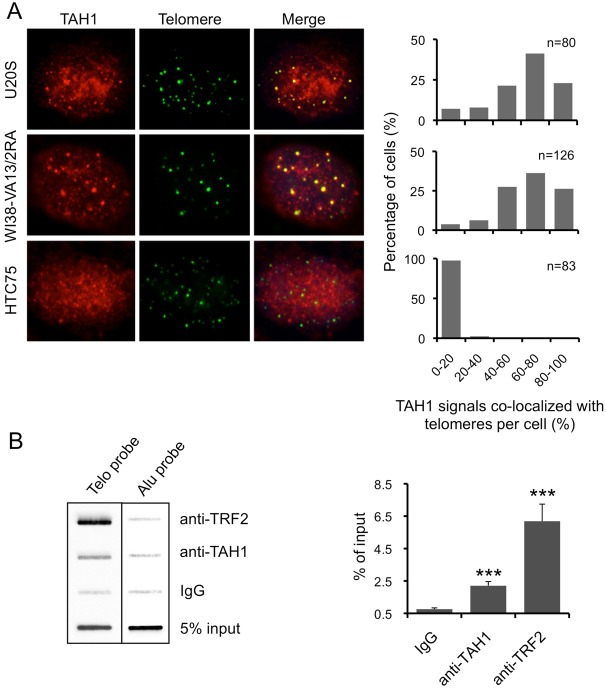
Exogenously expressed TAH1/HMBOX1 has been shown to localize to telomeres in both ALT and telomerase-positive cells (Déjardin and Kingston, 2009). To further study the role of TAH1 in telomere maintenance, we examined the telomeric localization of endogenous TAH1 in multiple cell lines. U2OS (a human osteosarcoma cell line) and WI38-VA13/2RA cells (a SV40-immortalized human lung cell line) use ALT pathway to maintain their telomeres, while HTC75 and HeLa cells are telomerase-positive. In both U2OS and WI38-VA13/2RA cells punctate immunostaining patterns of TAH1 appeared to overlap with ∼70% of telomere signals (Fig. 1A). Chromatin immunoprecipitation (ChIP) assays using anti-TAH1 antibodies also revealed specific enrichment of telomere DNA signals (∼1/3 of signals observed for TRF2) from U2OS cells (Fig. 1B), demonstrating the association of endogenous TAH1 with telomeres in ALT cells.
Fig. 1.
TAH1 localizes to telomeres more frequently in ALT cells than in telomerase-positive cells. (A) IF-FISH was carried out for endogenous TAH1 using the telomere PNA-TelC-FITC probe in U2OS, WI38-VA13/2RA and HTC75 cells. Quantifications of the percentage of cells containing varying degrees of TAH1–telomere co-localization are shown on the right. (B) Telomere ChIP analysis with the biotinylated telomere probe (TTAGGG)3 was performed using U2OS cells and anti-TAH1 antibodies. IgG served as IP control and an Alu probe was used for a loading control. A quantification of the data is shown on the right. For TRF2, n = 3. For TAH1 and IgG, n = 6. Error bars indicate standard error. ***P<0.001 compared with control.
In HTC75 cells, we could also observe co-localization of endogenous TAH1 with the telomeres, albeit at a much lower level compared with ALT cells (Fig. 1A). Similar results were also seen with HeLa cells (data not shown). The difference of telomere targeting in ALT cells versus telomerase-positive cells suggests that TAH1 may play a role in the ALT pathway.
TAH1 binds directly to telomeric DNA in a homeodomain-dependent manner
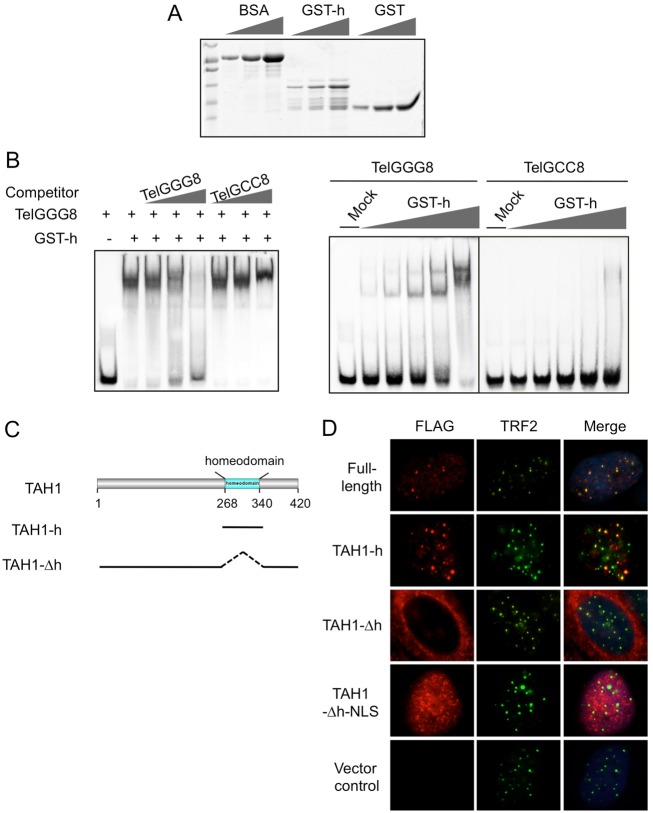
Homeodomains are known double-stranded DNA-binding modules (Berger et al., 2008); however, the specificity and activity of TAH1 homeodomain towards telomeric DNA has never been studied. To test whether TAH1 homeodomain could interact directly with telomeric DNA sequences, we carried out electrophoretic mobility shift assays (EMSA) using bacterially purified TAH1 homeodomain (Fig. 2A) and a biotinylated double-stranded telomeric DNA probe (biotin-TelGGG8). As shown in Fig. 2B, purified GST-tagged TAH1 homeodomain could slow the migration of the telomeric probe in a dose-dependent manner, but had no effect on the mutant probe (biotin-TelGCC8). Moreover, the retarded signals were reduced with increasing concentrations of the unlabeled TelGGG8 probe, but not the mutant probe (Fig. 2B, left). These data suggest that TAH1 can directly bind to telomeric DNA and the homeodomain likely mediates the telomeric targeting of TAH1.
Fig. 2.
TAH1 binds telomeric DNA in a homeodomain-dependent manner. (A) The amount of bacterially purified GST proteins (GST) and GST-tagged TAH1-homeodomain-only proteins (GST-h) was examined by Commassie Blue staining. A twofold increase in protein amount was loaded in successive lanes for each protein. BSA served as a loading control (1, 2 and 4 µg). (B) Left panel: unlabeled TelGGG8 or TelGCC8 probes were added at increasing concentrations (0.025, 0.25 and 2.5 µM) to the in vitro binding reactions containing GST-h (8 µM) and biotinylated TelGGG8 (25 nM). Right panel: different concentrations of bacterially expressed GST-h fusion proteins (0.5, 1, 2, 4 and 8 µM) were incubated with biotinylated TelGGG8 (25 nM) or TelGCC8 (25 nM) probes and examined by native PAGE. GST proteins (8 µM) were used as a negative control (Mock). (C) Schematic diagram of full-length TAH1 and its deletion mutants. (D) U2OS cells expressing FLAG-tagged full-length and mutant TAH1 were co-stained with anti-FLAG (red) and anti-TRF2 (green) antibodies. DAPI was used to stain the nuclei.
To test the dependence of TAH1 on its homeodomain for telomere localization, we generated two deletion mutants of TAH1 (Fig. 2C) for immunostaining analysis. As shown in Fig. 2D, both full-length TAH1 and the homeodomain only mutant (TAH1 hour) could localize to telomeres. In contrast, the homeodomain deletion mutant (TAH1-Δh) remained predominantly in the cytoplasm (Fig. 2D). When we attached an SV40 nuclear localization sequence (NLS) to its C terminus, TAH1-Δh-NLS displayed diffuse nuclear distribution but failed to form foci that co-stained with TRF2 (Fig. 2D). Taken together, our findings demonstrate that the homeodomain of TAH1 is necessary and sufficient for targeting TAH1 to telomeres through its direct interaction with telomere sequences.
TAH1 localizes to PML bodies in ALT cells
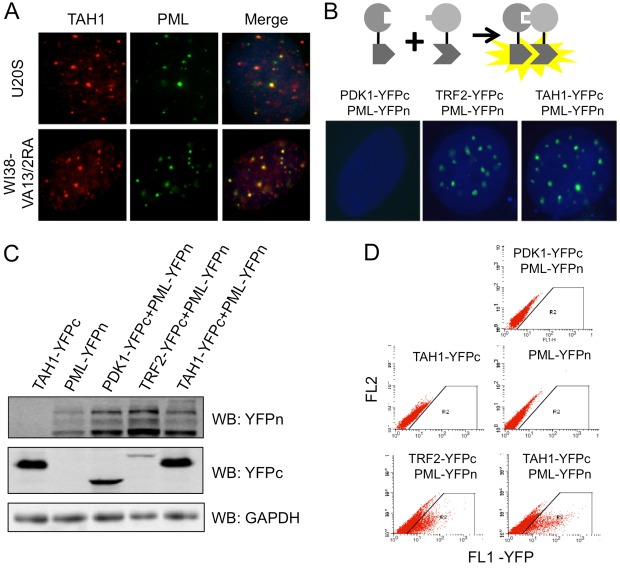
In the majority of U2OS and WI38-VA13/2RA cells (∼60%), endogenous TAH1 signals appeared to co-localize with >50% of PML foci (Fig. 3A), suggesting that TAH1 might reside in PML bodies and interact with PML. To further explore this possibility, we examined the interaction between TAH1 and PML using the bi-molecular fluorescence complementation (BiFC) assay. In BiFC assays, interactions between two proteins that are tagged with YFP fragments (YFPn or YFPc) will bring the two halves of YFP to close proximity for fluorescence complementation and detection (Hu et al., 2002; Ma et al., 2011) (Fig. 3B). When U2OS cells stably co-expressing PML and TAH1 respectively tagged with YFP fragments were examined, punctate YFP signals in the nucleus were apparent, whereas we detected no such signals in cells co-expressing PML with PDK1, a phosphoinositide-dependent kinase that does not localize to PML-containing nuclear bodies (PML NBs) (Fig. 3B). In TAH1 and PML co-expressing cells, both the pattern of nuclear BiFC signals and the percentage of YFP-positive cells were comparable to those observed in cells co-expressing TRF2 and PML (Fig. 3B–D). These observations indicate that TAH1 may interact with PML or be in the vicinity of PML.
Fig. 3.
TAH1 localizes to PML bodies in ALT cells. (A) U2OS cells and WI38-VA13/2RA cells were co-immunostained with antibodies against endogenous TAH1 (red) and PML (green). (B) BiFC assays were carried out in U2OS cells stably expressing PML-YFPn plus TAH1-YFPc. Cells expressing PML-YFPn plus PDK1-YFPc or PML-YFPn plus TFR2-YFPc were used as negative or positive controls, respectively. (C) Western blotting for protein expression in the indicated cell lines. (D) FACS analysis of fluorescence complementation in cells from C. U2OS cells expressing TAH1-YFPc only, PML-YFPn only, and PML-YFPn plus PDK1-YFPc were used as negative controls.
TAH1 regulates the number of APBs in ALT cells
The ability to associate with both telomeres and PML may enable TAH1 to link PML bodies with telomeres and impact the number of APBs in cells. To test this hypothesis, we knocked down TAH1 in U2OS cells with two independent short-hairpin RNAs (shRNAs), both of which achieved >70% knockdown efficiency (Fig. 4A,B). The knockdown cells were then examined for APBs, which should stain positive for both PML and TRF2 (Fig. 4C). We found extensive cell-to-cell variation in the number of APBs (white arrowheads), both in control and TAH1 knockdown cells. Reduced TAH1 expression did not appear to affect the total number of PML NBs (white arrows) (Fig. 4C). However, most TAH1 knockdown cells contained significantly fewer APBs compared to control cells (Fig. 4D, P<0.001). In fact, TAH1 depletion resulted in ∼40% decrease (from 42.7% to 25.6% and 28.1%) in the average ratio of APB:PML-NB per cell (Fig. 4E). APBs are enriched at G2/M phase. If TAH1 knockdown affects cell cycle progression, then the differences we observed in APB numbers might have arisen because of changes in cell cycle rather than a direct result of TAH1 inhibition. When cell cycle progression of TAH1 knockdown cells was monitored, however, we did not observe any differences between the knockdown and control cells (supplementary material Fig. S1A,B). Taken together, these results underline the importance of TAH1 in APB formation or persistence.
Fig. 4.
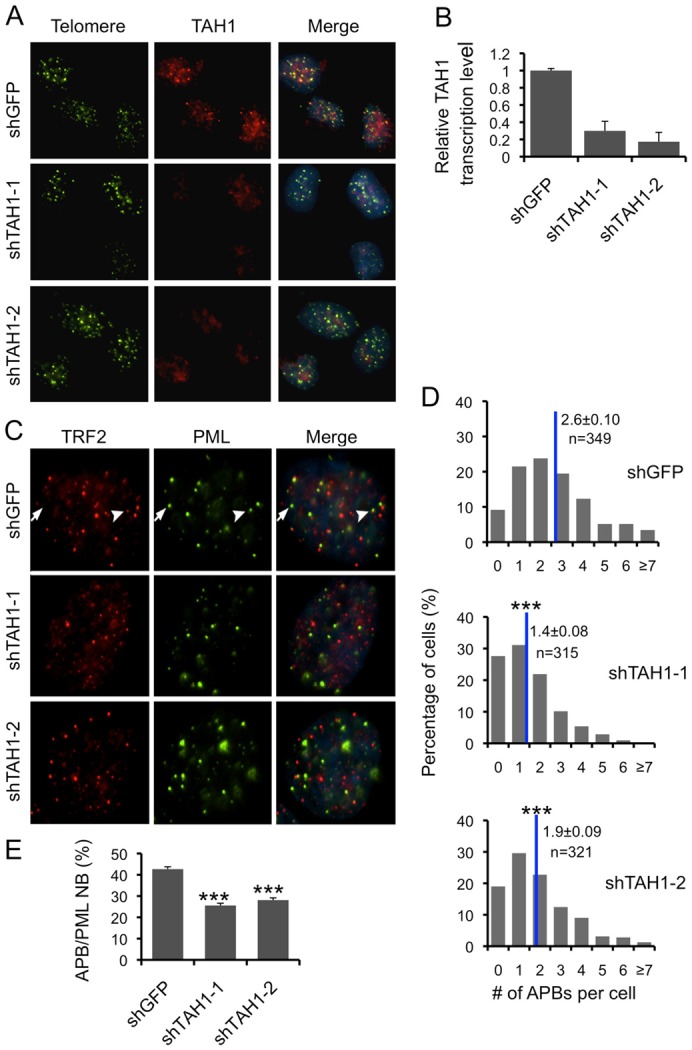
TAH1 regulates the number of APBs. Knockdown efficiency of TAH1 shRNAs was assessed by IF-FISH (A) and quantitative real-time PCR (B). shRNA sequences against GFP were used as control. (C) IF images of endogenous TRF2 (red) and PML (green) in TAH1 knockdown cells. Representative staining for PML NBs (arrows) and APBs (arrowheads) are indicated. (D) The number of APBs per cell was quantified and is shown in the graph, where n indicates the number of cells examined for each cell line, and blue lines mark the mean. (E) The ratio of APB:PML NB for each cell was calculated based on the total number of PML NBs and APBs per cell. The average APB:PML NB ratio for each cell line was then calculated and is shown in the graph. Error bars indicate standard errors (n = 3). ***P<0.001 compared with control.
TAH1 contributes to ALT activity without influencing telomere length
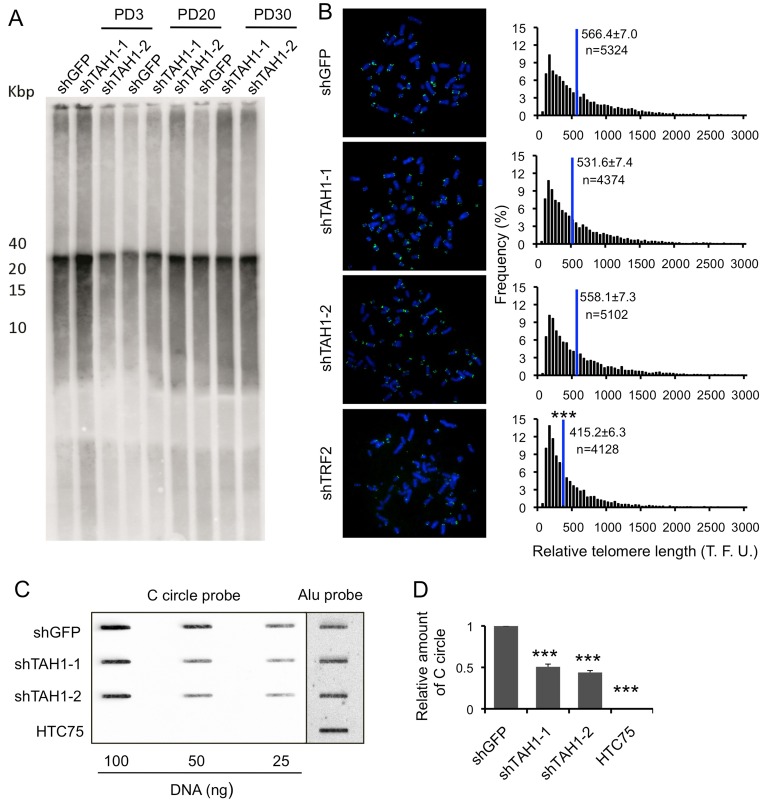
ALT cells can undergo drastic changes in telomere length, and depend on recombination for telomere maintenance where recombination byproducts such as signal-free ends may accumulate. To further probe the importance of TAH1 in the ALT pathway, we used telomeric fluorescence in situ hybridization (FISH) to detect telomere signal-free ends and end-to-end fusions, and telomere restriction fragment (TRF) analysis and telomere quantitative fluorescence in situ hybridization (Q-FISH) to examine telomere length. No significant changes in telomere signal-free ends or ratios of chromosomal fusions were apparent in TAH1 versus control knockdown cells (data not shown). As expected, significant shortening of telomeres occurred with TRF2 knockdown (Stagno D'Alcontres et al., 2007); however, TAH1 depletion had little effect on the average telomere length (Fig. 5A,B).
Fig. 5.
TAH1 contributes to ALT activity. (A) Telomere restriction fragment (TRF) analysis of control U2OS cells and those expressing shRNA sequences against TAH1. PD, population doubling. (B) Q-FISH analysis of control U2OS cells and those expressing shRNA sequences against TAH1. TRF2 knockdown cells were used as positive controls. Histograms show the distribution of relative telomere length expressed as fluorescence intensity (TFU, telomere fluorescence unit), and blue lines mark the mean. n indicates the number of telomere signals. Error bars indicate standard errors. ***P<0.001 compared with control. (C) Various amounts of genomic DNA from control and TAH1 knockdown U2OS cells was used for the CC assay. HTC75 cells were used as negative control. (D) Quantification of data from C. Intensity values from TAH1 knockdown cells were normalized to control cells for individual DNA concentration. The values were then combined and averaged for each cell line to determine the relative C-circle formation activity. Error bars indicate standard errors (n = 3). ***P<0.001 compared with control.
We then investigated whether TAH1 inhibition could impact the formation of C-circles, which are prevalent in ALT cells and possible templates or by-products of homologous recombination. Using the C-circle assay (CC assay) developed by Henson et al. (Henson et al., 2009), we compared the amount of C-circles in control versus TAH1 knockdown U2OS cells. As a positive control, we also examined U2OS cells depleted for SMC5 because of its critical role in C-circle information in ALT cells (Henson et al., 2009). As expected, knocking down SMC5 in U2OS cells reduced C-circle signals by ∼20% (supplementary material Fig. S2A–C). In comparison, TAH1 depletion led to a ∼50% reduction in C-circle signals (Fig. 5C,D), underlining the importance of TAH1 in the process of C-circle formation and possibly telomere recombination in ALT cells.
TAH1 regulates DNA damage responses at the telomeres in ALT cells
Next, we went on to examine whether TAH1 played a role in DNA damage responses at the telomeres by assessing the number of telomere-dysfunction-induced foci (TIF). When TRF2 was knocked down in U2OS cells, both the number of TIFs (as indicated by 53BP1–telomere co-localized foci) per cell and the percentage of TIF-positive cells increased (supplementary material Fig. S3A–D), consistent with the importance of TRF2 in telomere protection in these cells (Stagno D'Alcontres et al., 2007). Similarly, with TAH1 depletion, we also observed increases in both the number of TIFs per cell and the percentage of TIF-positive cells (Fig. 6A–C). In this case, TAH1 inhibition was accompanied by a ∼3 fold increase in the percentage of TIF-positive cells compared to control cells (Fig. 6C), a level comparable to TRF2 knockdown (supplementary material Fig. S3D). These results suggest that TAH1 inhibition could compromise the integrity of telomeres, and indicate that one major function of TAH1 in ALT cells may involve its regulation of DNA damage responses at telomeres.
Fig. 6.
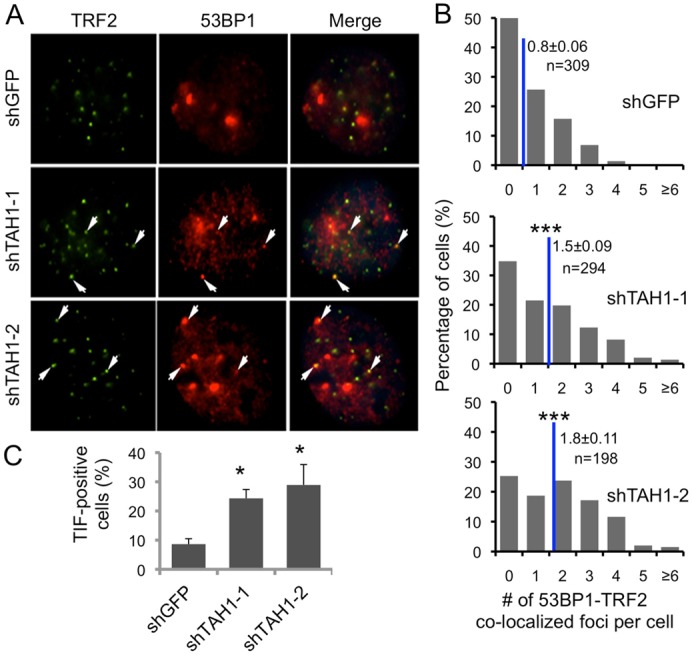
TAH1 regulates telomere DNA damage responses in ALT cells. (A) Representative images of the telomere-dysfunction-induced foci (TIF) analysis in control and TAH1 knockdown U2OS cells using anti-53BP1 (red) and TRF2 (green) antibodies. Arrows indicate superimposable foci. (B) 53BP1 and TRF2 co-staining foci (TIFs) were scored for individual cells from A. Blue lines mark the average number of TIFs per cell. Error bars indicate standard errors (n = 3). ***P<0.001 compared with control. (C) Percentages of TIF-positive cells were calculated based on data from A and B. Cells with ≧3 co-localization foci were scored as TIF positive. Error bars indicate standard errors (n = 3). *P<0.05 compared with control.
Discussion
In this study, we examined the telomere localization of TAH1 in ALT versus telomerase-positive cells. We found that the homeodomain of TAH1 could specifically bind telomere repeats in vitro and target its telomere localization in vivo. Furthermore, we provide evidence that TAH1 is important for the formation or stability of APBs, ALT activity, and telomere protection in ALT cells. We demonstrated that endogenous TAH1 was more frequently telomere targeted in ALT cells than in telomerase-positive cells. We were able to detect endogenous TAH1 telomere targeting in telomerase-positive cells, but at much lower frequency. Compared with telomerase-positive cells, ALT cells in general have exceedingly long telomeres (Henson et al., 2002), perhaps thus allowing more frequent binding of TAH1 and easier detection by immunostaining. Similar to the core telomere proteins TRF1 and TRF2, TAH1 can also bind telomeric double-stranded DNA. However, unlike TRF1 and TRF2, which can modulate telomere length in both telomerase-positive and ALT cells (Broccoli et al., 1997; Chong et al., 1995), TAH1 appears to function primarily in telomere protection in ALT cells. It is also possible that telomere shortening may only become apparent upon prolonged TAH1 knockdown, given the exceedingly long telomeres in ALT cells.
Functional APBs are specific sites of protein complex formation in ALT cells and important for ALT activity (Cesare and Reddel, 2010). For example, RNAi knockdown of PML, telomere proteins such as TRF1 and TRF2, and DNA repair and recombination proteins such as MRE11 and RAD50 led to disrupted APB formation (Jiang et al., 2007). With TAH1 knockdown in U2OS cells, we found a significant reduction in the number of APBs, even though the total number of PML NBs remained unaffected. Taken together with our data that TAH1 could interact with PML, these findings suggest that TAH1 may function to facilitate APB assembly by modulating the interaction between PML and telomeric DNA. We could readily detect fluorescence complementation between PML and TAH1 in BiFC assays, suggesting close proximity between the two YFP fragment-tagged proteins. However, GST-tagged TAH1 failed to co-immunoprecipitate (co-IP) with FLAG-tagged PML (data not shown). Both TRF1 and TRF2 have also been reported to associate with PML in BiFC assays (Brouwer et al., 2009), but we were likewise unable to co-precipitate PML with either TRF1 or TRF2 (data not shown). These data indicate that additional factors may be needed to bridge PML with TAH1 and shelterin/telosome members. It is also possible that co-IP may favor high-affinity interactions, whereas BiFC may be more sensitive at detecting low-affinity or transient associations.
Concurrent with APB reduction in TAH1 knockdown cells, we also observed an increase in telomeric DNA damage induced foci (TIFs). APBs contain numerous DNA damage repair proteins (Jiang et al., 2007), and a decrease in APBs (e.g. as a result of TAH1 knockdown) may lead to reduced ability for DNA damage repair and increased DNA damage signals at telomeres. Elevated TIFs in TAH1-depleted cells, however, were not accompanied by increased telomere signal-free ends, end-to-end fusions, or gross changes in telomere length. This is reminiscent of previous reports of spontaneous occurrences of DNA damage responses that were independent of telomere length in ALT cells (Cesare et al., 2009). The novel function of TAH1 warrants further investigation, as it may represent an essential link between DNA damage response machineries, PML NBs, and telomeres, and offer clues to how cancer cells bypass canonical telomere maintenance pathways for sustained growth and proliferation.
Materials and Methods
Constructs, cell lines and antibodies
Full-length or mutant TAH1 cDNAs were cloned into the pBabe retroviral vector for mammalian expression (Wan et al., 2009). The TAH1 homeodomain only mutant (TAH1 hour) encoding a.a. 236–341 was also cloned into the pDEST15 vector for bacterial expression. The region encompassing a.a. 266–340 was deleted to generate the homeodomain deletion mutant (TAH1-Δh). The SV40 (Simian virus 40) nuclear localization signal (NLS), PKKKRKVG, was appended to the C-terminus of TAH1-Δh to create TAH1-Δh-NLS. For immunofluorescence, TAH1 was tagged with FLAG at the C terminus. For bi-molecular fluorescence complementation (BiFC) assay, TAH1 was tagged at the C-terminus with the C-terminal fragment of YFP (YFPc, residues 156–239), whereas PML was C-terminally tagged with the N-terminal fragment of Venus YFP (YFPn, residues 1–155) (Ma et al., 2011). For RNAi, shRNA sequences were cloned into the pcl-mU6 retroviral vector between BamHI and HindIII sites (Xu et al., 2009). And siRNAs were reverse transfected into cells for 48 to 72 hours. The sequences are: shGFP: 5′-GTGGTACCAGCATCAGCCTT-3′; shTAH1-1: 5′-CCTGGAGAGTCATGGGATA-3′; shTAH1-2: 5′-GGACCTAGATGTAGATGAT-3′; shTRF2: 5′- GAGCATGGTTCCTAATAAT-3′; siTAH1-1: 5′-CCTGGAGAGTCATGGGATAdTdT-3′; siTAH1-2: 5′-GGACCTAGATGTAGATGATdTdT-3′; and siSMC5: 5′-GAAGCAAGAUGUUAUAGAAdTdT-3′.
HEK293T cells were transfected with constructs encoding various proteins or shRNA sequences for retrovirus production. U2OS cells were infected with the appropriate retroviruses, selected either in puromycin (1 µg/ml) for 3 days or G418 (500 ng/ml) for 7 days (YFPn construct) before analysis. For BiFC assay, cells stably co-expressing YFPn and YFPc tagged proteins were analyzed by microscopy and flow cytometry (Ma et al., 2011).
Rabbit polyclonal anti-TAH1 (Genetex), mouse monoclonal anti-TRF2 (Calbiochem), rabbit polyclonal anti-TRF2 (Abcam), rabbit polyclonal anti-FLAG (Abmart), rabbit polyclonal anti-53BP1 (Novus), mouse monoclonal anti-PML (Santa Cruz), and rabbit polyclonal anti-IgG (Sigma) antibodies were used in this study.
Indirect immunofluorescence (IF) and IF-fluorescent in situ hybridization (IF-FISH)
Indirect IF and IF for telomere dysfunction-induced foci (TIF) detection were essentially done as previously described (Wan et al., 2009). Briefly, cells that were plated on glass coverslips were fixed with 4% paraformaldehyde, permeablized in 0.5% Triton X-100 (in 1×PBS) before primary and secondary antibody incubation. For IF-FISH, an additional incubation with PNA-TelC-FITC probe (Panagene) was conducted at 37°C for two hours after secondary antibody incubation. Fluorescence microscopy was performed on a Nikon Ti microscope. For APB scoring, >300 cells were examined for each cell line. PML-positive nuclear foci that were also superimposable with TRF2 foci were counted as APBs. The average number of APBs per cell was then calculated for each cell line.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was essentially done as previously described (Ma, 2011). Briefly, cultured cells (in 150 mm dishes) were fixed with 1% formaldehyde, and collected by mechanical scraping. Sonicated lysate was then pre-cleared with protein A/G-agarose beads and control immunoglobulin (IgG) (2 µg), and incubated with appropriate antibodies (3 µg). The eluted DNA was purified using QIAquik PCR purification kit (Qiagen), dot-blotted onto Hybond-N+ membranes, and analyzed using biotin-labeled probes [Telo probe: 5′-Biotin-TTAGGGTTAGGGTTAGGGT; and Alu probe: 5′-Biotin-GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCA].
Protein purification and electrophoretic mobility shift assay (EMSA)
Bacterially expressed GST-tagged TAH1 homeodomain only protein (a.a. 236–341) was purified using glutathione-conjugated agarose beads, and eluted in elution buffer containing 10 mM reduced glutathione (GE) and 50 mM Tris (pH 8.0). The expression and purity of the recombinant proteins were verified by SDS-PAGE. For EMSA assay, different concentrations of purified GST-homeodomain proteins were incubated with biotin-labeled pre-annealed double-stranded DNA probes, TelGGG8 or TelGCC8 [TelGGG8: ds(TTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGT); TelGCC8: ds(TTAGCCTTAGCCTTAGCCTTAGCCTTAGCCTTAGCCTTAGCCTTAGCCT)], at room temperature for 20 minutes followed by electrophoresis, transfer to Nylon membranes, and blotting for biotin detection using the Chemiluminescent Nucleic Acid Detection Module (Thermo).
Real-time quantitative PCR (qPCR)
Real-time qPCR was carried out as described previously (Liang et al., 2008). Briefly, total RNA was isolated with the RNeasy mini Kit (QIAGENE), reverse-transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad), and then amplified using the ABI StepOnePlus real-time PCR system (Applied Biosystems). Cycling conditions are 40 cycles of 95°C for 15 s and 60°C for 60 s.
Telomere restriction fragment (TRF) assay
The TRF assay was performed using U2OS cells stably expressing shGFP or shTAH1 sequences as described previously with slight modification in telomere signal detection (Liu et al., 2004). Southern blotting was carried out using the biotin-labeled telomeric probe (5′-biotin-TTAGGGTTAGGGTTAGGGT), followed by biotin signal detection with the Chemiluminescent Nucleic Acid Detection Module (Thermo).
Telomere quantitative fluorescent in situ hybridization (Q-FISH)
U2OS cells were incubated with 0.5 µg/ml nocodazole for 6 h to enrich cells at metaphases. Metaphase-enriched cells were then hypotonically treated with 75 mM KCl solution, fixed with methanol∶glacial acetic acid (3∶1), and spread onto clean slides. Telomere FISH and quantification were performed as described previously (Huang et al., 2011), and the FITC-labeled (CCCTAA) peptide nucleic acid (PNA) probe was used in this study. Telomeres were denatured at 85°C for 4 min and hybridized with the telomere PNA probe (0.5 µg/ml) (Panagene, Korea). Chromosomes were stained with 0.5 µg/ml DAPI. Fluorescence from chromosomes and telomeres was digitally imaged on a Zeiss microscope with FITC/DAPI filters, using AxioCam and AxioVision software 4.8. For quantitative measurement of telomere length, telomere fluorescence intensity was integrated using the TFL-TELO program.
C-circle (CC) assay
CC assay was performed as described previously (Henson et al., 2009). Genomic DNA was digested with HinfI and RsaI (4 U/µg) plus RNaseA/T (25 ng/µg) (Dnase-free; Fementas). An aliquot of the digest was used as input, with the remaining diluted to 10 µl final volume that contained 25, 50, or 100 ng of DNA digest. The diluted digest (10 µl) was combined with 10 µl of reaction mixture (5 µg BSA, 0.1% Tween 20, 1 mM dATP, 1 mM dTTP, 1 mM dGTP, 2×Φ29 buffer, and 5 U Φ29 DNA polymerase (NEB)), and then incubated at 30°C for 4 hrs followed by 65°C for 20 min. For quantification, 10 µl final reaction product or input DNA was slot-blotted onto a 2×SSC-soaked Hybond-N+ membrane. Following UV crosslinking, the membrane was hybridized at 45°C with 5′-labeled TelC probes for Φ29 DNA polymerase amplified product or with Alu probes for input DNA [TelC probe: 5′-Biotin-CCCTAACCCTAACCCTAA; Alu probe: 5′-Biotin-GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCA], and then visualized using the Chemiluminescent Nucleic Acid Detection Module (Thermo).
Cell cycle profile analysis
Cultured cells were fixed in 70% ethanol, incubated in PBS with 50 µg/ml RNase A at 37°C for 10 min, and stained with 50 µg/ml propidium iodide before flow cytometry analysis.
Statistical analysis
Percentages were transformed using arcsine transformation. Percentage-transformed data and other data were analyzed by ANOVA and means compared by Fisher's protected least-significant difference (PLSD) using the StatView software from SAS Institute Inc. (Cary, NC, USA). Significant differences were defined as P<0.05 or lower.
Supplementary Material
Footnotes
Author contributions
X.F., J.H. and Z.S. designed the experiments. X.F., Z.L., and S.J. performed the experiments; F.L., X.H., Y.H., D.W., Y.Z. and W.M. gave technical support and conceptual advice. X.F., D.L., J.H., and Z.S. interpreted the data and wrote the manuscript.
Funding
This study was supported by the National Basic Research Program (973 Program) [grant numbers 2012CB911201, 2010CB945401]; the National Natural Science Foundation [grant numbers 31000611, 91019020 and 31171397]; Specialized Research Fund for the Doctoral Program of Higher Education grant number 20100171110028]; Introduced Innovative R&D Team of Guangdong Province [grant number 201001Y0104687244], Zhujiang Program of Science and Technology Nova in Guangzhou [grant number 2011J2200082]; and Fundamental Research Funds for the Central Universities [grant numbers 11lgpy22, 11lgjc08]. We would also like to acknowledge the support of the National Cancer Institute (NCI) [grant number CA133249]; National Institute of General Medical Sciences (NIGMS) [grant number GM095599]; the Welch Foundation [grant number Q-1673]; and the GRSA Shared Resource at the Dan L. Duncan Cancer Center [grant number P30CA125123]. The project described was also supported in part by Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (BCM IDDRC) [grant number 5P30HD024064] from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health. Deposited in PMC for release after 12 months.
Note added in proof
TAH1 was recently found to regulate telomerase activity in telomerase-positive cells (Kappei et al., 2013).
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.128512/-/DC1
References
- Berger M. F., Badis G., Gehrke A. R., Talukder S., Philippakis A. A., Peña-Castillo L., Alleyne T. M., Mnaimneh S., Botvinnik O. B., Chan E. T. et al. (2008). Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266–1276 10.1016/j.cell.2008.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H. (2001). Switching and signaling at the telomere. Cell 106, 661–673 10.1016/S0092-8674(01)00492-5 [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Greider C. W., Szostak J. W. (2006). Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12, 1133–1138 10.1038/nm1006-1133 [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997). Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 10.1038/ng1097-231 [DOI] [PubMed] [Google Scholar]
- Brouwer A. K., Schimmel J., Wiegant J. C., Vertegaal A. C., Tanke H. J., Dirks R. W. (2009). Telomeric DNA mediates de novo PML body formation. Mol. Biol. Cell 20, 4804–4815 10.1091/mbc.E09-04-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T. M., Reddel R. R. (1997). Telomere dynamics and telomerase activity in in vitro immortalised human cells. Eur. J. Cancer 33, 767–773 10.1016/S0959-8049(97)00065-8 [DOI] [PubMed] [Google Scholar]
- Bryan T. M., Englezou A., Dalla-Pozza L., Dunham M. A., Reddel R. R. (1997). Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3, 1271–1274 10.1038/nm1197-1271 [DOI] [PubMed] [Google Scholar]
- Cesare A. J., Griffith J. D. (2004). Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24, 9948–9957 10.1128/MCB.24.22.9948-9957.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare A. J., Reddel R. R. (2010). Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 10.1038/nrg2763 [DOI] [PubMed] [Google Scholar]
- Cesare A. J., Kaul Z., Cohen S. B., Napier C. E., Pickett H. A., Neumann A. A., Reddel R. R. (2009). Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 16, 1244–1251 10.1038/nsmb.1725 [DOI] [PubMed] [Google Scholar]
- Chen S., Saiyin H., Zeng X., Xi J., Liu X., Li X., Yu L. (2006). Isolation and functional analysis of human HMBOX1, a homeobox containing protein with transcriptional repressor activity. Cytogenet. Genome Res. 114, 131–136 10.1159/000093328 [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. (1995). A human telomeric protein. Science 270, 1663–1667 10.1126/science.270.5242.1663 [DOI] [PubMed] [Google Scholar]
- Cong Y. S., Wright W. E., Shay J. W. (2002). Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66, 407–425(Table of contents). 10.1128/MMBR.66.3.407-425.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- Déjardin J., Kingston R. E. (2009). Purification of proteins associated with specific genomic Loci. Cell 136, 175–186 10.1016/j.cell.2008.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M. A., Neumann A. A., Fasching C. L., Reddel R. R. (2000). Telomere maintenance by recombination in human cells. Nat. Genet. 26, 447–450 10.1038/82586 [DOI] [PubMed] [Google Scholar]
- Feldser D. M., Hackett J. A., Greider C. W. (2003). Telomere dysfunction and the initiation of genome instability. Nat. Rev. Cancer 3, 623–627 10.1038/nrc1142 [DOI] [PubMed] [Google Scholar]
- Heaphy C. M., Subhawong A. P., Hong S. M., Goggins M. G., Montgomery E. A., Gabrielson E., Netto G. J., Epstein J. I., Lotan T. L., Westra W. H. et al. (2011). Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 179, 1608–1615 10.1016/j.ajpath.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. D., Neumann A. A., Yeager T. R., Reddel R. R. (2002). Alternative lengthening of telomeres in mammalian cells. Oncogene 21, 598–610 10.1038/sj.onc.1205058 [DOI] [PubMed] [Google Scholar]
- Henson J. D., Cao Y., Huschtscha L. I., Chang A. C., Au A. Y., Pickett H. A., Reddel R. R. (2009). DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27, 1181–1185 10.1038/nbt.1587 [DOI] [PubMed] [Google Scholar]
- Hu C. D., Chinenov Y., Kerppola T. K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 10.1016/S1097-2765(02)00496-3 [DOI] [PubMed] [Google Scholar]
- Huang J., Wang F., Okuka M., Liu N., Ji G., Ye X., Zuo B., Li M., Liang P., Ge W. W. et al. (2011). Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 21, 779–792 10.1038/cr.2011.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. Q., Zhong Z. H., Henson J. D., Neumann A. A., Chang A. C. M., Reddel R. R. (2005). Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 25, 4334 10.1128/MCB.25.10.4334.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. Q., Zhong Z. H., Henson J. D., Reddel R. R. (2007). Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 26, 4635–4647 10.1038/sj.onc.1210260 [DOI] [PubMed] [Google Scholar]
- Jiang W. Q., Zhong Z. H., Nguyen A., Henson J. D., Toouli C. D., Braithwaite A. W., Reddel R. R. (2009). Induction of alternative lengthening of telomeres-associated PML bodies by p53/p21 requires HP1 proteins. J. Cell Biol. 185, 797–810 10.1083/jcb.200810084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappei D., Butter F., Benda C., Scheibe M., Draškovič I., Stevense M., Novo C. L., Basquin C., Araki M., Araki K., et al. (2013). HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J. 32, 1681–1701 10.1038/emboj.2013.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D. et al. (2008). Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 10.1038/ncb1736 [DOI] [PubMed] [Google Scholar]
- Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680 10.1038/ncb1142 [DOI] [PubMed] [Google Scholar]
- Ma W. (2011). Analysis of telomere proteins by Chromatin Immunoprecipitation (ChIP). Methods Mol. Biol. 735, 151–159 10.1007/978-1-61779-092-8_15 [DOI] [PubMed] [Google Scholar]
- Ma W., Kim H., Songyang Z. (2011). Studying of telomeric protein-protein interactions by Bi-molecular fluorescence complementation (BiFC) and peptide array-based assays. Methods Mol. Biol. 735, 161–171 10.1007/978-1-61779-092-8_16 [DOI] [PubMed] [Google Scholar]
- Nabetani A., Ishikawa F. (2009). Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 29, 703–713 10.1128/MCB.00603-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H. T. (2007). The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14, 581–590 10.1038/nsmb1259 [DOI] [PubMed] [Google Scholar]
- Scheel C., Schaefer K. L., Jauch A., Keller M., Wai D., Brinkschmidt C., van Valen F., Boecker W., Dockhorn-Dworniczak B., Poremba C. (2001). Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene 20, 3835–3844 10.1038/sj.onc.1204493 [DOI] [PubMed] [Google Scholar]
- Shay J. W., Bacchetti S. (1997). A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791 10.1016/S0959-8049(97)00062-2 [DOI] [PubMed] [Google Scholar]
- Stagno D'Alcontres M., Mendez-Bermudez A., Foxon J. L., Royle N. J., Salomoni P. (2007). Lack of TRF2 in ALT cells causes PML-dependent p53 activation and loss of telomeric DNA. J. Cell Biol. 179, 855–867 10.1083/jcb.200703020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Meng Z. J., Mason P. J., Veenstra T. D., Artandi S. E. (2008). Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–957 10.1016/j.cell.2008.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M., Qin J., Songyang Z., Liu D. (2009). OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 284, 26725–26731 10.1074/jbc.M109.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. C., Smogorzewska A., de Lange T. (2004). Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119, 355–368 10.1016/j.cell.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Wu G. K., Lee W. H., Chen P. L. (2000). NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275, 30618–30622 10.1074/jbc.C000390200 [DOI] [PubMed] [Google Scholar]
- Xin H., Liu D., Songyang Z. (2008). The telosome/shelterin complex and its functions. Genome Biol. 9, 232 10.1186/gb-2008-9-9-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Liao L., Qin J., Xu J., Liu D., Songyang Z. (2009). Identification of Flightless-I as a substrate of the cytokine-independent survival kinase CISK. J. Biol. Chem. 284, 14377–14385 10.1074/jbc.M807770200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager T. R., Neumann A. A., Englezou A., Huschtscha L. I., Noble J. R., Reddel R. R. (1999). Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59, 4175–4179 [PubMed] [Google Scholar]
- Zeng S. C., Xiang T., Pandita T. K., Gonzalez-Suarez I., Gonzalo S., Harris C. C., Yang Q. (2009). Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 11, 616–623 10.1038/ncb1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z. H., Jiang W. Q., Cesare A. J., Neumann A. A., Wadhwa R., Reddel R. R. (2007). Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 282, 29314–29322 10.1074/jbc.M701413200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.