Abstract
Ankylosing spondylitis is a common, highly heritable inflammatory arthritis affecting primarily the spine and pelvis. In addition to HLA-B*27 alleles, 12 loci have previously been identified that are associated with ankylosing spondylitis in populations of European ancestry, and 2 associated loci have been identified in Asians. In this study, we used the Illumina Immunochip microarray to perform a case-control association study involving 10,619 individuals with ankylosing spondylitis (cases) and 15,145 controls. We identified 13 new risk loci and 12 additional ankylosing spondylitis–associated haplotypes at 11 loci. Two ankylosing spondylitis–associated regions have now been identified encoding four aminopeptidases that are involved in peptide processing before major histocompatibility complex (MHC) class I presentation. Protective variants at two of these loci are associated both with reduced aminopeptidase function and with MHC class I cell surface expression.
Inflammatory arthritis in ankylosing spondylitis causes pain and stiffness and progressively leads to new bone formation and ankylosis (fusion) of affected joints. It affects 0.55% of populations of European ancestry (herein termed Europeans)1 and 0.23% of Chinese2, but is uncommon in Africans and Japanese, mostly owing to the low prevalence in these ancestry groups of HLA-B*27, the major genetic variant associated with ankylosing spondylitis. Whereas effective treatments are available that suppress inflammation and improve symptoms, there are not yet any treatments that have been shown to robustly slow the rate of ankylosis or induce disease remission.
Ankylosing spondylitis is highly familial (sibling recurrence risk ratio of >52)3 and heritable (h2 > 90%)4. It is two to three times more prevalent in men than in women, and men tend to be more severely affected. More than 80% of cases are positive for the HLA-B*27 allele, but only a minority of HLA-B*27 carriers develop ankylosing spondylitis (1–5%). The low proportion of HLA-B*27 carriers who develop ankylosing spondylitis reflects the fact that numerous other non–HLA-B*27 variants are likely to influence disease susceptibility3. In addition to HLA-B*27, 12 loci have previously been confirmed to be associated with ankylosing spondylitis in Europeans (ANTXR2, CARD9, ERAP1, IL12B, IL23R, KIF21B, PTGER4, RUNX3, TBKBP1, TNFRSF1A and chromosomes 2p15 and 21q22)5–7, and 2 loci have recently been reported in Han Chinese (HAPLN1-EDIL3 and ANO6)8.
The Immunochip Consortium has developed a custom microarray SNP genotyping chip (the Immunochip), the design of which has been informed by available genome-wide association study (GWAS) and deep sequencing data from various autoimmune and inflammatory diseases to provide a cost-effective platform for immunogenetic studies9,10. Genetic data from ankylosing spondylitis, psoriasis, Crohn's disease and ulcerative colitis, along with several classic autoimmune diseases, were used in the chip design, making it a powerful platform for studies of pleiotropic genetic effects in these related diseases. In this study, we aimed to identify new associations with ankylosing spondylitis and to dissect and refine the boundaries of known associated loci by performing a dense SNP genotyping study in 10,619 cases and 15,145 controls of European, East Asian and Latin American ancestry using the Immunochip.
Results
Primary association findings
After all sample quality control filters, the European cohort consisted of 9,069 cases and 13,578 controls, and the East Asian cohort consisted of 1,550 cases and 1,567 controls. The genomic inflation factor (λ) calculated using 1,922 SNPs included on the Immunochip from studies of reading and writing ability, psychosis and schizophrenia was 1.047 (λ1000 for an equivalent study of 1,000 cases and 1,000 controls = 1.0285), indicating minimal evidence of residual population stratification in the overall data set (quantile-quantile plots are presented in Supplementary Fig. 1).
Association at genome-wide significance (P < 5 × 10−8) was observed for 25 loci, including the MHC (Table 1 and Supplementary Fig. 2; genomic control–corrected results are shown in Supplementary Table 1). Suggestive association (P < 5 × 10−7) was observed at six additional loci (Supplementary Table 2). As with all GWAS, there is uncertainty as to the genes contributing to association at specific loci. At previously reported loci, association (P < 5 × 10−8) was seen with the most strongly associated previously reported SNPs at CARD9, ERAP1, IL12B, IL23R, KIF21B, RUNX3, NPEPPS-TBKBP1-TBX21, TNFRSF1A and chromosomes 2p15 and 21q22 (Table 1). At PTGER4, the previously associated SNP (rs10440635) also showed moderate association in the current study (P = 3.0 × 10−5; imputed). No SNPs at the ANTXR2 locus were included on the Immunochip.
Table 1. Non-MHc associations with ankylosing spondylitis susceptibility.
| Europeans | East Asians | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| SNP | Chr. | Positiona | Nearby gene(s) | Combined P | Risk/non-risk allele | Combined OR | RAF (case/control) | OR | P | RAF (case/control) | OR | P |
| Loci previously associated with ankylosing spondylitis at genome-wide significance | ||||||||||||
| rs6600247 | 1p36 | 25177701 | RUNX3 | 2.6 × 10−15 | C/T | 1.15 | 0.540/0.501 | 1.16 | 1.3 × 10−14 | 0.731/0.708 | 1.12 | 0.047 |
| rs11209026 | 1p31 | 67478546 | IL23R | 2.0 × 10−27 | G/A | 1.62 | 0.959/0.934 | 1.65 | 6.0 × 10−28 | 1.000/1.000 | NA | NA |
| rs41299637 | 1q32 | 199144473 | GPR25-KIF21B | 1.9 × 10−15 | T/G | 1.19 | 0.757/0.715 | 1.20 | 7.0 × 10−16 | 0.999/0.998 | 1.05 | 0.42 |
| rs6759298 | 2p15 | 62421949 | Intergenic | 4.9 × 10−47 | C/G | 1.29 | 0.447/0.378 | 1.31 | 3.6 × 10−41 | 0.437/0.374 | 1.28 | 1.6 × 10−6 |
| rs12186979 | 5p13 | 40560617 | PTGER4 | 4.3 × 10−6 | G/A | 1.08 | 0.516/0.498 | 1.09 | 5.4 × 10−6 | 0.202/0.191 | 1.06 | 0.26 |
| rs30187 | 5q15 | 96150086 | ERAP1 | 4.4 × 10−45 | T/C | 1.29 | 0.405/0.338 | 1.32 | 1.3 × 10−41 | 0.542/0.486 | 1.36 | 2.0 × 10−5 |
| rs6871626 | 5q33 | 158759370 | IL12B | 3.1 × 10−8 | A/C | 1.10 | 0.360/0.337 | 1.12 | 6.0 × 10−8 | 0.324/0.310 | 1.08 | 0.17 |
| rs1128905 | 9q34 | 138373660 | CARD9 | 7.0 × 10−9 | C/T | 1.10 | 0.529/0.503 | 1.12 | 1.6 × 10−9 | 0.316/0.315 | 1.00 | 0.99 |
| rs1860545 | 12p13 | 6317038 | LTBR-TNFRSF1A | 2.8 × 10−10 | C/T | 1.13 | 0.634/0.605 | 1.13 | 8.3 × 10−10 | 0.862/0.851 | 1.07 | 0.21 |
| rs9901869 | 17q21 | 42930205 | NPEPPS-TBKBP1-TBX21 | 6.0 × 10−15 | A/G | 1.14 | 0.548/0.516 | 1.15 | 2.3 × 10−12 | 0.667/0.626 | 1.18 | 0.002 |
| rs2836883 | 21q22 | 39388614 | Intergenic | 6.5 × 10−17 | G/A | 1.18 | 0.768/0.734 | 1.19 | 1.8 × 10−14 | 0.817/0.774 | 1.30 | 3.5 × 10−5 |
| New loci associated with ankylosing spondylitis at genome-wide significance | ||||||||||||
| rs4129267 | 1q21 | 152692888 | IL6R | 3.4 × 10−13 | C/T | 1.14 | 0.635/0.592 | 1.18 | 2.1 × 10−15 | 0.619/0.620 | 1.00 | 0.99 |
| rs1801274 | 1q23 | 159746369 | FCGR2A | 1.4 × 10−9 | T/C | 1.11 | 0.487/0.476 | 1.12 | 9.9 × 10−10 | 0.706/0.698 | 1.04 | 0.46 |
| rs12615545 | 2q31 | 181756697 | UBE2E3 | 1.0 × 10−9 | C/T | 1.12 | 0.451/0.421 | 1.11 | 2.3 × 10−7 | 0.710/0.673 | 1.20 | 8.5 × 10−4 |
| rs4676410 | 2q37 | 241212412 | GPR35 | 9.9 × 10−9 | T/C | 1.13 | 0.232/0.209 | 1.13 | 2.1 × 10−7 | 0.346/0.312 | 1.15 | 8.4 × 10−3 |
| rs17765610 | 6q15 | 90722494 | BACH2 | 5.3 × 10−8 | G/A | 1.15 | 0.131/0.118 | 1.17 | 3.3 × 10−8 | 0.017/0.018 | 1.00 | 0.96 |
| rs1250550 | 10q22 | 80730323 | ZMIZ1 | 1.5 × 10−9 | G/T | 1.11 | 0.678/0.652 | 1.11 | 5.8 × 10−7 | 0.583/0.539 | 1.20 | 3.9 × 10−4 |
| rs11190133 | 10q24 | 101268715 | NKX2-3 | 4.9 × 10−14 | C/T | 1.15 | 0.737/0.707 | 1.18 | 1.7 × 10−14 | 0.629/0.617 | 1.10 | 0.30 |
| rs11065898 | 12q24 | 110346958 | SH2B3 | 4.7 × 10−8 | T/C | 1.11 | 0.237/0.216 | 1.13 | 1.7 × 10−7 | 0.348/0.329 | 1.10 | 0.082 |
| rs11624293 | 14q31 | 87558574 | GPR65 | 1.5 × 10−10 | C/T | 1.20 | 0.106/0.087 | 1.23 | 1.8 × 10−10 | 0.158/0.145 | 1.11 | 0.14 |
| imm_16_ 28525386 | 16p11 | 28525386 | IL27-SULT1A1 | 2.6 × 10−9 | A/G | 1.11 | 0.421/0.393 | 1.11 | 1.4 × 10−7 | 0.258/0.232 | 1.16 | 0.012 |
| rs2531875 | 17q11 | 23172294 | NOS2 | 1.2 × 10−10 | G/T | 1.12 | 0.396/0.367 | 1.12 | 1.3 × 10−8 | 0.296/0.256 | 1.22 | 4.6 × 10−4 |
| rs35164067 | 19p13 | 10386181 | TYK2 | 3.4 × 10−10 | G/A | 1.14 | 0.819/0.796 | 1.16 | 6.5 × 10−9 | 0.574/0.549 | 1.11 | 0.039 |
| rs7282490 | 21q22 | 44440169 | ICOSLG | 6.2 × 10−9 | G/A | 1.11 | 0.411/0.390 | 1.10 | 1.4 × 10−6 | 0.581/0.543 | 1.18 | 1.3 × 10−3 |
Locus plots for reported associations are shown in supplementary Figure 5. Chr., chromosome; RAF, risk allele frequency; NA, not available.
NCBI Build 36 human genome coordinates.
We observed little evidence of association with the two previously reported loci in Han Chinese, either in Europeans, East Asians (Chinese, Taiwanese and Koreans) or in the combined data set (P > 0.05)8. To increase power for these variants, we genotyped a total of 2,998 East Asian cases and 5,547 East Asian controls. No association was seen (P > 0.05) for either rs4552569 (chromosome 5q14, between HAPLN1 and EDIL3) or rs17095830 (chromosome 12q12, ANO6) (Supplementary Table 3). rs4552569 showed only nominal significance in Europeans (P = 0.02). rs17095830 was not directly typed on the Immunochip, but, in a previous GWAS6, no association was observed with this SNP (P > 0.1).
Genome-wide significance was seen at 13 loci not previously known to be associated with ankylosing spondylitis (Table 1). The strongest association at each locus was with a common variant (minor allele frequency (MAF) > 5%), but several associations were also seen with rare variants (MAF < 1%) at these loci, including in the genes CARD9, IL23R, LNPEP and TYK2. Both rare variants in IL23R were nonsynonymous coding variants, whereas the CARD9, TYK2 and LNPEP variants were located at exon-intron boundaries and were predicted to influence splicing.
In total, 24.4% of the heritability of ankylosing spondylitis is now explained: 4.3% from loci other than HLA-B and 20.1% due to HLA-B*27 itself.
IL-23 pathway genes
Genetic studies provided the first evidence that interleukin (IL)-23 is involved in the pathogenesis of ankylosing spondylitis, and variants in several genes involved in the IL-23 proinflammatory cytokine pathway have been shown to be associated with the disease. This study adds to that list, with loci containing TYK2, IL6R and IL27 achieving genome-wide significance.
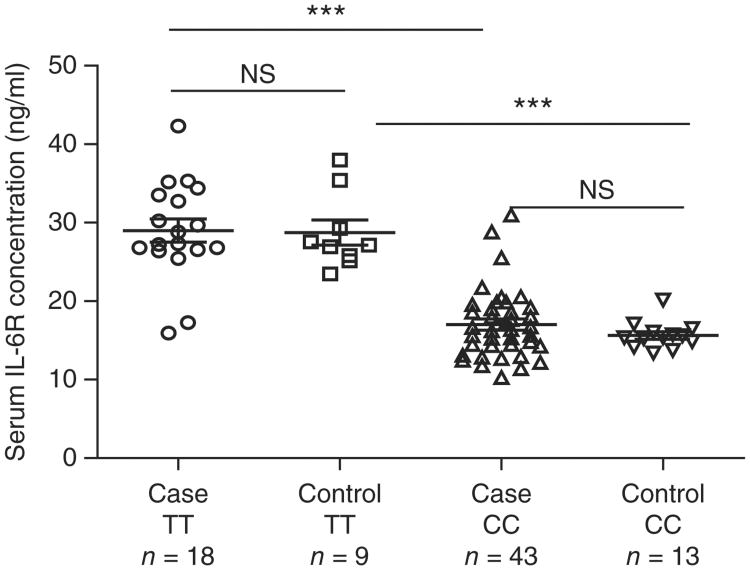
IL-6 signaling through IL-6R has diverse proinflammatory effects. rs4129267, the most strongly associated IL6R SNP in this study, is also associated with asthma11 but with the opposite direction of association to ankylosing spondylitis. The allele associated with risk of ankylosing spondylitis at this SNP is strongly associated with lower serum concentrations of the soluble form of the IL-6 receptor (sIL-6R), with each allele associated with a 1.4-fold variation in serum IL-6R conc-netrations12. We found that sIL-6R concentrations varied strongly by rs4129267 genotype, both overall and separately, in cases and controls (Fig. 1). Overall, homozygous carriers of the T allele at rs4129267 had sIL-6R concentrations 73% higher than homozygous carriers of the C allele (28.9 versus 16.7 ng/ml; P = 7.8 × 10−17). This SNP has previously been associated with serum C-reactive protein (CRP) concentrations at genome-wide significance; in the current study, serum CRP concentrations were 30% higher in cases homozygous for the T allele than in cases homozygous for the C allele (19.2 versus 14.8 mg/l).
Figure 1.
IL6R polymorphism alters IL-6R serum concentrations. IL-6R concentrations were determined in cases and controls who were homozygous for either the T or C allele at the rs4129267 SNP. In both cases and controls, individuals homozygous for the C allele showed significantly lower concentrations of circulating IL-6R. *** P < 0.0001; NS, not significant. Bars represent mean ± s.e.m.
At IL23R, we previously identified two independent disease-associated haplotypes6 tagged by rs11209026 and rs11209032. Here, after conditioning on rs1 1209026, the strongest association was with rs12141575 (odds ratio (OR) = 1.15; P = 9.4 × 10−11); this SNP is in strong linkage disequilibrium (LD) with rs1 1209032 (r2 = 0.993) and also with rs1495965, which has been reported to be associated with Behçet's disease13,14. Behçet's disease is complicated by sacroiliitis resembling ankylosing spondylitis in up to 10% of cases15.
Six loci showed suggestive association (5 × 10−8 < P < 5 × 10−7), including a region on chromosome 2q11 encoding IL1R2 and IL1R1. Conditional analyses showed that there were two separate signals at this locus (Table 2), one in each gene. Both genes encode receptors for the cytokine IL-1, which, among diverse proinflammatory functions, also promotes T helper 17 (TH17) lymphocyte differentiation.
Table 2. Secondary signals in Europeans at loci known to be associated with ankylosing spondylitis.
| Chr. | SNP | Position | Nearby gene(s) | Risk/non-risk allele | Conditional SNP | P | OR | RAF (case/control) | LD (r2/D′) with conditional SNP |
|---|---|---|---|---|---|---|---|---|---|
| 1p31 | rs12141575 | 67520024 | IL23R | A/G | rs11209026 | 9.4 × 10−11 | 1.15 | 0.370/0.330 | 0.034/0.983 |
| 1q23 | rs2039415 | 159121069 | FCGR2A | C/T | rs1801274 | 7.4 × 10−5 | 1.09 | 0.702/0.682 | 0.002/0.062 |
| 2q12 | rs2192752 | 102135805 | IL1R2-IL1R1 | C/A | rs4851529 | 4.1 × 10−6 | 1.11 | 0.239/0.222 | 0.007/0.192 |
| 5q15 | rs10045403 | 96173489 | ERAP1-ERAP2 | A/G | rs30187 and rs2910686 | 5.8 × 10−14 | 1.20 | 0.783/0.730 | 0.178/0.958 0.091/0.429 |
| 5q15 | rs2910686 | 96278345 | ERAP1-ERAP2 | C/T | rs30187 | 4.5 × 10−17 | 1.17 | 0.450/0.440 | 0.153/0.617 |
| 5q33 | rs6556416 | 158751323 | IL12B | C/A | rs6871626 | 4.4 × 10−6 | 1.11 | 0.704/0.675 | 0.048/0.443 |
| 6q15 | rs639575 | 91047852 | BACH2 | A/T | rs17765610 | 8.6 × 10−5 | 1.08 | 0.624/0.609 | 0.000/0.042 |
| 12p13 | rs7954567 | 6361386 | LTBR-TNFRSF1A | A/G | rs1860545 | 1.2 × 10−7 | 1.11 | 0.363/0.341 | 0.002/0.068 |
| 16p11 | rs35448675 | 28236248 | IL27-SULT1A1 | A/G | imm_16_28525386 | 2.4 × 10−4 | 1.24 | 0.007/0.006 | 0.003/0.955 |
| 17q11 | rs2297518 | 23120724 | NOS2 | A/G | rs2531875 | 6.3 × 10−7 | 1.13 | 0.212/0.190 | 0.000/0.048 |
| 19p13 | rs6511701 | 10486067 | TYK2 | A/C | rs35164067 | 1.4 × 10−4 | 1.10 | 0.220/0.218 | 0.162/0.419 |
Aminopeptidase genes
We previously identified a strong association between ERAP1 and ankylosing spondylitis5 that is restricted to HLA-B*27–positive disease6. This observation was replicated here (Table 3 and Supplementary Figs. 3 and 4), with strong interaction observed in the European cases between HLA-B*27 and both independently associated ERAP1 haplotype–tagging SNPs (rs30187: β = 0.390, P = 2.9 × 10−11; rs10045403: β = −0.282, P = 9.3 × 10−6; Supplementary Table 4). We observed no evidence of epistasis between HLA-B*27 and rs30187 (or other SNPs in ERAP1) in the East Asian samples, probably because of inadequate statistical power. In this analysis, we had 35% power to detect an association at a significance level of P < 0.05 with the observed allele frequency of rs30187 in controls and the effect size observed in the overall analysis. The low power to detect a major effect at rs30187 in HLA-B*27–positive East Asian samples suggests that the power to detect an interaction would be quite small. The observed OR (95% confidence interval (CI)) in HLA-B*27–positive subjects at rs30187 was 0.73 (0.53–1.03) compared to 1.20 (0.89–1.61) in HLA-B*27–negative subjects. Although these analyses did not detect significant interaction (P > 0.05), they are consistent with an interaction between rs30187 and HLA-B*27, where rs30187 is only associated in HLA-B*27–positive disease. This finding warrants further exploration with larger sample sizes in East Asian populations.
Table 3. Association analysis of rs30187 and rs2910686 haplotypes in samples positive and negative for HLA-B*27.
| Sample | SNP (ERAP1) | Haplotype counts (controls/cases) | SNP (ERAP2) | Haplotype | Haplotype counts (controls/cases) | OR (95% CI) | rs2910686 association P value |
|---|---|---|---|---|---|---|---|
| rs30187[T] | 790/5,641 | rs2910686[T] | TT | 662/4,595 | 1.18 (0.96–1.44) | 0.12 | |
| rs2910686[C] | TC | 128/1,046 | |||||
| HLA-B*27 positive | |||||||
| rs30187[C] | 1,580/7,979 | rs2910686[T] | CT | 646/2,923 | 1.20 (1.07–1.34) | 1.5 × 10−3 | |
| rs2910686[C] | CC | 934/5,056 | |||||
| rs30187[T] | 8,298/763 | rs2910686[T] | TT | 6,902/629 | 1.05 (0.87–1.28) | 0.64 | |
| rs2910686[C] | TC | 1,396/134 | |||||
| HLA-B*27 negative | |||||||
| rs30187[C] | 16,240/1,515 | rs2910686[T] | CT | 6,842/548 | 1.28 (1.15–1.43) | 7.7 × 10−6 | |
| rs2910686[C] | CC | 9,398/967 | |||||
Association was assessed by 1-degree-of-freedom χ2 test.
LNPEP and ERAP2 are both members of the endoplasmic reticulum (ER) aminopeptidase family and have substantial sequence homology with ERAP1. They are both encoded on chromosome 5q15, immediately centromeric to the ERAP1 locus. A previous study, which did not control for the association of ERAP1 with ankylosing spondylitis, observed no association of an ERAP2 loss-of-function variant with ankylosing spondylitis16. Other studies have identified associations with ERAP2 but have not dissected them from the known associations of ERAP1 SNPs5,17. Here, controlling for the association of ERAP1 with ankylosing spondylitis, two functionally important SNPs in ERAP2 were found to be associated with ankylosing spondylitis: rs2549782, which leads to a change in ERAP2 catalytic activity18, and rs2248374, where the protective G allele causes complete loss of ERAP2 mRNA and absence of ERAP2 protein19. In the European samples, controlling for the association with ERAP1, we identified SNPs in ERAP2 and LNPEP that were associated with ankylosing spondylitis (lead SNP, rs2910686: OR = 1.2, P = 4.5 × 10−17; Supplementary Fig. 3b). Because the association of ERAP1 variants was restricted to HLA-B*27–positive ankylosing spondylitis, analyzing ERAP2 and LNPEP SNPs in HLA-B*27–negative cases and controls produced similar results to analyses of the combined (HLA-B*27–positive and HLA-B*27–negative) cases and controls when the association with ERAP1 was controlled for. Thus, we observed association with ERAP2 SNPs in HLA-B*27–negative ankylosing spondylitis cases (rs2910686: OR = 1.19, P = 2.13 × 10−5). To investigate the possibility that this finding might be an artifact caused by the strong LD between the ERAP2 locus and HLA-B*27 in cases, we tested association at this locus in a multivariate analysis, controlling in the one analysis for association of the two ankylosing spondylitis–associated ERAP1 hap-lotypes (tagged by rs30187 and rs10045403), their interaction with HLA-B*27 and HLA-B*27 itself. In this analysis, association with rs2910686 was robust (P = 6.6 × 10−8), suggesting that ERAP2-LNPEP is independently associated with ankylosing spondylitis. Haplotype counts for rs2910686 and rs30187 in both HLA-B*27–positive and HLA-B*27–negative cases are shown in Table 3.
In addition to these ERAP1 and ERAP2 associations, we observed genome-wide significant association of SNPs on chromosome 17q21 around the gene NPEPPS, which encodes puromycin-sensitive aminopeptidase (rs9901869: OR = 1.14, P = 6.0 × 10−15) (Supplementary Fig. 5). The NPEPPS protein localizes to the cytoplasm and is thought to be involved in processing proteasome-derived peptides before their transport to the endoplasmic reticulum and presentation by human leukocyte antigen (HLA) class I molecules20. Association had previously been reported at this locus and was ascribed to TBKBP1 or TBX21. Here SNPs mapping to NPEPPS and TBX21 were independently associated with ankylosing spondylitis, and conditional analysis suggested that there are at least two independent signals at this locus. When conditioning on rs9901869, SNP rs1 1657479 in the 3′ UTR of TBX21 remained significantly associated with ankylosing spondylitis (OR = 1.09; P = 1.8 × 10−3). The data did not allow us to determine whether TBKBP1 or TBX21 was primarily associated with ankylosing spondylitis, but both represent attractive candidates. TBKBP1 is a component of the tumor necrosis factor (TNF) signaling pathway, and TBX21 is a transcription factor that influences the differentiation of T helper 1 (TH1) and natural killer (NK) cells21.
Genes influencing lymphocyte activation and differentiation
It has recently been shown that cell type–specific trimethylation of histone H3 at lysine 4 (H3K4me3) chromatin marks can inform the fine mapping of associated SNPs to identify causal variation22. We therefore tested all ankylosing spondylitis-associated SNPs against H3K4me3 chromatin marks in different cell lines from the Encyclopedia of DNA Elements (ENCODE) Project23. This analysis showed a strong enrichment of disease-associated SNPs associated with H3K4me3 chromatin marks in cells of immune origin (Supplementary Fig. 6).
Because of this association and taking into account the likely pathogenic role of T lymphocytes in ankylosing spondylitis and the involvement of several genes associated with ankylosing spondylitis in T-lymphocyte differentiation, we tested association of the SNPs for ankylosing spondylitis with CD4+ and CD8+ T cell counts in a previously published GWAS data set24. Association (P < 0.005) was seen between CD8+ lymphocyte counts and ankylosing spondylitis– associated SNPs in the loci harboring the genes IL7R, RUNX3 and ZMIZ1 (Supplementary Table 5). Association was also observed between SNPs in EOMES and CD8+ lymphocyte counts, but these were not the same EOMES SNPs that were associated with ankylosing spondylitis. We also showed association of the genes SH2B3 and BACH2 with both ankylosing spondylitis and CD4+ lymphocyte counts (Supplementary Table 5). We previously showed that CD8+ lymphocyte counts are lower in ankylosing spondylitis cases than in healthy age- and sex-matched controls6. In contrast, in this study, we found that ankylosing spondylitis cases not on biological therapy had similar CD4+ lymphocyte counts as age-matched controls (Supplementary Fig. 7).
HLA Region
After SNP imputation in the MHC region, rs1 16488202 was found to tag HLA-B*27 more accurately in both Europeans and Asians than our previously reported tagging SNP rs4349859 and also rs13202464, reported to tag HLA-B*27 in Asian populations8 (Supplementary Table 6). The expected strong association was observed with HLA-B*27 (OR = 46; P < 1 × 10−100) (Supplementary Fig. 8). Risk of ankylosing spondylitis was further increased in HLA-B*27 homozygotes; HLA-B*27 homozygosity was more prevalent in HLA-B*27-positive cases than in HLA-B*27-positive controls (OR = 2.07; P = 0.0025).
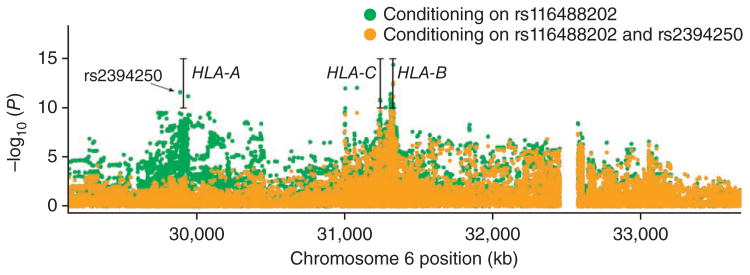
Controlling for association with HLA-B*27, there was residual signal with SNPs near HLA-A and HLA-B (Fig. 2). The residual signal at HLA-B may reflect either imperfect HLA-B*27 tagging by rs1 16488202 or association of other HLA-B alleles with ankylosing spondylitis. No other individual non–HLA-B*27 allele was associated with ankylosing spondylitis, although this may represent imperfect HLA-B imputation using single SNPs.
Figure 2.
Ankylosing spondylitis susceptibility associations in the MHC region conditioning on the HLA-B*27–tagging SNP rs116488202 and further conditioning on the HLA-A*02–tagging SNP rs2394250. The 85-kb gap between positions 32,465 kb and 32,550 kb corresponds to an assembly correction between NCBI Genome Builds 36 and 37 of the human genome.
The most strongly associated SNP near HLA-A, rs2394250, tags the classical allele HLA-A*0201 (Supplementary Table 7). Association of the HLA-A*0201 allele was independent of HLA-B*27 genotype, present in both HLA-B*27–positive (OR = 1.21, P = 6.5 × 10−12; conditioning on rs1 16488202) and HLA-B*27–negative (OR = 1.36, P = 3.2 × 10−13) disease (Supplementary Table 7b). No significant correlation was noted between HLA-B*27 and SNPs tagging HLA-A*0201 (r2 < 0.01), and, thus, this association is not a manifestation of LD with HLA-B*27.
Overlap with other immune-mediated diseases
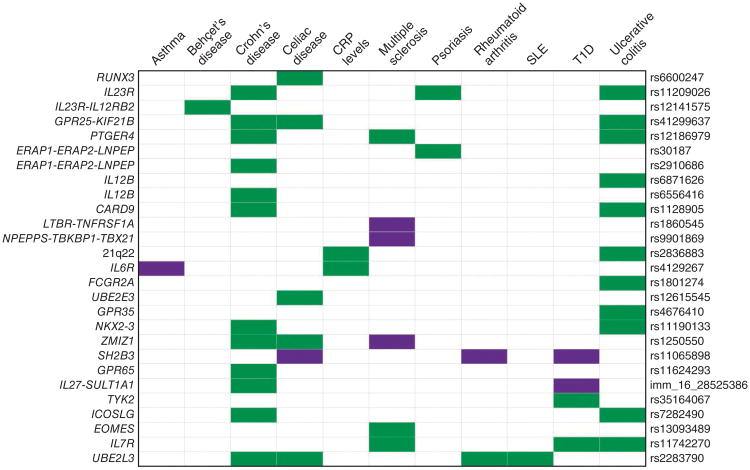
Considering the loci associated with ankylosing spondylitis in this study, we found substantial overlap with other immune-mediated diseases (Fig. 3 and Supplementary Table 8), notably, inflammatory bowel disease (either Crohn's disease or ulcerative colitis) and celiac disease. Ankylosing spondylitis–associated loci were associated with the same SNP in the same direction of association at 12 loci shared with Crohn's disease, at 11 loci shared with ulcerative colitis and at 6 of 7 loci shared with celiac disease. Overlap of associated loci with other diseases was not as marked, including for rheumatoid arthritis (one concordant, one discordant), psoriasis (two concordant), multiple sclerosis (three concordant, three discordant) and type 1 diabetes (two concordant, two discordant).
Figure 3.
Ankylosing spondylitis genetic susceptibility loci overlap with those of other autoimmune diseases. Diseases are represented in columns, and ankylosing spondylitis susceptibility loci are represented in rows. Shared susceptibility loci are colored green if effect size is concordant and purple if effect size is discordant. Data are shown in supplementary table 8. SLE, systemic lupus erythematosus; T1D, type 1 diabetes.
Refinement of disease associations and secondary signals
In the design of the Immunochip, eight loci already known to be associated with ankylosing spondylitis were selected for fine mapping. Compared with available ankylosing spondylitis GWAS data, the current study had greater marker density at these loci and larger sample size (from 3,023 cases and 8,779 controls to 10,619 cases and 15,145 controls). Nonetheless, for most loci studied, the disease-associated region was not substantially narrowed; less than 10% narrowing of the region was observed for four of the eight loci that were fine mapped (Supplementary Table 9). This suggests that, for many loci associated with common variants, the extent of LD at the locus will be too great to permit substantial refinement of the locus using sample sizes and marker densities of the magnitude employed here.
Two or more independent signals were identified at 12 of the 25 genome-wide significant loci (P < 5 × 10−4; Table 2 and Supplementary Fig. 9), including 1 locus (ERAP1 on chromosome 5q15) with 3 associated haplotypes (Supplementary Fig. 3). This is a similar proportion to that found in celiac disease (13 of 36 loci)10. Taken together, these secondary signals contribute 0.75% of the heritability of ankylosing spondylitis (Supplementary Table 10).
Rare variants
Because of the likelihood of population stratification affecting rare variant associations, rare variant associations (MAF < 1%) were only tested on a reduced sample subset of individuals from the UK (7,447 cases and 11,479 controls). Considering loci with common variant associations achieving genome-wide significance, we identified six that harbored rare SNPs that were disease associated (P < 5 × 10−3; Supplementary Table 11). These associations remained significant after conditioning on the common variants, and variants at three loci (IL23R, TYK2 and KEAP1) remained significant after controlling for the number of variants studied per locus. Four loci had exonic rare variant associations in the absence of a common variant association (P < 5 × 10−5) (KLKB1, RAD50, PRDM1 and DYRK4; Supplementary Table 11b). The rare variant with the largest effect was a predicted splice-site variant in TYK2 (rs280518: OR = 7.7, P = 0.002).
Comparisons across ancestry groups
The power of our East Asian case-control cohort was much lower than for our European cohort; nonetheless, at least nominal association (P < 0.01) was detected in East Asians at 13 of the 23 loci for which we identified associations at genome-wide significance in the overall data set (Table 1).
At some loci, association was seen in both East Asians and Europeans but with different SNPs. At IL23R, the primary associated variant in Europeans, rs11209026, was not polymorphic in East Asians, as we and others have previously reported25. However, association was observed at IL23R with a low-frequency nonsynonymous SNP in East Asians (rs76418789: p.Gly149Arg, OR = 1.5, P = 8.2 × 10−4). The minor, protective allele was predicted to be deleterious by both SIFT26 and PolyPhen analysis27. The same SNP was also nominally associated with ankylosing spondylitis in Europeans (P = 0.01). The MAF of rs76418789 was ∼10 times greater in East Asians than in Europeans (East Asians, 3.7%; Europeans, 0.34%). Of the other loci associated with ankylosing spondylitis in Europeans but not in East Asians, only at BACH2 was the key associated variant present at a much lower frequency in East Asians (rs17765610; MAF of 11.8% in Europeans and 1.8% in East Asians), suggesting that, at most loci with discordant association between ancestry groups, this was not due to differences in the population frequency of the associated SNP.
At PTGER4, association in Europeans peaked 155 kb 5′ of the gene (peak associated SNP, rs12186979: OR = 1.1, P = 5.4 × 10−6), whereas the peak of association in East Asians was in intron 2 of the gene (rs13354346: OR = 1.3, P = 1.5 × 10−5) (Supplementary Fig. 5).
Discussion
This study confirmed the association of 12 of the 13 previously reported loci associated with ankylosing spondylitis in Europeans and identified 13 additional loci at genome-wide significance. We found no independent support for two loci previously reported to be associated with ankylosing spondylitis in Han Chinese, suggesting that the original associations of these loci may have been false positives. Additional associated haplotypes were identified at 13 loci, increasing the total number of distinct ankylosing spondylitis associations to 43.
These findings highlight the role of some major biological pathways in the pathogenesis of ankylosing spondylitis, including the IL-23 pathway, gut immunity, T-lymphocyte differentiation or activation, and peptide processing before HLA class I presentation.
We identified three new ankylosing spondylitis–associated genes (TYK2, IL27 and IL6R) with known effects on the IL-23 pathway. TYK2 is a member of the Janus kinase family of intracellular signaling proteins and is involved in signal transduction from IL-23R, as well as other cytokine receptors, including those in interferon (IFN)-α, IFN-β, IL-6, IL-10 and IL-12 signaling. Common TYK2 variants are also associated with Crohn's disease28 and psoriasis29. A different, rare TYK2 variant, rs34536443, is associated with multiple sclerosis30. IL-6 signaling through IL-6R has diverse proinflammatory effects. In combination with transforming growth factor (TGF)-β, it influences the ratio of TH17 to regulatory T (Treg) cells, promoting the differentiation of TH17 cells from naive T cells and inhibiting TGF-β–induced differentiation into Treg cells31. Previous studies have reported no increase in TH17 lymphocyte counts in ankylosing spondylitis, suggesting that the IL6R association with ankylosing spondylitis operates through mechanisms other than effects on TH17 lymphocytes32,33. Whether IL-6 has a role in the differentiation or activation of non-canonical cellular sources of IL-17 such as γδ T cells, NK cells, neutrophils and mast cells, which have been implicated in ankylosing spondylitis, is unclear. IL-27 potentiates the differentiation of CD4+ TH1 cells, while suppressing the differentiation of T helper 2 (TH2) and TH17 cells. IL27 has previously been associated with both Crohn's disease and type 1 diabetes (Supplementary Table 8), with the association in type 1 diabetes being in the opposite direction to that observed in both ankylosing spondylitis and Crohn's disease.
At IL23R, we identified rare variant associations with ankylosing spondylitis in addition to the two known common variant haplotypes at this locus. Although one of these haplotypes has been shown to be due to association with the rs11209026 coding SNP6,25, it is not clear from these genetic studies whether the second haplotype tagged by the intergenic SNP rs12141575 influences ankylosing spondylitis through effects on IL23R or IL12RB2. IL12RB2 encodes one of the two subunits of IL-12R, the stimulation of which drives CD4+ lymphocyte differentiation toward the TH1 lineage and away from the TH17 lymphocyte phenotype.
We identified six ankylosing spondylitis–associated genes that were also associated either with variation in CD8+ lymphocyte counts (EOMES, IL7R, RUNX3 and ZMIZ1) or CD4+ lymphocyte counts (BACH2 and SH2B3). EOMES encodes eomesodermin, a transcription factor involved in CD8+ T cell differentiation whose expression is induced by RUNX3 (refs. 34–36). Where eomesodermin is deficient, CD8+ T cells have been shown to express IL-17 (ref. 37). IL-7 acts through IL-7R to induce RUNX3 expression in developing T cells, in turn favoring differentiation toward the CD8+ T cell lineage38. ZMIZ1 is a transcriptional coactivator of the protein inhibitor of activated STAT (PIAS)-like family and thus may have effects on STAT-mediated cytokine signaling. ZMIZ1 has recently been shown to cooperate with activating NOTCH1 mutations in inducing T cell acute lymphoblastic leukemia, consistent with it having a role in T cell differentiation39. Whether these genes affect risk of ankylosing spondylitis directly through effects on CD8+ T cell differentiation is unclear. For example, although the risk haplotype at RUNX3 is associated with lower CD8+ T cell counts, at IL7R, the opposite phenotype is observed, suggesting that the mechanisms involved are more complex than a simple effect on CD8+ T cell counts. IL-7 treatment has been shown to increase TH17 lymphocyte counts, and it may be that the association of IL7R with ankylosing spondylitis operates directly through such an effect40.
One of the genes associated with both ankylosing spondylitis and CD4+ lymphocyte counts, BACH2, encodes a B cell–specific transcription factor with diverse effects on B cell differentiation and function41; association of ankylosing spondylitis with CD4+ lymphocyte counts may thus be an indirect effect mediated by B cells. This is particularly noteworthy given the recent evidence suggesting that rituximab, a B cell–targeted therapy, may have beneficial effects in ankylosing spondylitis42. SH2B3 (also known as LNK) encodes an adaptor protein involved in T cell receptor signaling43. CD8+ lymphocytes are activated by the interaction of MHC class I peptides with their T cell receptors and may in turn become cytotoxic or memory T cells.
The association of four aminopeptidases involved in peptide trimming before HLA class I presentation is particularly noteworthy. We have shown here and previously that genetic variants associated with reduced function of ERAP1 and loss of expression of ERAP2 are protective for ankylosing spondylitis. Whether LNPEP is also involved is uncertain, and identification of the key associated variants at NPEPPS will require further studies. It is possible that these genes operate in ankylosing spondylitis through a quantitative effect on HLA class I peptide presentation or a qualitative effect on the peptide repertoire presented. Downregulation of ERAP1 (refs. 44,45) and ERAP2 (ref. 19) expression has been shown to reduce cell surface expression of HLA class I molecules. ERAP1 preferentially cleaves hydrophobic amino acids, whereas ERAP2 preferentially cleaves basic residues. ERAP1-ERAP2 heterodimers may thus act in concert, particularly in cleaving longer peptides45. It has been suggested that misfolding of nascent HLA-B*27 in the ER, leading to ER stress, may be involved in the pathogenesis of ankylosing spondylitis46. It is also possible that, by influencing the quantity of peptide available during HLA-B*27 folding, ERAP1 and ERAP2 variants associated with disease risk slow the rate of this folding, thereby increasing ER stress.
In this study, over one-third of loci with common variant associations were found to harbor more than one disease-associated haplotype. Identifying these additional haplotypes increased the proportion of genetic variance explained in ankylosing spondylitis and, more notably, led to valuable biological insights. For example, association of ankylosing spondylitis with SNPs on chromosome 12p13 has previously been reported, although it was not clear whether the association was primarily with TNFRSF1A or LTBR, both plausible candidate genes6,7,47. The current study shows that there are two signals at this locus, one in TNFRSF1A and the other in LTBR. The primary associated SNP at TNFRSF1A (rs1860545) is in strong LD with a multiple sclerosis–associated SNP, rs1800696 (r2 = 0.96, D′ = 0.98) but with the opposite direction of association. rs1800696 has recently been shown to lead to the splicing out of exon 6 of TNFR1, resulting in loss of the transmembrane domain48. The resulting protein acts as a soluble decoy receptor for TNF, akin to the TNF inhibitor drug etanercept. TNF inhibitors are highly effective therapeutic drugs in ankylosing spondylitis, but their use can lead to induction or exacerbation of multiple sclerosis. The association with ankylosing spondylitis suggests the possibility that disease activity and response to TNF inhibitor therapy may be affected by this SNP.
Comparison of the genetic associations of ankylosing spondylitis with other diseases reinforces the considerable overlap with Crohn's disease, ulcerative colitis and ankylosing spondylitis. Ankylosing spondylitis frequently complicates inflammatory bowel disease (both Crohn's disease and ulcerative colitis), and increased cofamiliality with inflammatory bowel disease has been demonstrated49,50, suggesting shared etiopathogenesis. Overlapping genetic susceptibility between ankylosing spondylitis and inflammatory bowel disease has previously been reported51,52. The genes involved include many with effects on the IL-23 pathway, supporting the notion of this being a key pathway in the pathogenesis of these conditions, most likely through effects on gut mucosal immunology. However, the major loci for each disease are not shared, with ankylosing spondylitis showing no association with NOD2 or ATG16L1 and neither Crohn's disease nor ulcerative colitis showing association with HLA-B*27. This suggests that these disease-specific loci contribute to the organ and tissue specificity of the diseases with which they are associated, whereas the IL-23 pathway is involved in the core immunological pathway underlying all these conditions.
We also identified associations with three loci encoding G protein–coupled receptors, including GPR35, GPR37 and GPR65, and a fourth (GPR25) is close to KIF21B, an established ankylosing spondylitis locus where the key associated variants are not yet defined. The functions of these genes and their ligands are not well established. GPR35 is reported to act as a receptor for 2-acyl lysophosphatidic acid, and GPR65 is reported to be a receptor for glycosphingolipids and protons; the ligands for GPR25 and GPR37 are not known. GPR65 has previously been associated with Crohn's disease53 and multiple sclerosis54. The mouse homolog of GPR65, T cell death–associated gene 8, inhibits proinflammatory cytokine production (including of TNF-α and IL-6) in acidic conditions55, suggesting a potential mechanism in diverse autoimmune diseases. However, it also has anti-apoptotic effects and an ability to activate not only cyclic AMP (cAMP) intracellular signaling but also other pathways, including mitogen-activated protein kinase (MAPK) and MEK/ERK signaling and thus is likely to have multiple functions56. Further research is needed into the functions of these genes and their roles in autoimmune diseases.
It has long been suspected that associations in the MHC region with ankylosing spondylitis are not completely explained by HLA-B*27. The association of HLA-A*0201 with ankylosing spondylitis at genome-wide significance in both HLA-B*27–positive and HLA-B*27–negative cases confirms that suspicion. HLA-A*02 has previously been reported to be associated with anterior uveitis complicating ankylosing spondylitis57 and is a risk factor for vitiligo58. HLA-A*0201 has a protective effect in multiple sclerosis54 and is a risk allele for type 1 diabetes59, but no HLA-A association has previously been reported with ankylosing spondylitis itself.
In conclusion, we have increased the number of ankylosing spondylitis–associated loci to 31, identifying 13 new loci and 12 additional ankylosing spondylitis–associated haplotypes at 11 loci, bringing the total number of genetic signals independently associated with anky-losing spondylitis to 43. These loci reinforce the mounting evidence that aberrant peptide processing before MHC class I presentation and alterations of the IL-23 pathway are key elements in the pathogenesis of ankylosing spondylitis.
URL. Haploxt, http://genome.sph.umich.edu/wiki/Haploxt.
Online Methods
Samples
All cases had definite ankylosing spondylitis according to the modified New York criteria60. Written informed consent was obtained from all cases with approval from the relevant research ethics authorities at each participating center. A total of 12,252 DNA samples passed genotyping control filters. The case collection consisted of 10,417 individuals of European ancestry, 1,560 of Asian ancestry and 275 from Latin America (Colombia and Mexico). Of these, 2,425 cases of European ancestry have previously been reported in GWAS.
We obtained 12,338 controls of European ancestry, 1,570 of East Asian ancestry and 445 from Latin America. These included shared controls from the UK 1958 Birth Cohort, the UK Blood Services Common Controls and the United States and from participating centers from France, The Netherlands, Norway, Spain, Mexico, Colombia, China, Taiwan and Korea.
Genotyping
Samples were genotyped using the Immunochip, an Illumina Infinium platform, according to the manufacturer's recommendations. Control samples from the UK were genotyped at the Sanger Centre (Hinxton, Cambridge, UK) and at the University of Virginia (Charlottesville, Virginia, USA), control samples from the United States were genotyped at the Feinstein Institute (New York, New York, USA), and all other controls and all cases were genotyped at the University of Queensland Diamantina Institute (Brisbane, Queensland, Australia). Bead intensity data were processed and normalized for each sample in GenomeStudio; data for successfully genotyped samples were extracted, and genotypes were called within collections using optiCall61. NCBI Build 36 (hg18) mapping was used (Illumina manifest file Immuno_BeadChip_11419691_B.bpm).
SNPs rs4552569, rs13210693 and rs 17095830 were genotyped in 2,998 East Asian cases and 5,547 East Asian controls using TaqMan probes according to the manufacturer's instructions.
The genotyping accuracy of seven disease-associated SNPs with MAF < 1% was confirmed by custom TaqMan genotyping assays. Taking into consideration the five successfully developed assays, genotypes were completely concordant with Immunochip genotyping. Assays could not be developed for two SNPs (rs280518 in TYK2 and rs75430612 in IL2R2). However, given the clean clustering achieved for these SNPs in microarray genotyping and the fact that they had previously been reported, it is likely that they represent true positives.
Data quality control
We first excluded, for each of the collections separately, SNPs with call rate below 95% or with a Hardy-Weinberg equilibrium P value of <1 × 10−6 in controls, as well as samples with call rate below 95%. For the overlapping SNPs, we performed pairwise missingness tests between the collections and removed all SNPs with differential missingness (P < 1 × 10−7). After merging data sets, SNPs with call rate below 98% and samples with call rate below 98% were removed. A total of 128,935 SNPs were analyzed. Cluster plots were visually inspected for all SNPs used to inform conclusions.
The origins of samples of European and Asian ancestry were confirmed by principal-component analysis. Immunochip data were merged with genotype information from seven HapMap 3 populations (CEU, TSI, YRI, MEX, JPT, CHD and CHB), and samples were identified as European or East Asian on the basis of their projection onto the first five principal components of genetic variation (Supplementary Figs. 10–12). Ancestry outliers were removed by assigning samples to an ancestry group using a model-based unsupervised clustering approach62. A second round of principal-component analysis was performed on the European and Asian populations independently to better resolve ancestry differences within the cohorts. In both cases, only the first principal component was correlated with the places of origin of the samples, and only in the European collection was the principal component marginally correlated with case-control status (P = 0.016). The projection of the samples analyzed onto the first two principal components is shown in Supplementary Figures 3 and 4.
Duplicated samples (intentional or unintentional) and those showing cryptic relatedness were assessed for the European and Asian cohorts separately by calculating identity by descent using PLINK (v1.07)63. For each pair of related samples (PI_HAT > 0.2), the sample with the lower call rate was removed, and, where the pair involved a case and a control with similar call rates (both above 98%), cases were preferentially selected for inclusion.
Association analysis
Association analysis and population stratification control was performed using linear mixed models as implemented in FaST-LMM (v1.05)64. For each chromosome, a relationship matrix was constructed with all SNPs, excluding those in the chromosome being analyzed and in the MHC region. Conditional analysis for secondary signal detection was performed by fitting the primary SNP as a fixed effect as implemented in FaST-LMM and using the same SNP set for the relationship matrix.
The decision to apply linear mixed models rather than principal-component analysis for population stratification was informed by a comparison of logistic regression with principal components as covariates against linear mixed models, including different strategies for computing the kinship (or similarity) matrix. In this analysis, we concluded that linear mixed models, when using a kinship matrix, outperformed logistic regression with principal components as covariates, as assessed by the genomic inflation factor in all SNPs (data not shown). We also noticed that including all SNPs when computing the kinship matrix resulted in further loss of power, particularly matrices calculated including HLA-B*27–tagging SNPs, which are a good proxy for phenotype status in a case-control study such as ours. Thus, including SNPs tagging HLA-B*27 in the kinship matrix would have the effect of controlling for affected status, reducing statistical power in a case-control study.
Transformation of SNP effects from the linear 0–1 scale to the liability scale is described in the Supplementary Note. Association plots were generated using LocusZoom, with recombination rates estimated from the HapMap CEU panel (Utah residents of Northern and Western European ancestry) and pairwise LD r2 values estimated from the set of control samples65.
Low-frequency SNP association analysis
A collection of 7,447 cases and 11,479 controls were selected for low-frequency SNP association analysis. These samples were of UK European origin according to principal-component analysis (Supplementary Fig. 11) and recruitment center informaiton. Association analysis was performed with Fisher's exact test and with logistic regression conditioning on common variant association within the locus.
Imputation
We imputed genotypes in candidate regions for the European and Asian cohorts using the EUR and ASN reference panels, respectively, from the 1000 Genomes Project (Phase 1, 2010–2011 data freeze)66. Genotype data were phased with MACH, and genotypes were imputed with Minimac67. SNPs with low imputation quality (r2 < 0.5) were excluded. Association analysis in imputed genotypes was assessed with probabilistic genotypes, correcting for population stratification with the first five principal components as covariates, using logistic regression as implemented in mach2dat.
HLA-B*27–tagging SNP
Imputed SNPs in the MHC region were tested for association and for tagging of HLA-B*27. Performance as a tagging SNP was assessed with a cohort of samples previously genotyped at this locus (Supplementary Table 6b), including 754 controls from the 1958 Birth Cohort population68, 542 ankylosing spondylitis cases from the UK and Australia and 104 ankylosing spondylitis cases and 5 controls from China. SNP rs116488202 was used to tag HLA-B*27 in all analyses.
Overlap with other autoimmune diseases
For all genome-wide significant and suggestive loci for ankylosing spondylitis susceptibility, we searched in a chromosomal window of 0.5 cM around the lead SNP for associations with other autoimmune diseases in the National Human Genome Research Institute (NHGRI) GWAS catalog (accessed 5 June 2012). We then computed the LD between the ankylosing spondylitis–associated SNP and the reported SNP using phased data from the 1000 Genomes Project. If the ankylosing spondylitis–associated SNP was not found, we searched for the next most significant SNP in the locus. When a pair of SNPs was in LD (either r2 > 0.40 or D′ > 0.40), a connection between the two diseases was noted. Positively correlated alleles were then compared for their risk or protective effect on the two diseases to determine whether the directionali-ties of effect were concordant or discordant. LD was computed with Haploxt.
HLA-B*27 experiment-wide interactions
Testing for interaction between HLA-B*27–tagging SNP rs116488202 and all other non-MHC SNPs was performed in samples of European ancestry by logistic regression fitting a dominant term for the HLA-B*27–tagging SNP and an additive term for each test SNP, including a multiplicative interaction term, and five principal components for ancestry correction:
where y is the log odds of disease, βo is the intercept, x1 is a SNP variable reflecting an HLA-B*27 dominant effect (0 or 1), x2 is a SNP variable coded to reflect an underlying additive effect (0, 1 or 2), PCi codes for the projection of samples onto the ith principal component and β terms are regression coefficients estimated from the data.
CD4+ lymphocyte counts in ankylosing spondylitis cases and controls
Peripheral blood was obtained from 20 individuals with active ankylosing spondylitis (erythrocyte sedimentation rate of >25 mm/h and CRP concentration of > 10 mg/l) who were naive for TNF inhibitor and 20 age-matched healthy controls. Peripheral blood mononuclear cells (PBMCs) were extracted using standard density gradient centrifugation over Ficoll-Paque Plus (GE Healthcare). Extracted PBMCs were frozen in FBS with 10% DMSO until needed. Frozen PBMCs were thawed into RPMI supplemented with 20% FBS and were washed once in RPMI supplemented with 10% FBS. Cells were rested in RPMI supplemented with 10% FBS for approximately 1 h at 37 °C at 5% CO2 before further use. Cells were stained with CD3 ECD (UCHT1, Beckman Coulter) and CD4 Pacific Blue (13B8.2, Beckman Coulter). Antibodies were used at final dilutions of 1:100 of antibody stocks. Dead cells were excluded using a Live/Dead Fixable Dead Cell Stain kit (Invitrogen). Cells were acquired on a Gallios flow cytometer (Beckman Coulter), and staining was analyzed using Kaluza software (Beckman Coulter).
IL-6R measurements in serum
Serum was collected from ankylosing spondylitis cases and controls of European ancestry attending the Princess Alexandra Hospital Brisbane Ankylosing Spondylitis Specialist Clinic who were either homozygous for the T or C allele of rs4129267. Serum concentrations of IL-6R were measured using an IL-6R Quantikine ELISA (R&D Systems), and optical density (OD) was determined on a Synergy 2 Microplate reader (BioTek). Serum was diluted 1:100, and ELISAs were performed according to the manufacturer's instructions. All data shown in graphs represent mean ± s.e.m., and differences between groups were analyzed using non-parametric one-way ANOVA; Kruskal-Wallis test with Dunn's multiple-comparison post-hoc test in GraphPad Prism 5 software. Statistical significance was accepted at a significance level of P < 0.05.
Supplementary Material
Acknowledgments
We thank all participating subjects with ankylosing spondylitis and healthy individuals who provided the DNA and clinical information necessary for this study. The Wellcome Trust Case Control Consortium 2 project is funded by the Wellcome Trust (083948/Z/07/Z). We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council (G0000934) and the Wellcome Trust (068545/Z/02), and of the UK National Blood Service controls, funded by the Wellcome Trust. We thank J.C. Barrett for contributing the design of the Immunochip and for helpful analytical discussion, as well as E. Gray, S. Bumpstead, D. Simpkin and the staff of the Wellcome Trust Sanger Institute Sample Management and Genotyping teams for their genotyping and analytical contributions. The Australo-Anglo-American Spondyloarthritis Consortium (TASC) study was funded by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants P01-052915 and R01-AR046208. Funding was also received from University of Texas at Houston Clinical and Translational Science Award (CTSA) UL1RR024188, Cedars-Sinai General Clinical Research Center (GCRC) grant MO1-RR00425, the Intramural Research Program, NIAMS, US National Institutes of Health and the Rebecca Cooper Foundation (Australia). This study was funded, in part, by Arthritis Research UK (grants 19536 and 18797), by the Wellcome Trust (grant 076113) and by the Oxford Comprehensive Biomedical Research Centre ankylosing spondylitis chronic disease cohort (theme A91202). We thank A. Harrison (University of Otago) for his contribution to the New Zealand ankylosing spondylitis cohort. H.X. was funded by National Natural Science Foundation of China grants 81020108029 and 30872339. Portuguese sample collection was performed by COnhecer a Realidade PORtuguesa sobre a Espondilite Anquilosante (CORPOREA Study Group), coordinated by F.M.P.-S. and supported by Bolsa Investigação da Sociedade Portuguesa de Reumatologia/Schering-Plough 2007. The Spanish ankylosing spondylitis case collection was supported by Spanish grant FICYT PC-10-70-Fondos FEDER European Union. The Spondyloarthritis Research Consortium of Canada (SPARCC) was funded by a National Research Initiative Award from the Arthritis Society (Canada). French sample collection was performed by the Groupe Française d'Etude Génétique des Spondylarthrites, coordinated by R. Said-Nahal and funded by Agence Nationale de Recherche GEnetics, Microbiota, Inflammation and Spondyloarthritis (GEMISA) grant ANR-10-MIDI-0002. We thank the Norwegian Bone Marrow Donor registry for providing data from healthy Norwegian controls. W.P.M. is a Medical Scientist of Alberta Innovates–Health Solutions. The Psoriatic Arthritis Program is supported by the Krembil Foundation and the Arthritis Society. P.C.R. is funded by the National Health and Medical Research Council (Australia) (NHMRC) and Arthritis Australia. J.Y. is supported by NHMRC grants 613672 and 1011506. M. Ward is supported by the Intramural Research Program, NIAMS, US National Institutes of Health. D.E. is supported by the research council of Ghent University and by the Fund for Scientific Research Flanders. M.A.B. is funded by a National Health and Medical Research Council (Australia) Senior Principal Research Fellowship, and support for this study was received from a National Health and Medical Research Council (Australia) program grant (566938) and project grant (569829) and from the Australian Cancer Research Foundation and the Rebecca Cooper Medical Research Foundation. We thank A. Gardiner and the Brisbane Convention and Exhibition Centre for their assistance in preparing the manuscript. We are also very grateful for the invaluable support received from the National Ankylosing Spondylitis Society (UK) and the Spondyloarthritis Association of America in case recruitment. Additional financial and technical support for subject recruitment was provided by the NIHR Oxford Musculoskeletal Biomedical Research Unit and NIHR Thames Valley Comprehensive Local Research and by an unrestricted educational grant from Abbott Laboratories.
Footnotes
Note: Supplementary information is available in the online version of the paper.
Author Contributions: J. Hadler, K.C., K.P. and J. Harris performed genotyping. A.C., P.C.R., T.K., P.L., J.Y., M.A.B. and D.M.E. performed statistical analyses. J.P.P., S.L., K.B.J., S.-C.S., M. Weisman, M. Ward, X.Z., H.-J.G., G.C., J.N., B.A.L., Ø.F., J.T., K.L., L.J., Y.L., X.W., L.A.B., D.E., R.B.-V., S.S., L.A., C.F., J.L., N.H., J. Mulero, J.L.F.-S., M.A.G.-G., C.L.-L., P. Deloukas, P. Donnelly, P.B., K.G., H.G., D.D.G., P.R., W.P.M., H.X., J.B.A.C., I.E.v.d.H.-B., C.-T.C., R.V.-O., C.R.-S., I.M.H., F.M.P.-S., R.D.I., V.V., J. Martin, M.B., J.D.R. and T.-H.K. all contributed to subject recruitment and study design. A.C., M.A.B., D.M.E. and B.P.W. wrote the manuscript, and all authors contributed to manuscript drafting and reviewed the final manuscript. T.J.K. performed cell count and IL-6R studies in ankylosing spondylitis cases and controls. M.A.F. performed GWAS of cell counts in controls.
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Braun J, Listing J, Sieper J. Overestimation of the prevalence of ankylosing spondylitis in the Berlin study: comment on the article by Braun et al—Reply. Arthritis Rheum. 2005;52:4049–4050. doi: 10.1002/art.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ng SC, et al. Epidemiology of spondyloarthritis in the People's Republic of China: review of the literature and commentary. Semin Arthritis Rheum. 2007;37:39–47. doi: 10.1016/j.semarthrit.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Brown MA, Laval SH, Brophy S, Calin A. Recurrence risk modelling of the genetic susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2000;59:883–886. doi: 10.1136/ard.59.11.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MA, et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 1997;40:1823–1828. doi: 10.1002/art.1780401015. [DOI] [PubMed] [Google Scholar]
- 5.Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reveille JD, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42:123–127. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Z, et al. A genome-wide association study in Han Chinese identifies new susceptibility loci for ankylosing spondylitis. Nat Genet. 2012;44:73–77. doi: 10.1038/ng.1005. [DOI] [PubMed] [Google Scholar]
- 9.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira MA, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuki N, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet's disease susceptibility loci. Nat Genet. 2010;42:703–706. doi: 10.1038/ng.624. [DOI] [PubMed] [Google Scholar]
- 14.Remmers EF, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet's disease. Nat Genet. 2010;42:698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ait Badi MA, et al. Skeletal manifestations in Behcet's disease. A report of 79 cases. Rev Med Interne. 2008;29:277–282. doi: 10.1016/j.revmed.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Harvey D, et al. A common functional variant of endoplasmic reticulum aminopeptidase 2 (ERAP2) that reduces major histocompatibility complex class I expression is not associated with ankylosing spondylitis. Rheumatology. 2011;50:1720–1721. doi: 10.1093/rheumatology/ker199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsui FW, et al. Association of an ERAP1-ERAP2 haplotype with familial ankylosing spondylitis. Ann Rheum Dis. 2010;69:733–736. doi: 10.1136/ard.2008.103804. [DOI] [PubMed] [Google Scholar]
- 18.Evnouchidou I, et al. A common single nucleotide polymorphism in Endoplasmic Reticulum Aminopeptidase 2 induces a specificity switch that leads to altered antigen processing. J Immunol. 2012;189:2383–2392. doi: 10.4049/jimmunol.1200918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrés AM, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lévy F, et al. The final N-terminal trimming of a subaminoterminal proline-containing HLA class I–restricted antigenic peptide in the cytosol is mediated by two peptidases. J Immunol. 2002;169:4161–4171. doi: 10.4049/jimmunol.169.8.4161. [DOI] [PubMed] [Google Scholar]
- 21.Xia Z, et al. A 17q12 allele is associated with altered NK cell subsets and function. J Immunol. 2012;188:3315–3322. doi: 10.4049/jimmunol.1102775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira MA, et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson SI, et al. Association of ERAP1, but not IL23R, with ankylosing spondylitis in a Han Chinese population. Arthritis Rheum. 2009;60:3263–3268. doi: 10.1002/art.24933. [DOI] [PubMed] [Google Scholar]
- 26.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato K, et al. Strong evidence of a combination polymorphism of the tyrosine kinase 2 gene and the signal transducer and activator of transcription 3 gene as a DNA-based biomarker for susceptibility to Crohn's disease in the Japanese population. J Clin Immunol. 2009;29:815–825. doi: 10.1007/s10875-009-9320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ban M, et al. Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur J Hum Genet. 2009;17:1309–1313. doi: 10.1038/ejhg.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 32.Kenna TJ, et al. Enrichment of circulating interleukin-17–secreting interleukin-23 receptor–positive γ/δ T cells in patients with active ankylosing spondylitis. Arthritis Rheum. 2012;64:1420–1429. doi: 10.1002/art.33507. [DOI] [PubMed] [Google Scholar]
- 33.Appel H, et al. Analysis of IL-17+ cells in facet joints of patients with spondyloarthritis suggests that the innate immune pathway might be of greater relevance than the Th17-mediated adaptive immune response. Arthritis Res Ther. 2011;13:R95. doi: 10.1186/ar3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 35.Yagi R, et al. The transcription factor GATA3 actively represses RUNX3 protein–regulated production of interferon-γ. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JH, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rakowski LA, et al. Convergence of the ZMIZ1 and NOTCH1 pathways at C-MYC in acute T lymphoblastic leukemias. Cancer Res. 2013;73:930–941. doi: 10.1158/0008-5472.CAN-12-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartgring SA, Willis CR, Bijlsma JW, Lafeber FP, van Roon JA. Interleukin-7 aggravated joint inflammation and tissue destruction in collagen-induced arthritis is associated with T-cell and B-cell activation. Arthritis Res Ther. 2012;14:R137. doi: 10.1186/ar3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muto A, et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song IH, et al. Different response to rituximab in tumor necrosis factor blocker–naive patients with active ankylosing spondylitis and in patients in whom tumor necrosis factor blockers have failed: a twenty-four-week clinical trial. Arthritis Rheum. 2010;62:1290–1297. doi: 10.1002/art.27383. [DOI] [PubMed] [Google Scholar]
- 43.Huang X, Li Y, Tanaka K, Moore KG, Hayashi JI. Cloning and characterization of Lnk, a signal transduction protein that links T-cell receptor activation signal to phospholipase Cγ1, Grb2, and phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 1995;92:11618–11622. doi: 10.1073/pnas.92.25.11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evnouchidou I, et al. Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by infuencing basic enzymatic properties of the enzyme. J Immunol. 2011;186:1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saveanu L, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 46.Colbert RA, et al. HLA-B27 misfolding activates the IL-23/IL-17 axis via the unfolded protein response in transgenic rats: evidence for a novel mechanism of inflammation. Arthritis Rheum. 2007;1283:S515. [Google Scholar]
- 47.Karaderi T, et al. Evidence of genetic association between TNFRSF1A encoding the p55 tumour necrosis factor receptor, and ankylosing spondylitis in UK Caucasians. Clin Exp Rheumatol. 2012;30:110–113. [PubMed] [Google Scholar]
- 48.Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemminki K, Li X, Sundquist K, Sundquist J. Familial association of inflammatory bowel diseases with other autoimmune and related diseases. Am J Gastroenterol. 2010;105:139–147. doi: 10.1038/ajg.2009.496. [DOI] [PubMed] [Google Scholar]
- 50.Thjodleifsson B, Geirsson AJ, Bjornsson S, Bjarnason I. A common genetic background for inflammatory bowel disease and ankylosing spondylitis: a genealogic study in Iceland. Arthritis Rheum. 2007;56:2633–2639. doi: 10.1002/art.22812. [DOI] [PubMed] [Google Scholar]
- 51.Danoy P, et al. Association of variants at 1q32 and STAT3 with ankylosing spondylitis suggests genetic overlap with Crohn's disease. PLoS Genet. 2010;6:e1001195. doi: 10.1371/journal.pgen.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laukens D, et al. Evidence for significant overlap between common risk variants for Crohn's disease and ankylosing spondylitis. PLoS ONE. 2010;5:e13795. doi: 10.1371/journal.pone.0013795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confrmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Onozawa Y, et al. Activation of T cell death–associated gene 8 regulates the cytokine production of T cells and macrophages in vitro. Eur J Pharmacol. 2012;683:325–331. doi: 10.1016/j.ejphar.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Ryder C, McColl K, Zhong F, Distelhorst CW. Acidosis promotes Bcl-2 family mediated evasion of apoptosis: involvement of acid-sensing G protein–coupled receptor GPR65 signaling to MEK/ERK. J Biol Chem. 2012;287:27863–27875. doi: 10.1074/jbc.M112.384685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan MA, Kushner I, Braun WE. Association of HLA-A2 with uveitis in HLA-B27 positive patients with ankylosing spondylitis. J Rheumatol. 1981;8:295–298. [PubMed] [Google Scholar]
- 58.Liu JB, et al. Association of vitiligo with HLA-A2: a meta-analysis. J Eur Acad Dermatol Venereol. 2007;21:205–213. doi: 10.1111/j.1468-3083.2006.01899.x. [DOI] [PubMed] [Google Scholar]
- 59.Noble JA, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59:2972–2979. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27:361–368. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 61.Shah TS, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–1603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 63.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lippert C, et al. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 65.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wellcome Trust Case Control Consortium. Genomewide association study of 14,000 cases of seven common diseases and 3000 controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.