Abstract
Fluoroquinolone antibiotics have been a mainstay in the treatment of bacterial diseases. The most notable representative, ciprofloxacin, possesses potent antimicrobial activity; however, a rise in resistance to this agent necessitates development of novel derivatives to prolong the clinical lifespan of these antibiotics. Herein we have synthesized and analyzed the antimicrobial properties of a library of N-acylated ciprofloxacin analogues. We find that these compounds are broadly effective against Gram-positive and Gram-negative bacteria, with many proving more effective than the parental drug, and several possessing MICs ≤ 1.0 µg/ml against methicillin-resistant Staphylococcus aureus and Bartonella species. An analysis of spontaneous mutation frequencies reveals very low potential for resistance in MRSA compared to existing fluoroquinolones. Mode of action profiling reveals that modification of the piperazinyl nitrogen by acylation does not alter the effect of these molecules towards their bacterial target. We also present evidence that these N-acylated compounds are highly effective at killing intracellular gbacteria, suggesting the suitability of these antibiotics for therapeutic treatment.
Keywords: N-Acyl ciprofloxacins, Staphylococcus aureus, MRSA, Antimicrobial activity, Antibiotic resistance
Over the last 50 years, the fluoroquinolone antibiotics have been a mainstay in the treatment of bacterial diseases. The most notable member of this family, ciprofloxacin, possesses potent antimicrobial activity against a broad spectrum of Gram-negative and Gram-positive pathogens and, despite recent evidence of bacterial strains having fluoroquinolone resistance, remains one of the foremost lines of defense against pathogenic bacteria.1 Broadly, quinolone-based antibiotics inhibit bacterial DNA replication by interfering with the ability of DNA gyrase and topoisomerase IV to reseal nicked DNA prior to strand passage.2 Acquired resistance to the fluoroquinolones is mediated through chromosomal mutations in bacterial genes encoding these enzymes at specific domains, known as the Quinolone Resistance-Determining Regions (QRDR). Resistance can also occur via mutations that affect import and/or export of the drug, via non-specific efflux mechanisms.2a,3 More recently there have been reports of plasmid-mediated resistance mechanisms, including the quinolone resistance proteins Qnr, Aac(6’) Ib-cr and QepA.4
In an attempt to overcome pathways of bacterial drug-resistance, we set out to explore ways to enhance bioactivity of ciprofloxacin. Given the mode of action and well-characterized structure-activity requirements of this drug, we viewed the best opportunity to accomplish this would be through attachment of lipophilic acyl residues to the nitrogen of the piperazinyl ring. Recent studies have indicated that increased bulkiness of alkyl substituents at this site enhances protection from efflux exporter proteins, and decreases bacterial drug-resistance.5 The antimicrobial properties of a small number of N-acylated ciprofloxacins have previously been described in the patent literature.6 As of yet, there have been no detailed investigations into how well these (and related) analogues may function against drug-resistant bacteria, or whether there might be perturbations to their mode of action as antibacterial agents. In this report we describe the first such studies on N-acylated ciprofloxacin analogues and their microbiological properties against representative pathogenic bacteria, including multidrug resistant strains, and investigate whether they act by interfering with the existing and known mechanisms of action for this class of compounds.
A focused library of 18 lipophilic, N-acylated ciprofloxacin derivatives 2a-r were synthesized using the published procedure of Azema, by treating ciprofloxacin with the requisite acyl chloride or acid anhydride7 in the presence of triethylamine at room temperature (Figure 1). The desired N-acylated products were obtained in 32–96% yields after chromatography, and suitably characterized by proton and carbon NMR spectroscopy.
Figure 1.
Synthesis of N-acylated ciprofloxacin derivatives 2a-r
The antimicrobial properties of these N-acylated ciprofloxacin derivatives were evaluated against several key Gram-positive and Gram-negative bacteria by Kirby-Bauer disk diffusion assay, determination of the minimum inhibitory concentration (MIC), DNA gyrase activity assay, spontaneous mutation frequency assay, and intracellular viability assay.
The N-acyl ciprofloxacins were tested for in vitro bioactivity against three separate Gram-positive bacteria, including Staphylococcus aureus, Bacillus anthracis and Enterococcus faecalis. For the staphylococci, methicillin-susceptible Staphylococcus aureus (MSSA) and methicillin-resistant Staphylococcus aureus (MRSA) strains were examined for comparison.
Several strains of S. aureus were used in testing the N-acyl ciprofloxacins 2a-r. The clinical isolate CBD-635 (MRSA, USA100) was used for initial disk diffusion assays, and ATCC strain 43300 (MRSA), the laboratory strain SH1000 (MSSA) and CBD-635 (MRSA) were employed for the minimum inhibitory concentration assays.8
Disk diffusion assays were performed in triplicate, as previously described, with the average zones of bacterial growth inhibition of each compound shown in Table 1.8 All but four (2c, 2g, 2k, and 2r) of the N-acylated ciprofloxacin derivatives we tested had greater anti-MRSA activity than ciprofloxacin, with the most active of the analogs being N-hexanoyl derivative 2e.
Table 1. Results of Kirby-Bauer testing of N-acylated ciprofloxacins against MRSA USA 100.
Data is shown in millimeters and represents the average diameter of the zone of inhibition from three independent experiments. Each assay was performed with 50 µg of drug per disk. For those compounds that displayed no activity, a zone of 6 mm is shown, which corresponds to the diameter of the disk.
| Compound | R | MRSA (CBD-635) |
|---|---|---|
| 2a | methyl | 24 |
| 2b | ethyl | 20 |
| 2c | propyl | 6 |
| 2d | butyl | 30 |
| 2e | pentyl | 36 |
| 2f | hexyl | 30 |
| 2g | heptyl | 6 |
| 2h | octyl | 22 |
| 2i | nonyl | 34 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 26 |
| 2k | CH(CH2CH2CH3)2 | 6 |
| 2l | CH(CH2CH3)2 | 30 |
| 2m | CH(CH3)CH2CH2CH3 | 33 |
| 2n | C(CH3)2CH2CH3 | 31 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 35 |
| 2p | CH2CH(CH3)2 | 28 |
| 2q | C(CH3)3 | 22 |
| 2r | phenyl | 6 |
| Ciprofloxacin | 6 |
The minimum inhibitory concentrations of the N-acyl ciprofloxacins were evaluated against Staphylococcus aureus SH1000 and the multidrug-resistant MRSA strain CBD-635 according to previous published procedures.8 None of the derivatives exhibited discernible inhibitory activity toward CBD-635 below a concentration of 100 ug/ml (data not shown). Consequently, we elected to use another more common MRSA strain (ATCC 43300), which shows only limited resistance to antibiotics beyond β-lactam compounds. All the antimicrobial assays were performed in triplicate, with the averaged MIC values shown in Table 2. Ciprofloxacin was used as a positive control. Against the MSSA strain, derivatives 2a, 2d, 2h, 2i, 2j, 2k, 2m, 2n, and 2q were all as active as ciprofloxacin, while 2d, 2n, and 2q showed slightly better activity. With regards to the MRSA strain, 2a, 2d, 2h, 2i, 2l, 2m, and 2n gave MIC values lower than that of ciprofloxacin. Curiously, compounds 2d, 2l, 2m, and 2n all showed enhanced bioactivity towards the MRSA than the MSSA.
Table 2. Minimum inhibitory concentrations of N-acyl ciprofloxacins 2a-r against MSSA and MRSA.
Data shown is in ug/ml of antibiotic compound, tested in triplicate and averaged.
| Compound | R | MSSA (SH1000) |
MRSA (ATCC 43300) |
|---|---|---|---|
| 2a | methyl | 10 | 10 |
| 2b | ethyl | 40 | 100+ |
| 2c | propyl | 100+ | 100+ |
| 2d | butyl | 7.5 | 1 |
| 2e | pentyl | 20 | 25 |
| 2f | hexyl | 25 | 100+ |
| 2g | heptyl | 25 | 25 |
| 2h | octyl | 10 | 10 |
| 2i | nonyl | 10 | 10 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 10 | 100+ |
| 2k | CH(CH2CH2CH3)2 | 10 | 100+ |
| 2l | CH(CH2CH3)2 | 100+ | 1 |
| 2m | CH(CH3)CH2CH2CH3 | 10 | 1 |
| 2n | C(CH3)2CH2CH3 | 5 | 1 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 25 | 100+ |
| 2p | CH2CH(CH3)2 | 100+ | 100+ |
| 2q | C(CH3)3 | 7.5 | 100+ |
| 2r | phenyl | 25 | 100+ |
| Ciprofloxacin | 10 | 15 |
Given the ease with which Saureus develops resistance to antimicrobial agents, we undertook spontaneous mutation frequency assays with selected compounds from our library (Table 3).8 For this we chose three representatives (2a, 2i and 2m), which each had MICs of 10 µg/ml in our MSSA assay, and 2b, which had an MIC of 40 µg/ml. In addition, we also included ciprofloxacin as a control agent for these studies. As such, agar containing 2a, 2i and 2m at 1x-, 1.5x-, 2.0x- and 2.5x MIC was prepared, alongside media containing ciprofloxacin at 2.5x MIC. When inoculated with overnight cultures of MSSA we found that all four concentrations of 2i produced lawns of growth, suggesting that resistance is readily developed for this compound. We also obtained a lawn of growth for 2m at 1x MIC; however, we obtained significantly fewer colonies at higher concentrations, with none even being detectable at 2.5x MIC From all tests, we obtained eleven 2m-resistant colonies from a total inoculum of 1.2 × 10−10. This yielded a spontaneous mutation rate of 1.08 × 10−9 for this agent. Testing with compound 2a yielded resistant colonies for each of the concentrations tested, apart from 2.5x MIC, which failed to produce growth. In total we isolated 232 colonies for 2a, from a combined inoculum of 1.7 × 1010, yielding a mutation rate of 7.3 ×10−7. Given the elevated MIC of 2b, we chose the single, and commonly employed, concentration of 2.5 x MIC for analysis. Despite repeating this assay six times, using a combined bacterial inoculum of 3.67 × 1010, we were unable to obtain any mutant colonies. In contrast to these findings, when using a combined inoculum of 5.38 × 108 on agar containing 2.5x MIC of ciprofloxacin, we obtained 551 colonies from five individual tests. This results in a spontaneous mutation frequency of 1.02 × 10−6 for the parent drug. As such, this is a more than 71-fold increase in mutation frequency when compared to 2a, and a more than 1000-fold increase when compared to 2m. This significance is further heightened by the observation that no resistance to either 2a or 2m was observed at the 2.5x MIC concentrations used for ciprofloxacin.
Table 3. Spontaneous mutation frequencies for selected N-acylated ciprofloxacin analogues.
Numbers in the upper row of the table refer to fold increase of the MIC. Lawn refers to a complete covering of the plate with bacterial cells, ND = Not Determined. The values indicate the total colonies obtained for at least 3 independent replicates per compound.
| Compound | 1.0x | 1.5x | 2.0x | 2.5x |
|---|---|---|---|---|
| 2a | 90 | 41 | 101 | 0 |
| 2b | ND | ND | ND | 0 |
| 2i | lawn | lawn | lawn | lawn |
| 2m | lawn | 7 | 4 | 0 |
| Ciprofloxacin | ND | ND | ND | 551 |
N-acyl ciprofloxacins 2a-r were also tested against B. anthracis (Sterne)9 and E. faecalis (DS16)10 using the previously described Kirby-Bauer disk diffusion assay (Table 4). The N-acyl ciprofloxacins performed well in this assay, with only one compound (2r) failing to surpass ciprofloxacin in bioactivity against B. anthracis. Derivatives 2a, 2b, 2c, 2d, 2e, 2f, 2m, 2n,4 2p, and 2q all fared better than the positive control against E. faecalis. For both bacteria, bioactivity dropped as the length or lipophilicity of the acyl chain increased.
Table 4. Kirby-Bauer assay of N-acylated ciprofloxacins 2a-r against B. anthracis and E. faecalis.
Data is shown in millimeters and represents the average diameter of the zone of inhibition from three independent experiments. Each assay was performed with 50 µg of drug per disk. For those compounds that displayed no zone of growth inhibition, a value of 6 mm is shown, which corresponds to the diameter of the disk.
| Compound | R |
B. anthracis (Sterne) |
E. faecali (DS16) |
|---|---|---|---|
| 2a | methyl | 89 | 48 |
| 2b | ethyl | 86 | 46 |
| 2c | propyl | 86 | 40 |
| 2d | butyl | 80 | 39 |
| 2e | pentyl | 77 | 38 |
| 2f | hexyl | 71 | 36 |
| 2g | heptyl | 65 | 29 |
| 2h | octyl | 55 | 23 |
| 2i | nonyl | 6 | 6 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 65 | 28 |
| 2k | CH(CH2CH2CH3)2 | 72 | 26 |
| 2l | CH(CH2CH3)2 | 81 | 22 |
| 2m | CH(CH3)CH2CH2CH3 | 67 | 33 |
| 2n | C(CH3)2CH2CH3 | 57 | 45 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 80 | 25 |
| 2p | CH2CH(CH3)2 | 70 | 32 |
| 2q | C(CH3)3 | 80 | 44 |
| 2r | phenyl | 49 | 6 |
| Ciprofloxacin | 42 | 31 |
N-Acyl ciprofloxacin compounds 2a-r were also tested and found to be effective against Bartonella and Escherichia coli, clinically-significant Gram-negative microbes. Disk diffusion assays were evaluated against four species of Bartonella, including B. henselae (ATCC 49882)11, B. quintana (ATCC VR358), B. elizabethae (F9251)12, and B. vinsonii (ATCC VR152). Minimum inhibitory concentration assays were performed only with the B. henselae strain. All assays were performed in triplicate. While the N-acyl ciprofloxacins showed inhibitory activity against the four Bartonella species tested (Table 5 and Table 6), most showed diminished growth inhibition zone sizes compared to ciprofloxacin. A general relationship between lipophilicity and activity was observed with the more hydrophobic compounds yielding smaller growth inhibition zones.
Table 5. Kirby-Bauer assay of N-acylated ciprofloxacins 2a-r against B. henselae and B. quintana.
Data is shown in millimeters and represents the average diameter of the zone of inhibition from three independent experiments. Each assay was performed with 20 µg of drug per disk. For *those compounds that displayed no activity, a zone of 6 mm is shown, which corresponds to the diameter of the disk.
| Compound | R |
B.henselae (ATCC49882) |
B. Quintana (ATCC VR358) |
|---|---|---|---|
| 2a | methyl | 52 | 6 |
| 2b | ethyl | 56 | 58 |
| 2c | propyl | 34 | 29 |
| 2d | butyl | 38 | 35 |
| 2e | pentyl | 31 | 30 |
| 2f | hexyl | 28 | 20 |
| 2g | heptyl | 16 | 14 |
| 2h | octyl | 6 | 11 |
| 2i | nonyl | 10 | 11 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 12 | 6 |
| 2k | CH(CH2CH2CH3)2 | 14 | 6 |
| 2l | CH(CH2CH3)2 | 33 | 17 |
| 2m | CH(CH3)CH2CH2CH3 | 37 | 24 |
| 2n | C(CH3)2CH2CH3 | 41 | 27 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 15 | 7 |
| 2p | CH2CH(CH3)2 | 62 | 36 |
| 2q | C(CH3)3 | 40 | 33 |
| 2r | phenyl | 21 | 7 |
| Ciprofloxacin | 56 | 6 |
Table 6. Kirby-Bauer assay of N-acylated ciprofloxacins 2a-r against B. elizabethae and B. vinsonii.
Data is shown in millimeters and represents the average diameter of the zone of inhibition from three independent experiments. Each assay was performed with 20 µg of drug per disk. ND = Not Determined. For those compounds that displayed no activity, a zone of 6 mm is shown, which corresponds to the diameter of the disk.
| Compound | R |
B.elizabethae (F9251) |
B. vinsonii (ATCCVR152) |
|---|---|---|---|
| 2a | methyl | 58 | ND |
| 2b | ethyl | 57 | 60 |
| 2c | propyl | 42 | 44 |
| 2d | butyl | 35 | 57 |
| 2e | pentyl | 30 | 21 |
| 2f | hexyl | 20 | 22 |
| 2g | heptyl | 14 | 15 |
| 2h | octyl | 12 | 14 |
| 2i | nonyl | 9 | 9 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 12 | 15 |
| 2k | CH(CH2CH2CH3)2 | 6 | 6 |
| 2l | CH(CH2CH3)2 | 26 | 25 |
| 2m | CH(CH3)CH2CH2CH3 | 32 | 30 |
| 2n | C(CH3)2CH2CH3 | 32 | 33 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 8 | 10 |
| 2p | CH2CH(CH3)2 | 37 | 40 |
| 2q | C(CH3)3 | 36 | 36 |
| 2r | phenyl | 13 | 12 |
| Ciprofloxacin | 58 | 58 |
The minimum inhibitory concentrations of the N-acyl ciprofloxacins were evaluated against B. henselae. Ciprofloxacin was used as a positive control with the averaged MIC values shown in Table 7. Compounds 2b, 2c, and 2n were as active as ciprofloxacin, whereas 2p and 2r displayed higher activity than the control.
Table 7. Minimum inhibitory concentration assay of N-acylated ciprofloxacins 2a-r against B. henselae.
Determined by agar dilution, data shown is in ug/ml of antibiotic compound, tested in triplicate and averaged. ND = not determined.
| Compound | R |
B. henselae (ATCC 49882) |
|---|---|---|
| 2a | methyl | ND |
| 2b | ethyl | 0.5 |
| 2c | propyl | 0.5 |
| 2d | butyl | 5.0 |
| 2e | pentyl | 5.0 |
| 2f | hexyl | 10.0 |
| 2g | heptyl | 10.0 |
| 2h | octyl | 10.0 |
| 2i | nonyl | 5.0 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | ND |
| 2k | CH(CH2CH2CH3)2 | 5.0 |
| 2l | CH(CH2CH3)2 | 5.0 |
| 2m | CH(CH3)CH2CH2CH3 | 0.8 |
| 2n | C(CH3)2CH2CH3 | 0.5 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 1.0 |
| 2p | CH2CH(CH3)2 | 0.2 |
| 2q | C(CH3)3 | ND |
| 2r | phenyl | 0.2 |
| Ciprofloxacin | 0.5–113 |
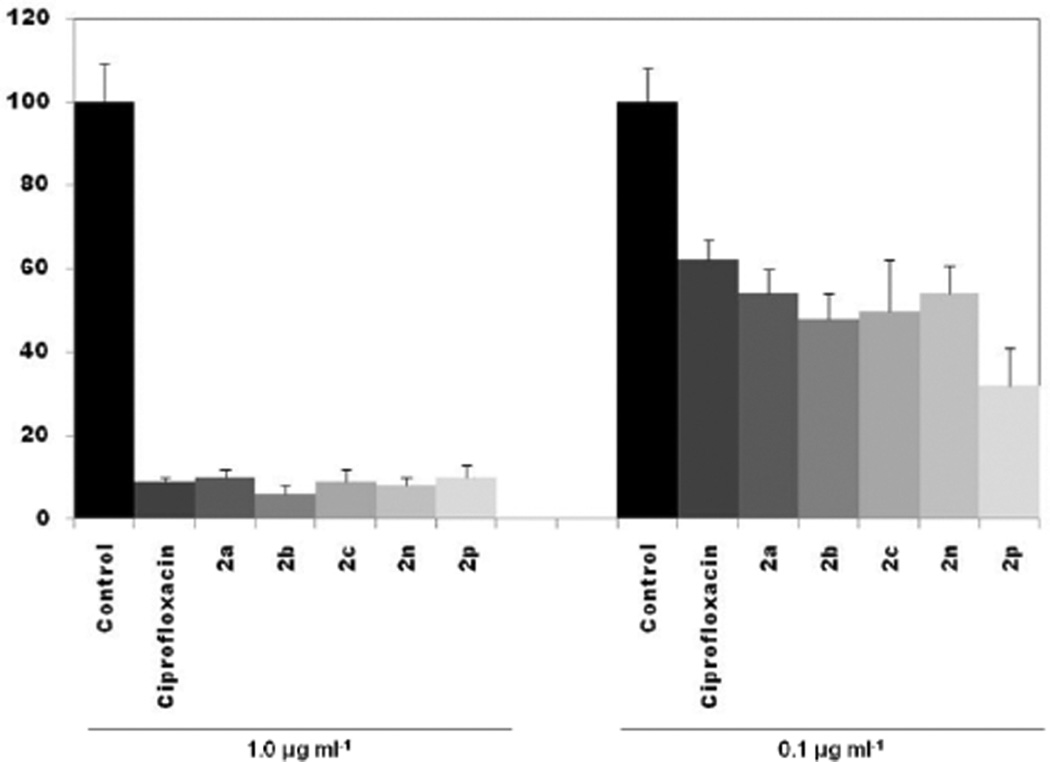
In order to assess the activity of representative compounds against the facultative intracellular bacterium B. henselae, and bioavailability inside the cell, we performed cell infection assays using immortalized microvascular cell line HMEC-1 as described previously.14 After extracellular bacteria were killed with gentamicin, infected cells were incubated for 96 hours with select test compounds. At a concentration of 1.0 µg/ml, all compounds tested were effective at reducing the number of intracellular bacteria to levels ≤10% of those found in infected cells exposed only to solvent controls (Figure 2). At 0.1 µg/ml, the number of surviving bacteria found in cell lysates was higher, ranging from 32% for compound 2p, to 54% for compounds 2a and 2n.
Figure 2. Assay for intracellular antimicrobial activity of selected N-acylated ciprofloxacin compounds against Bartonella henselae.
HMEC cells were infected with B. henselae for 4h, before being incubated with select compounds at the concentration specified for 96h. Values are shown as percentage of colony forming units in comparison to control, which contained media only. Error bars are shown as ± SD.
The Kirby-Bauer assay was used to determine the N-acyl ciprofloxacins antimicrobial assay against the D5Hα strain of E. coli.15 Several of the derivatives proved to be more potent than ciprofloxacin. 2a, 2b, 2c, 2d, 2f, and 2q all yielded larger zones than the positive control as shown in Table 8.
Table 8. Disk diffusion assay of N-acylated ciprofloxacins 2a-r against E. coli.
Data is shown in millimeters and represents the average diameter of the zone of inhibition from three independent experiments. Each assay was performed with 50 µg of drug per disk. For those compounds that displayed no activity, a zone of 6 mm is shown, which corresponds to the diameter of the disk.
| Compound | R |
E. coli (D5Hα) |
|---|---|---|
| 2a | methyl | 60 |
| 2b | ethyl | 57 |
| 2c | propyl | 48 |
| 2d | butyl | 47 |
| 2e | pentyl | 30 |
| 2f | hexyl | 45 |
| 2g | heptyl | 6 |
| 2h | octyl | 30 |
| 2i | nonyl | 6 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 6 |
| 2k | CH(CH2CH2CH3)2 | 6 |
| 2l | CH(CH2CH3)2 | 39 |
| 2m | CH(CH3)CH2CH2CH3 | 33 |
| 2n | C(CH3)2CH2CH3 | 38 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 6 |
| 2p | CH2CH(CH3)2 | 36 |
| 2q | C(CH3)3 | 51 |
| 2r | phenyl | 6 |
| Ciprofloxacin | 43 |
A major requirement for any potential antibiotic targeted towards bacterial species is that it must have a prokaryotic target that is selective and distinct from any eukaryotic counterpart. Therefore, in order to assess the relative toxicity of our library towards eukaryotic cells we repeated our disk diffusion studies using Saccharomyces cerevisiae. Encouragingly, none of the compounds tested were toxic to this organism at concentrations used for the antimicrobial testing (data not shown).
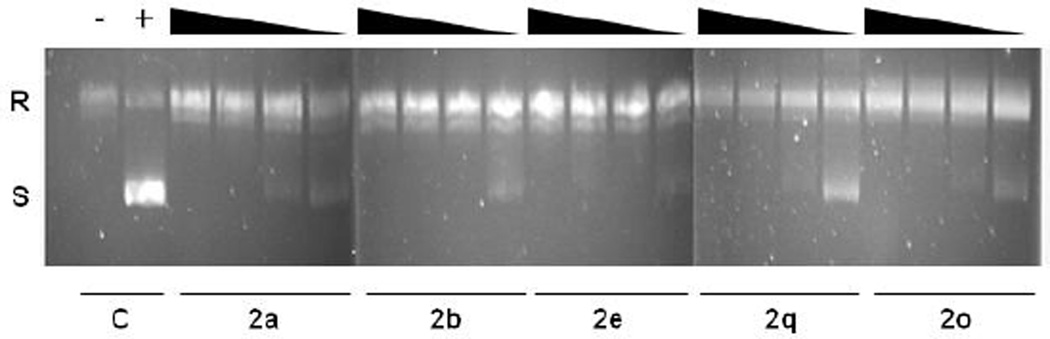
As fluoroquinolones target enzymes that mediate DNA supercoiling, we employed a classic biochemical assay to measure DNA gyrase activity in the presence of select compounds.16 All compounds tested (2a, 2b, 2q, 2e and 2o) exhibited a clear ability to inhibit the supercoiling activity of purified E. coli DNA gyrase when tested with pUC19 at concentrations ranging from 1.0 to 100 µg/ml (Figure 3). Of note, tests with compound 2e appeared to demonstrate minimal supercoiling of the plasmid at even very low concentrations of the antibiotic (1.0 µg/ml). Thus, all five test compounds have inhibitory activity against DNA gyrase, indicating that N-acylation of ciprofloxacin does not abrogate gyrase inhibitory capabilities.
Figure 3. The effects of selected N-acyl ciprofloxacin compounds on DNA Gyrase activity.
Relaxed circular (R) pUC19 DNA was incubated in the presence of E. coli DNA gyrase and decreasing concentrations (100 µg/mL, 50 µg/mL, 10 µg/mL and 1.0 µg/ml) of select compounds. Gyrase conversion of pUC19 to its supercoiled (S) form was inhibited by increasing concentrations of each compound. Control samples without compound (C), in the absence (−) or presence (+) of DNA gyrase, are also shown.
In addition to this biochemical approach, we also undertook DNA sequencing analyses of our spontaneously generated 2a and 2m MSSA mutants. Accordingly, we sequenced the Quinolone Resistance-Determining Regions (QRDR) of the genes encoding DNA gyrase (gyrA and gyrB) and topoisomerase IV (grlA and grlB)17 from representative mutant isolates. We also conducted this analysis in parallel on our parental MSSA strain, and used the data derived to identify mutations arising in these regions. The sequence data for all mutants revealed identical mutations in each strain in the gyrA and grlA genes, regardless of compound used to generate them. Specifically, S84L and E88K mutations were observed in gyrA, and S80Y and E84G mutations in grlA. Interestingly, whilst the former three mutations are well characterized for ciprofloxacin resistance in S. aureus, the latter has only been documented rarely. Indeed, E84G mutation of grlA is more commonly associated in S. aureus with resistance to derivatives of ciprofloxacin, such as trovafloxacin, norfloxacin and besofloxacin.18 No mutations were observed in the QRDR of gyrB or grlB of the mutant strains.
Prior structure-activity studies on the fluoroquinolones starting in the 1970’s have enabled substantial improvements in their potency, spectrum of activity, and in vivo efficacy. Essentially every site on the quinolone framework has been chemically derivatized and evaluated for antibacterial activities, leading to a well-defined understanding of the optimal groups for each site in terms of electrostatics, size, and shape. Included in this list are the C6 fluoro substituent, and the C7 piperazinyl side chain found in ciprofloxacin, and its related structural analogues. Data from the present study indicates that N-acylation of ciprofloxacin not only affects, but can improve, the antibacterial activity of this drug against a variety of bacterial species. When compared to the parental compound, we observed a general increase in efficacy for the derivative compounds. Indeed, it appears that N-acylation of ciprofloxacin significantly improves antibacterial activity towards Gram-positive organisms. Specifically, with regards to the Kirby-Bauer assays, only four derivatives proved less effective than ciprofloxacin when tested against the MRSA strain CBD-635 (2c, 2g, 2k and 2r), eight against E. faecalis (2g, 2h, 2i, 2j, 2k, 2l, 2o and 2r), and only one (2i) had decreased activity compared to ciprofloxacin against B. anthracis. Additionally, a number of compounds showed improvement in activity over ciprofloxacin against Gram-negative organisms, when tested against E. coli and B. quintana.
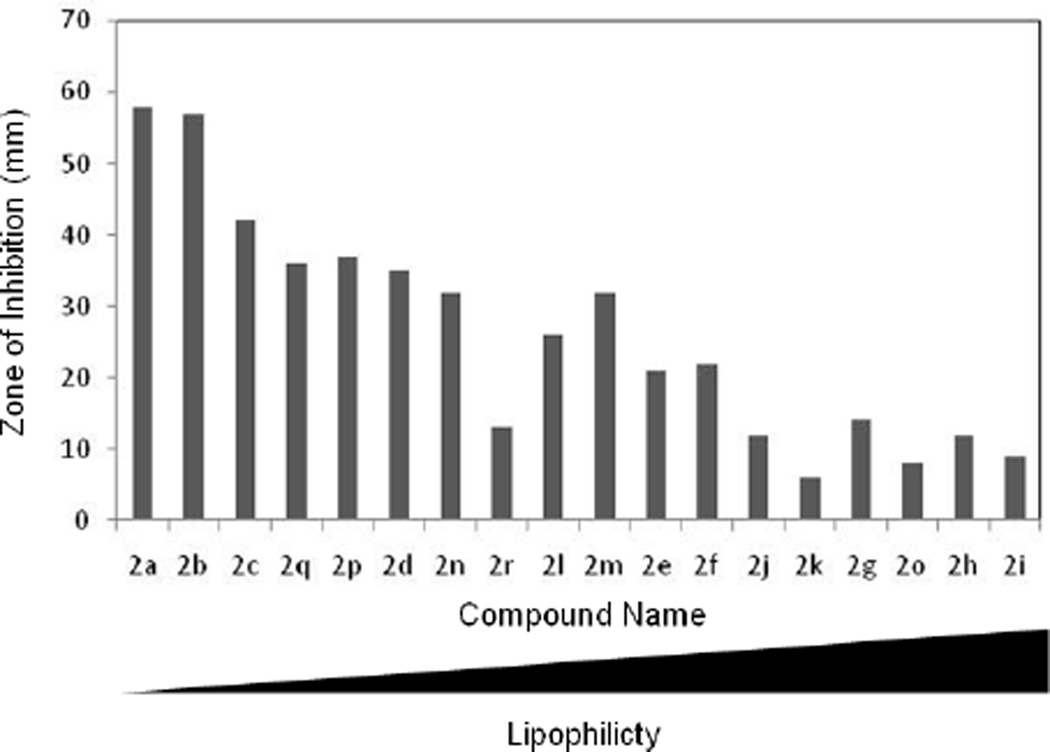
Interestingly, N-acylated ciprofloxacins appear to alter the growth and survival of Gram-positive and Gram-negative bacteria in different Ways . For examples, in the case of the Gram-negative organisms E. coli and Bartonella species, there is a general trend of decreasing bioactivity for the compounds as the acyl chain length increases, which coincides with increasing lipophilicity. The calculated values for logP, a measurement of a compound´s lipophilic character, are provided in Table 9, and plotted out versus anti-Bartonella bioactivity in Figure 4.
Table 9. N-Acyl ciprofloxacins in increasing order of lipophilicity, as determined by their calculated logP values.
(ChemDraw, version 7.0).
| Compound | R | Calculated LogP |
|---|---|---|
| 2a | methyl | 0.17 |
| 2b | ethyl | 0.70 |
| 2c | propyl | 1.23 |
| 2q | C(CH3)3 | 1.41 |
| 2p | CH2CH(CH3)2 | 1.63 |
| 2d | butyl | 1.76 |
| 2n | C(CH3)2CH2CH3 | 1.96 |
| 2r | phenyl | 1.99 |
| 2l | CH(CH2CH3)2 | 2.07 |
| 2m | CH(CH3)CH2CH2CH3 | 2.07 |
| 2e | pentyl | 2.29 |
| 2f | hexyl | 2.81 |
| 2j | CH(CH2CH3)CH2CH2CH2CH3 | 3.12 |
| 2k | CH(CH2CH2CH3)2 | 3.12 |
| 2g | heptyl | 3.34 |
| 2o | CH2CH(CH3)CH2(CH3)3 | 3.48 |
| 2h | octyl | 3.87 |
| 2i | nonyl | 4.40 |
Figure 4. Antimicrobial activity of N-acyl ciprofloxacin derivatives against Gram-negative bacteria is inversely proportional to their lipophilicity.
Values shown are the zones of inhibition against Bartonella elizibethae (in mm), versus the compound listed in order of increasing logP.
This trend is most clear for Bartonella elizabethae, but is also seen with other Bartonella species, and, to a lesser extent, E. coli and B. anthracis (data not shown). Conversely, there is no apparent lipophilicity-activity correlation for MRSA and only a very weak trend for E. faecalis (data not shown). It is interesting that Azema and colleagues reported an analogous observation for ciprofloxacin derivatives bearing lipophilic N-side chains in their antitumor properties against five human cancer cell lines.7
Using a biochemical assay, we were able to demonstrate that select representatives of our library efficiently inhibited the ability of purified E. coli DNA gyrase to super-coil the plasmid pUC19. Furthermore, when analyzing strains of S. aureus having developed resistance to compounds 2a and 2m, we observed point mutations in both gyrA and grlA genes of DNA gyrase, and topoisomerase IV, respectively. As such, it would appear that modification of the piperazinyl nitrogen by acylation does not alter the manner in which these molecules act toward their bacterial target. With regards to the potential for resistance, we demonstrate that, whilst spontaneous mutation was readily obtained for one of the compounds (2i), resistance to others was a far less frequent occurrence. Specifically, we obtained a cumulative mutation frequency of 7.3 ×10−7 for 2a and 1.2 × 10−10 for 2m. In contrast we herein show a spontaneous mutation frequency of 1.02 × 10−6 for ciprofloxacin at 2.5x MIC. As such, this is a more than 71-fold increase in resistance frequency when compared to 2a, and a more than 1000-fold increase when compared to 2m. This clearly demonstrates that the N-acylated ciprofloxacin derivatives in our library have vastly lower mutation frequencies than for ciprofloxacin, which is of particular importance given that ciprofloxacin is rarely used in treating S. aureus infections due to relatively high resistance rates.19 By and large, the mutations obtained within our resistant strains were classical for this type of antimicrobial agent. Specifically, the S84L and E88K mutations in gyrA, and S80Y mutation in grlA have previously been reported for ciprofloxacin. With regards to the E84G mutation of grlB, this is far less common, and is more frequently associated with resistance to trovafloxacin, norfloxacin and besofloxacin.18 As such, the more favorable mutation frequency, coupled with an unusual collection of point mutations required to achieve resistance, suggests the potential suitability of these compounds for treating S. aureus infections.
The N-acylated ciprofloxacin derivatives were even more effective against Gram-negative bacteria, and having lower MICs than those for S. aureus. Most Gram-negative bacteria, and Bartonella species in particular, are sensitive to quinolones, however, newer drugs have been reported to exhibit greater activity than ciprofloxacin.20 In addition, both naturally occurring mutations and laboratory generated mutations in the QRDR of gyrA have been reported and associated with fluoroquinolone resistance in Bartonella species.21 Accordingly, the development of novel quinolone derivatives, such as those presented in this study, is desirable. This contention is enhanced by the finding that select members of our library were able to efficiently kill intracellular B. henselae. Previously, it was reported that levofloxacin (MIC = 0.84 µg/ml) was better than ciprofloxacin (MIC = 15.2 µg/ml), sparfloxacin (MIC = 6.4 µg/ml) and ofloxacin (MIC = 5.6 µg/ml) at killing intracellular B. henselae in infected Vero cells,14b suggesting that the newer antibiotic variants may possess better intracellular activity. As such, the observation that the N-acylated compounds from our library induced 50–70% killing of intracellular bacteria at 0.1 µg/ml, and almost complete killing at 1.0 µg/ml, suggests very real enhancements in activity for these derivatives over existing fluoroquinolones. Indeed, the development of fluoroquinolones with enhanced intracellular activity is not only critical for treating infections caused by Bartonella species, but also infections caused by obligate intracellular bacteria as well. These findings, coupled with the absolute lack of toxicity to the eukaryote S. cerevisiae, further suggest the physiological relevance and suitability of these compounds as a potential treatment option.22
Acknowledgments
This study was supported in part by grant 1R01AI080626-01A2 (LNS) from the National Institute of Allergies and Infectious Diseases, and DOD/DARPA grant HR0011-08-1-0087 (BA and ET).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.a Chin NX, Neu HC. Ciprofloxacin, a quinolone carboxylic acid compound active against aerobic and anaerobic bacteria. Antimicrob. Agents Chemother. 1984;25(3):319. doi: 10.1128/aac.25.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wise R, Andrews JM, Edwards LJ. In vitro activity of Bay 09867, a new quinoline derivative, compared with those of other antimicrobial agents. Antimicrob. Agents Chemother. 1983;23(4):559. doi: 10.1128/aac.23.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Molec. Biol. Rev. 1997;61(3):377. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hoshino K, Kitamura A, Morrissey I, Sato K, Kato J, Ikeda H. Comparison of inhibition of Escherichia coli topoisomerase IV by quinolones with DNA gyrase inhibition. Antimicrob. Agents Chemother. 1994;38(11):2623. doi: 10.1128/aac.38.11.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Gellert M, Mizuuchi K, O’Dea MH, Itoh T, Tomizawa J-I. Nalidixic acid resistance: A second genetic character involved in DNA gyrase activity. Proc. Nat’l. Acad. Sci. 1977;74(11):4772. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hooper DC. New uses for new and old quinolones and the challenge of resistance. Clin. Infect. Dis. 2000;30(2):243. doi: 10.1086/313677. [DOI] [PubMed] [Google Scholar]; c Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Nat’l. Acad. Sci. 1977;74(11):4767. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Asai T, Sato C, Masani K, Usui M, Ozawa M, Ogino T, Aoki H, Sawada T, Izumiya H, Watanabe H. Epidemiology of plasmid-mediated quinolone resistance in Salmonella enterica serovar typhimurium isolates from food-producing animals in Japan. Gut. 2010;2(1):17. doi: 10.1186/1757-4749-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, Medalla F, Chiller TM, Hooper DC. Plasmid-mediated quinolone resistance in non-typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 2006;43(3):297. doi: 10.1086/505397. [DOI] [PubMed] [Google Scholar]; c Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351(9105):797. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]; d Sjolund-Karlsson M, Howie R, Rickert R, Krueger A, Tran TT, Zhao S, Ball T, Haro J, Pecic G, Joyce K, Fedorka-Cray PJ, Whichard JM, McDermott PF. Plasmid-mediated quinolone resistance among non-typhi Salmonella enterica isolates, USA. Emerg. Infect. Dis. 2010;16:1789. doi: 10.3201/eid1611.100464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Beyer R, Pestova E, Millichap JJ, Stosor V, Noskin GA, Peterson LR. A convenient assay for estimating the possible involvement of efflux of fluoroquinolones by Streptococcus pneumoniae and Staphylococcus aureus: evidence for diminished moxifloxacin, sparfloxacin, and trovafloxacin efflux. Antimicrob. Agents Chemother. 2000;44(3):798. doi: 10.1128/aac.44.3.798-801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Davies TA, Kelly LM, Pankuch GA, Credito KL, Jacobs MR, Appelbaum PC. Antipneumococcal activities of gemifloxacin compared to those of nine other agents. Antimicrob. Agents Chemother. 2000;44(2):304. doi: 10.1128/aac.44.2.304-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Pestova E, Mllichap JJ, Noskin GA, Peterson LR. Intracellular targets of moxifloxacin: a comparison with other fluoroquinolones. J. Antimicrob. Chemother. 2000;45(5):583. doi: 10.1093/jac/45.5.583. [DOI] [PubMed] [Google Scholar]; d Costa S, Falcao C, Viveiros M, Machado D, Martins M, Melo-Cristino J, Amaral L, Couto I. Exploring the contribution of efflux on the resistance to fluoroquinolones in clinical isolates of Staphylococcus aureus . BMC Microbiology. 2011;11(1):241. doi: 10.1186/1471-2180-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen UGK, Kiihle E, Zeiler H, Metzger KG. US patent 4559341. 1984 [Google Scholar]
- 7.Azéma J, Guidetti B, Dewelle J, Le Calve B, Mjatovic T, Korolyov A, Vaysse J, Malet-Martino M, Martino R, Kiss R. 7-((4-Substituted)piperazin-1-yl) derivatives of ciprofloxacin: Synthesis and in vitro biological evaluation as potential antitumor agents. Bioorg. Med. Chem. 2009;17(15):5396. doi: 10.1016/j.bmc.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 8.Burda W, Fields K, Gill J, Burt R, Shepherd M, Zhang X, Shaw L. Neutral metallated and meso-substituted porphyrins as antimicrobial agents against Gram-positive pathogens. European J. Clin. Microbiol. Infect. Dis. 2012:327. doi: 10.1007/s10096-011-1314-y. [DOI] [PubMed] [Google Scholar]
- 9.Sterne M. The effects of different carbon dioxide concentrations on the growth of virulent anthrax strains. Onderstepoort. J. Vet. Sci. Anim. Ind. 1937;9:49. [Google Scholar]
- 10.Gawron-Burke C, Clewell DB. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982;300(5889):281. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- 11.Regnery RL, Anderson BE, Clarridge JE, Rodriguez-Barradas MC, Jones DC, Carr JH. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J. Clin. Microbiol. 1992;30(2):265. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Steigerwalt AG, Weaver RE, Daneshvar MI, O’Connor SP. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 1993;31(4):872. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dörbecker C, Sander A, Oberle K, Schülin-Casonato T. In vitro susceptibility of Bartonella species to 17 antimicrobial compounds: comparison of Etest and agar dilution. J. Antimicrob. Chemother. 2006;58(4):784. doi: 10.1093/jac/dkl341. [DOI] [PubMed] [Google Scholar]
- 14.a Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, Lawley TJ. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J. Investig. Dermatol. 1992;99(6):683. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]; b Ives TJ, Marston EL, Regnery RL, Butts JD. In vitro susceptibilities of Bartonella and Rickettsia spp. to fluoroquinolone antibiotics as determined by immunofluorescent antibody analysis of infected Vero cell monolayers. Int. J. Antimicrob. Agents. 2001;18(3):217. doi: 10.1016/s0924-8579(01)00388-0. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. T. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 16.Maxwell A, Burton NP, O’Hagan N. High-throughput assays for DNA gyrase and other topoisomerases. Nucl. Acids Res. 2006;34(15):e104. doi: 10.1093/nar/gkl504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horii T, Suzuki Y, Monji A, Morita M, Muramatsu H, Kondo Y, Doi M, Takeshita A, Kanno T, Maekawa M. Detection of mutations in quinolone resistance-determining regions in levofloxacin- and methicillin-resistant Staphylococcus aureus: effects of the mutations on fluoroquinolone MICs. Diag. Microbiol. Infect. Dis. 2003;46(2):139. doi: 10.1016/s0732-8893(03)00037-3. [DOI] [PubMed] [Google Scholar]
- 18.a Fitzgibbon JE, John JF, Delucia JL, Dubin DT. Topoisomerase mutations in trovafloxacin-resistant Staphylococcus aureus . Antimicrob. Agents Chemother. 1998;42(8):2122. doi: 10.1128/aac.42.8.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Haas W, Pillar CM, Hesje CK, Sanfilippo CM, Morris TW. Bactericidal activity of besifloxacin against staphylococci, Streptococcus pneumoniae and Haemophilus influenzae . J. Antimicrob. Chemother. 2010;65(7):1441. doi: 10.1093/jac/dkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kosmidis C, DeMarco CE, Frempong-Manso E, Seo SM, Kaatz GW. In silico genetic correlations of multidrug efflux pump gene expression in Staphylococcus aureus . Int. J. Antimicrob. Agents. 2010;36(3):222. doi: 10.1016/j.ijantimicag.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Blumber HM, Rimland D, Carroll DJ, Terry P, Wachsmuth IK. Rapid development of ciprofloxacin resistance in methicillin-susceptiable and -resistant Staphylococcus aureus. J. Infect. Dis. 1991;163(6):1279. doi: 10.1093/infdis/163.6.1279. [DOI] [PubMed] [Google Scholar]
- 20.Rolain JM, Brouqui P, Koehler JE, Maguina C, Dolan MJ, Raoult D. Recommendations for treatment of human infections caused by Bartonella species . Antimicrob. Agents Chemother. 2004;48(6):1921. doi: 10.1128/AAC.48.6.1921-1933.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a Angelakis E, Biswas S, Taylor C, Raoult D, Rolain J-M. Heterogeneity of susceptibility to fluoroquinolones in Bartonella isolates from Australia reveals a natural mutation in gyrA. J. Antimicrob. Chemother. 2008;61(6):1252. doi: 10.1093/jac/dkn094. [DOI] [PubMed] [Google Scholar]; b Minnick MF, Wilson ZR, Smitherman LS, Samuels DS. gyrA Mutations in ciprofloxacin-resistant Bartonella bacilliformis strains obtained in vitro . Antimicrob. Agents Chemother. 2003;47(1):383. doi: 10.1128/AAC.47.1.383-386.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Experimental procedures and data on the N-acyl ciprofloxacinsBacterial strains and growth conditions.E. coli, S. aureus, E. faecalis, B. anthracis and S. cerevisiae strains were grown as described previously.8 Bartonella strains were cultured on chocolate agar prepared from heart infusion agar base supplemented with 5% bovine hemoglobin at 37 °C in 5% CO2.Disk diffusion sensitivity assays.Disk diffusion assays for S. aureus, B. anthracis, E. faecalis, E. coli and S. cerevisiae were performed as described previously.8 Owing to the fastidious nature of Bartonella spp., standardized susceptibility testing guidelines (CLSI or EUCAST) are not available. As such, these assays were performed as described previously, with the following modifications.13 Twenty µl of relevant antibiotics, at a concentration of 1mg/ml, were spotted onto the center of 6 mm paper disks (BBL) on a sheet of aluminum foil, in a biological safety cabinet. Disks were allowed to dry for 20 minutes, and then stored in a sealed bag with desiccant at 4 °C. Growth from 4 day old plates was resuspended in 1.0 ml sterile Heart Infusion Broth, and turbidity was adjusted to a McFarland 2.0 by visual inspection. The bacterial suspension was spread over the surface of a chocolate agar plate using a swab. The inoculum was allowed to dry into the agar in a biological safety cabinet for 15 minutes. Disks were then placed in the center of plates, which were inverted and incubated at 37 °C in a 5% CO2 incubator for one week. For all organisms, the zone of inhibition was measured by recording the diameter, to the nearest mm, for each disk.Minimum inhibitory concentration determination.The minimum inhibitory concentration of compounds against MRSA and MSSA strains was determined as described previously.8 MICs for Bartonella strains were determined via agar dilution methods. Briefly, strains were tested for growth on chocolate agar plates containing antibiotics at 10.0 µg/ml, 1.0 µg/ml, and 0.1 µg/ml. Compounds inhibiting growth at ≤ 1.0 µg/ml were further tested to determine the more precise MIC using 2-fold dilutions at and below 1.0 µg/ml. Agar plates containing DMSO (without compound) as a control were prepared at the highest dilution to assess any antibacterial activity associated with the solvent. Growth from four day old chocolate agar plates was collected for each Bartonella strain tested. The growth was suspended into 0.5 ml of sterile Heart Infusion broth. The turbidity was adjusted to a McFarland standard of 2.0 by visual comparison to turbidity standards. Twenty five µl of each bacterial suspension was spotted onto plates containing varying concentrations of drug. Chocolate agar plates with no antibiotics were used as controls to confirm viability. Inoculation drops were allowed to briefly dry into the agar. Plates were inverted and incubated at 37°C with 5% CO2 for 7 days. Growth was recorded as + or − for each strain on duplicate plates.Derivation of spontaneous mutation frequencies.TSB agar (TSA) was prepared containing N-acyl ciprofloxacin derivatives 2a, 2i or 2m at concentrations equivalent to 1x, 1.5x, 2.0x and 2.5x the experimentally-determined MIC for MSSA. For 2b, TSA plates were prepared at a concentration equivalent to 2.5x MIC for MSSA. Overnight broth cultures of MSSA were prepared as described previously8, with 1 ml aliquots extracted, and cells harvested by centrifugation. Supernatants were removed, and pellets resuspended in 100 µl of fresh TSB. These preparations were then used to inoculate the N-acyl ciprofloxacin-containing agar, and spread using sterile glass beads. The colony forming units (cfu) per ml of the inoculating culture was determined via serial dilution into TSA containing no antibiotic compound. Spontaneous mutation frequencies were calculated by dividing the number of colonies obtained by the total bacterial load inoculated.Sequence analysis of quinolone binding domains for spontaneously resistant strains.DNA was extracted from spontaneously resistant MSSA mutants using a DNeasy kit (Qiagen), according to the manufacturer’s instructions. Samples were subject to DNA sequencing reactions (MWG) using primers specific for the Quinolone Resistance-Determining Regions (QRDR) of the gyrAB and grlAB genes of S. aureus, as described previously by Horii et al.17Assay for intracellular activity against Bartonella henselae.The HMEC-1 human microvascular endothelial cell line was maintained in MCDB131 medium supplemented with 10% FBS, 5% L-glutamine, 10 ng/ml EGF, and 1 µg/ml hydrocortisone.1 HMEC-1 were infected with the Houston-1 strain of B. henselae at an MOI of 100 for 4 hours as previously described.22 After infection, the cells were washed 2x with PBS, then treated with gentamicin (200 µg/ml) for 1 hour to kill extracellular adherent bacteria. Infected cells were washed as before and media with test antibiotics were added at concentrations of 0.1 µg/ml and 1.0 µg/ml. After addition of test antibiotics, infected cells were incubated for 96 hours. Following incubation, the antibiotics were removed, the infected cells were washed as before, and lysed with 0.1% saponin. Lysates were plated on chocolate agar and incubated for 7 days. After incubation, the CFU / ml were counted to determine the number of viable bacteria.DNA gyrase activity assay.The activity of select compounds against DNA gyrase was tested using relaxed circular pUC19 DNA in the presence of E. coli DNA gyrase and antibiotics at concentrations of 1.0 µg/ml, 5.0 µg/ml, 10 µg/ml, and 25 µg/ml. Samples were incubated at 37 °C for 1 hour then analyzed by gel electrophoresis to quantify the amount of relaxed and supercoiled DNA, as previously described.16Synthetic procedures.All the chemicals used for the synthesis of the N-acylated ciprofloxacins were purchased from Aldrich Chemical Company and used without further purification. Thin layer chromatography was performed using Silica Gel 60 F254 purchased from EMD Chemicals. A UVG-11 Minera light lamp was used to visualize the TLC plates. The NMR spectra were recorded in deuterated chloroform using an Inova 400 MHz instrument.General methods for the synthesis of N-acyl ciprofloxacins 2a-r.Method A: Ciprofloxacin (500 mg, 1.5 mmol) and triethylamine (300 µl, 2 mmol) were stirred in 20 mL of methylene chloride at 0 °C for 15 min. The desired acyl chloride (2.25 mmol) was added dropwise. The suspension was allowed to stir at room temperature until a clear solution was observed. To this solution, hexane was added drop wise until a white precipitate formed. The precipitate was then filtered off and dried. If further purification was needed, the desired compound was isolated via flash chromatography using 20% methanol in dichloromethane as the eluent.Method B: Ciprofloxacin (500 mg, 1.5 mmol) and triethylamine (300 uL, 2 mmol) were stirred in 20 mL of methylene chloride at 0 °C for 15 min. The desired acid anhydride (3 mmol) was added dropwise. The suspension was allowed to stir at room temperature until a clear solution was observed. To this solution, hexane was added drop wise until a white precipitate formed. The precipitate was then filtered off and dried. If further purification was needed, the desired compound was isolated via flash chromatography using 20% methanol in dichloromethane as the eluent.7-(4-Acetylpiperazin-1-yl)-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2a).Obtained 460 mg (81%) as an off-white solid. Melting Point: >260 °C 1H NMR (400 MHz, CDCl3) δ ppm 8.70 (s, 1 H) 7.96 (d, J=12.8 Hz, 1 H) 7.34 (d, J=6.6 Hz, 1 H) 3.77 (m, 4 H) 3.53 (br. s., 1 H) 3.31 (m, 4 H) 2.14 (s, 3 H) 1.38 (d, J=5.4 Hz, 2 H) 1.18 (br. s., 2 H) 13C NMR (101 MHz, CDCl3) δ ppm 177.0 (d, J=3.0 Hz), 169.1, 166.8, 153.6 (d, J=250.0 Hz), 147.5, 145.4 (d, J=10.7 Hz), 139.0, 120.2 (d, J=7.6 Hz), 112.5 (d, J=23.0 Hz), 108.1, 50.1, 49.4, 46.0, 41.0, 35.3, 21.3, 8.27-(4-propionylpiperazin-1-yl)-1-Cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2b).Obtained 530 mg (91%) as an off-white solid. Melting Point: >260 °C 1H NMR (400 MHz, CDCl3) δ ppm 8.69 (s, 1 H) 7.95 (d, J=12.8 Hz, 1 H) 7.33 (d, J=6.6 Hz, 1 H) 3.77 (m, 4 H) 3.53 (br. s., 1 H) 3.31 (m, 4 H) 2.39 (q, J=7.4 Hz, 2 H) 1.37 (d, J=5.0 Hz, 2 H) 1.17 (m, 5 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.9 (d, J=3.1 Hz), 172.4, 166.7, 153.6 (d, J=251.6 Hz), 147.4, 145.4 (d, J=10.9 Hz), 138.9, 112.4 (d, J=23.2Hz), 108.1, 105.0 (d, J=3 Hz), 50.1, 49.3, 45.1, 41.1, 35.3, 26.4, 9.3, 8.27-(4-Butyryl-piperazin-1-yl)-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2c).Obtained 589 mg (97%) as an off-white solid. Melting Point: >260 °C 1H NMR (400 MHz, CDCl3) δ ppm 8.73 (s, 1 H) 8.00 (d, J=12.9 Hz, 1 H) 7 34(d, J=7.0 Hz, 1 H) 3.79 (m, 4 H) 3.53 (m, 1 H) 3.30 (m, 4 H) 2.35 (m, 2 H) 1.69 (m, 2 H) 1.38 (d, J=6.6 Hz, 2 H) 1.18 (d, J=2.7 Hz, 2 H) 0.98 (t, J=7.4 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ ppm 177.0, 171.6, 166.8, 154.8 (d, J=251.5 Hz), 147.5, 145.4 (d, J=10.7 Hz), 138.97, 120.3 (d, J=9.1 Hz) 112.6 (d, J=24.5 Hz), 108.2, 105.0 (d, J=3.0 Hz), 50.3, 49.4, 41.0, 35.2, 35.1, 18.6, 13.9, 8.21-Cyclopropyl-6-fluoro-4-oxo-7-(4-pentanoyl-piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid (2d).Obtained 423 mg (68%) as an off-white solid. Melting Point: >260 °C 1H NMR (400 MHz, CDCl3) δ ppm 8.56 (br. s., 1 H) 7.79 (d, J=12.0 Hz, 1 H) 7.29 (d, J=7.4 Hz, 1 H) 3.76 (m, 4 H) 3.54 (m, 1 H) 3.30 (m, 4 H) 2.34 (t, J=8.0 Hz, 2 H) 1.59 (quin, J=7.5 Hz, 2 H) 1.34 (m, 4 H) 1.16 (m, 2 H) 0.89 (t, J=7.2 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.7, 171.8, 166.5, 153.5 (d, J=250.1 Hz), 147.3, 145.3 (d, J=11.0 Hz), 138.9, 119.6, 112.0 (d, J=23.4 Hz), 107.7, 105.0 (d, J=4.1 Hz), 50.1, 49.3, 45.3, 41.0, 35.3, 32.9, 27.3, 22.5, 13.8, 8.11-Cyclopropyl-6-fluoro-7-(4-hexanoyl-piperazin-1-yl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2e).Obtained 531 mg (83%) as an off-white solid. Melting Point: 204–206 °C. 1H NMR (400 MHz, CDCl3) δ ppm 0.80 (t, J=6.3 Hz, 3 H) 1.10 (m, 2 H) 1.28 (m, 6 H) 1.55 (br. s., 2 H) 2.29 (t, J=7.4 Hz, 2 H) 3.23 (m, 4 H) 3.52 (br. s., 1 H) 3.72 (m, 4 H) 7.24 (br. s., 1 H) 7.66 (d, J=12.9 Hz, 1 H) 8.46 (br. s., 1 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.5, 171.8, 166.5, 153.3 (d, J=251.6 Hz), 147.2, 145.3 (d, J=7.6 Hz), 138.8, 119.4 (d, J=9.9 Hz), 111.7 (d, J=20.7 Hz), 107.5, 105.0 (d, J=3.2 Hz), 49.9, 49.3, 45.2, 41.0, 35.3, 33.1, 24.8, 22.3, 13.8, 8.11-Cyclopropyl-6-fluoro-7-(4-heptanoyl-piperazin-1-yl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2f).Obtained 560 mg (84%) as an off-white solid. Melting Point: 162–164 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.66 (s, 1 H) 7.92 (d, J=12.9 Hz, 1 H) 7.32 (d, J=7.0 Hz, 1 H) 3.77 (m, 4 H) 3.53 (tt, J=7.0, 3.7 Hz, 1 H) 3.31 (m, 4 H) 2.35 (t, J=8.0 Hz, 2 H) 1.63 (quin, J=7.5 Hz, 2 H) 1.33 (m, 8 H) 1.17 (m, 2 H) 0.86 (t, J=8.0 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.9, 171.8, 166.7, 153.5 (d, J=252.6 Hz), 147.4, 145.4 (d, J=9.3 Hz), 138.9, 120.0, 112.3 (d, J=24.9 Hz), 108.0, 105.0 (d, J=3.4 Hz), 50.1, 49.3, 45.3, 41.0, 35.2, 33.2, 31.5, 29.1, 25.2, 22.5, 14.0, 8.21-Cyclopropyl-6-fluoro-7-(4-octanoyl-piperazin-1-yl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2g).Obtained 656 mg (95%) as an off-white solid. Melting Point: 154–156 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.70 (s, 1 H) 7.96 (d, J=12.9 Hz, 1 H) 7.33 (d, J=7.0 Hz, 1 H) 3.77 (m, 4 H) 3.53 (br. s., 1 H) 3.31 (m, 4 H) 2.36 (t, J=7.8 Hz, 2 H) 1.65 (s, 2 H) 1.32 (m, 12 H) 0.86 (t, J=7.0 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ ppm 177.0, 171.8, 166.7, 153.6 (d, J=250.3 Hz), 147.4, 145.4 (d, J=10.7 Hz), 138.9, 120.1, 112.5 (d, J=23.0 Hz), 108.1, 105.0 (d, J=3.4 Hz), 50.2, 49.4, 45.3, 41.0, 35.2, 33.2, 31.6, 29.4, 29.0, 25.2, 22.5, 14.0, 8.21-Cyclopropyl-6-fluoro-7-(4-nonanoyl-piperazin-1-yl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2h).Obtained 693 mg (96%) as an off-white solid. Melting Point: 136–142 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.73 (s, 1 H) 8.00 (d, J=12.9 Hz, 1 H) 7.34 (d, J=7.0 Hz, 1 H) 3.80 (m, 4 H) 3.52 (m, 1 H) 3.30 (m, 4 H) 2.36 (t, J=7.4 Hz, 2 H) 1.64 (m, 2 H) 1.28 (m, 14 H) 0.86 (t, J=7.0 Hz, 3 H)13C NMR (101 MHz, CDCl3) δ ppm 177.0, 171.8, 166.7, 153.5 (d, J=248.7 Hz), 147.5, 145.4 (d, J=10.8 Hz), 138.9, 120.3, 112.6 (d, J=21.6 Hz), 108.2, 105.0 (d, J=2.9 Hz), 50.3, 49.4, 45.3, 41.0, 35.2, 33.2, 31.7, 29.4, 29.3, 29.1, 25.2, 22.6, 14.0, 8.21-Cyclopropyl-7-(4-decanoyl-piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2i).Obtained 233 mg (32%) as an off-white solid. Melting Point: 130–136 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.63 (s, 1 H) 7.88 (d, J=5.1 Hz, 1 H) 7.31 (d, J=6.6 Hz, 1 H) 3.76 (m, 4 H) 3.53 (br. s., 1 H) 3.30 (m, 4 H) 2.35 (t, J=7.6 Hz, 2 H) 1.62 (m, 2 H) 1.29 (m, 16 H) 0.84 (dd, J=7.0, 5.5 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.8, 171.9, 166.7, 153.4 (d, J=251.6 Hz), 147.4, 145.4 (d, J=9.2 Hz), 138.9, 119.9, 112.3 (d, J=23.0 Hz), 107.9, 105.1 (d, J=3.1 Hz), 50.2, 49.4, 45.4, 41.1, 35.3, 33.2, 31.8, 29.3, 25.3, 22.6, 14.1, 8.21-Cyclopropyl-7-[4-(2-ethyl-hexanoyl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2j).Obtained 646 mg (93%) as an off-white solid. Melting Point: 138–150 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.61 (s, 1 H) 7.84 (d, J=13.3 Hz, 1 H) 7.30 (d, J=7.0 Hz, 1 H) 3.86 (m, 4 H) 3.53 (br. s., 1 H) 3.30 (m, 4 H) 2.58 (m, 1 H) 1.64 (m, 2 H) 1.34 (m, 10 H) 0.85 (m, 5 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.8 (d, J=3.1 Hz),174.9, 166.7, 153.5 (d, J=251.6 Hz), 147.4, 145.3 (d, J=10.7 Hz), 138.9, 119.8 (d, J=7.7 Hz), 112.2 (d, J=23.0 Hz), 107.9, 50.4, 49.6, 45.4, 42.5, 41.2, 35.3, 32.3, 29.8, 259, 228, 14.0, 12 1 8.21-Cyclopropyl-6-fluoro-4-oxo-7-[4-(2-propyl-pentanoyl)-piperazin-1-yl]-1,4-dihydroquinoline-3-carboxylic acid (2k).Obtained 331 mg (48%) as an off-white solid. Melting Point: 178–179 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.54 (s, 1 H) 7.74 (d, J=12.9 Hz, 1 H) 7.27 (d, J=4.7 Hz, 1 H) 3.79 (m, 4 H) 3.53 (br. s., 1 H) 3.28 (m, 4 H) 2.65 (m, 1 H) 1.59 (m, 2 H) 1.25 (m, 10 H) 0.83 (t, J=7.2 Hz, 6 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.7, 175.0, 166.7, 153.4 (d, J=250.1 Hz), 147.3, 145.3 (d, J=10.7 Hz), 138.9, 119.6 (d, J=7.7 Hz), 112.0 (d, J=27.6 Hz), 107.7, 105.0 (d, J=3.0 Hz), 50.3, 49.5, 45.4, 41.2, 40.4, 35.4, 35.1, 20.8, 14.1, 8.11-Cyclopropyl-7-[4-(2-ethyl-butyryl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2l).Obtained 389 mg (58%) as an off-white solid. Melting Point: 248–254 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.68 (s, 1 H) 7.94 (d, J=12.9 Hz, 1 H) 7.33 (d, J=7.0 Hz, 1 H) 3.85 (m, 4 H) 3.53 (dd, J=7.0, 3.5 Hz, 1 H) 3.31 (m, 4 H) 2.54 (tt, J=8.2, 5.3 Hz, 1 H) 1.66 (m, 2 H) 1.50 (m, 2 H) 1.38 (q, J=6.5 Hz, 2 H) 1.18 (m, 2 H) 0.88 (t, J=7.4 Hz, 6 H) 13C NMR (101 MHz, CDCl3) δ ppm 176.9, 174.6, 166.8, 153.5 (d, J=251.6 Hz), 147.5, 145.4 (d, J=10.7 Hz), 139.0, 120.1, 112.4 (d, J=24.6 Hz), 108.1, 105.0 (d, J=3.0 Hz), 50.5, 49.6, 45.4, 44.1, 41.3, 35.3, 25.5, 12.1, 8.21-Cyclopropyl-6-fluoro-7-[4-(2-methyl-pentanoyl)-piperazin-1-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2m).Obtained 504 mg (78%) as an off-white solid. Melting Point: 182–184 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.7 (s, 1 H) 8.0 (d, J=13.0 Hz, 1 H) 7.3 (d, J=7.1 Hz, 1 H) 3.8 (m, 4 H) 3.5 (br. s., 1 H) 3.3 (d, J=16.7 Hz, 4 H) 2.7 (m, 1 H) 1.3 (m, 8 H) 1.1 (d, J=6.8 Hz, 3 H) 0.9 (t, J=6.8 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ 176.9, 175.4, 166.8, 153.5 (d, J=253.1 Hz), 147.5, 145.4 (d, J=10.7 Hz), 139.0, 120.0, 112.4 (d, J=23.0 Hz) 108, 108.0, 105.0 (d, J=3.1 Hz), 50.4, 49.5, 45.3, 41.2, 36.2, 35.1, 20.6, 17.5, 14.1, 8.21-Cyclopropyl-7-[4-(2,2-dimethyl-butyryl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2n).Obtained 343 mg (53%) as an off-white solid. Melting Point: 182–184 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.77 (s, 1 H) 8.05 (d, J=12.5 Hz, 1 H) 7.34 (d, J=7.8 Hz, 1 H) 3.88 (m, 4 H) 3.51 (m, 1 H) 3.30 (m, 4 H) 1.66 (q, J=8.0 Hz, 2 H) 1.39 (m, 2 H) 1.27 (m, 6 H) 1.19 (m, 2 H) 0.91 (t, J=7.8 Hz, 3 H) 13C NMR (101 MHz, CDCl3) δ 177.1, 175.8, 166.9, 153.6 (d, J=251.6 Hz), 147.6, 145.4 (d, J=10.7 Hz), 139.0, 138.9, 120.4, 112.7 (d, J=23.0 Hz), 108.3, 104.9, 50.0, 49.9, 44.7, 43.0, 35.3, 33.3, 26.5, 9.5, 8.31-Cyclopropyl-6-fluoro-4-oxo-7-[4-(3,5,5-trimethyl-hexanoyl)-piperazin-1-yl]-1,4-dihydroquinoline-3-carboxylic acid (2o).Obtained 386 mg (54%) as an off-white solid. Melting Point: 204–206 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.7 (s, 1 H) 7.9 (d, J=12.5 Hz, 1 H) 7.3 (d, J=6.3 Hz, 1 H) 3.8 (m, 4 H) 3.5 (m, 1 H) 3.3 (m, 4 H) 2.3 (m, 2 H) 2.1 (m, 1 H) 1.4 (d, J=6.3 Hz, 2 H) 1.2 (m, 4 H) 1.0 (d, J=6.3 Hz, 3 H) 0.9 (m, 9 H) 13C NMR (101 MHz, CDCl3) δ 176.9, 171.2, 166.7, 153.5 (d, J=251.6 Hz), 147.4, 145.4 (d, J=10.7 Hz), 139.0, 120.0, 112.3 (d, J=23.0 Hz), 108.0, 105.0 (d, J=3.0 Hz), 50.8, 50.3, 49.4, 45.5, 42.5, 41.0, 35.3, 31.1, 30.0, 27.1, 22.9, 8.21-Cyclopropyl-7-[4-(2,2-dimethyl-propionyl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2p).Obtained 462 mg (68%) as an off-white solid. Melting Point: >260 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.7 (s, 1 H) 7.9 (d, J=12.9 Hz, 1 H) 7.3 (d, J=7.0 Hz, 1 H) 3.8 (m, 4 H) 3.5 (br. s., 1 H) 3.3 (m, 4 H) 2.3 (d, J=7.0 Hz, 2 H) 2.1 (dt, J=13.5, 6.5 Hz, 1 H) 1.4 (d, J=6.6 Hz, 2 H) 1.2 (br. s., 2 H) 1.0 (d, J=6.6 Hz, 6 H) 13C NMR (101 MHz, CDCl3) δ ppm 177.0, 171.1, 166.7, 153.5 (d, J=250.0 Hz), 147.5, 145.4 (d, J=10.7 Hz), 139.0, 120.1, 112.5 (d, J=24.6 Hz), 108.1, 105.1 (d, J=3.0 Hz), 50.3, 49.5, 45.6, 42.0, 41.0, 35.3, 25.7, 22.5, 8.21-Cyclopropyl-7-[4-(2,2-dimethyl-propionyl)-piperazin-1-yl]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2q).Obtained 530 mg (85%) as an off-white solid. Melting Point: >260 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.70 (s, 1 H) 7.96 (d, J=12.9 Hz, 1 H) 7.33 (d, J=7.0 Hz, 1 H) 3.87 (m, 4 H) 3.52 (m, 1 H) 3.31 (m, 4 H) 1.38 (m, 2 H) 1.30 (s, 9 H)1.19 (m, 2 H) 13C NMR (101 MHz, CDCl3) δ 177.0, 176.6, 166.8, 153.6 (d, J=250.0 Hz), 147.5, 145.4 (d, J=10.7 Hz), 139.0, 120.1, 112.5 (d, J=24.6 Hz), 108.1, 105.0 (d, J=3.0 Hz), 49.9, 49.8, 44.8, 38.7, 35.3, 28.4, 8.27-(4-Benzoyl-piperazin-1-yl)-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3carboxylic acid (2r).Obtained 572 mg (87%) as an off-white solid. Melting point: >260 °C. 1H NMR (400 MHz, CDCl3) δ ppm 8.8 (s, 1 H) 8.0 (d, J=12.5 Hz, 1 H) 7.5 (m, 6 H) 3.9 (m, 4 H) 3.6 (br. s., 1 H) 3.4 (m, 4 H) 1.4 (d, J=6.6 Hz, 2 H) 1.2 (br. s., 2 H) 13C NMR (101 MHz, CDCl3) δ 176.0, 153.9 (d, J=253.1 Hz), 148.1, 145.9 (d, J=9.2 Hz), 139.4, 132.9, 131.1, 128.9, 127.1, 119.1, 116.2, 113.4, 112.4 (d, J=24.5 Hz), 106.9, 105.3 (d, J=3.1 Hz), 50.1, 49.0, 47.8, 42.5, 36.1, 8.2