Abstract
Attempts to formulate a protective HIV-1 vaccine through classic vaccine design strategies have not been successful. Elicitation of HIV-1-specific broadly neutralizing antibodies (bnAbs) at high titers that are present before exposure might be required to achieve protection. Recently, the application of new technologies has facilitated the study of clonal lineages of HIV-1 envelope (Env) antibodies, which have provided insights into HIV-1 antibody development during infection and upon vaccination. Strategies are being developed for the analysis of infection and vaccine candidate-induced antibodies, their gene usage, and their maturation pathways such that this information can be used to attempt to guide rational vaccine design.
Keywords: HIV-1 vaccine, broadly neutralizing antibodies, immunogen design
Hurdles to overcome for the development of a preventive HIV-1 vaccine
As of 2010 it was estimated that 34 million people were living with HIV-1 worldwide [1]. The HIV-1 pandemic remains a global emergency for which there is currently no cure. Vaccination has historically been the most effective measure for controlling the transmission of infectious agents [2]; therefore, the development of a protective HIV-1 vaccine is a global public health priority [3].
Many licensed vaccines (e.g., smallpox, hepatitis B, measles, pertussis, polio, rabies, and yellow fever) induce specific antibodies that are correlates of protection [2, 4, 5]. Since the introduction of vaccination against smallpox, there have been scientific and technical advances that have led to the development of vaccines against numerous human diseases [6, 7]. Despite these advances, there are pathogens, such as HIV-1 [8, 9] and influenza [8], for which vaccination has not led to broad and long-lasting protection. For HIV-1 and influenza, this is due in part to genetic diversity of the pathogen [10], difficulty in eliciting broadly neutralizing antibodies (bnAbs) to epitopes conserved among different strains [8, 11, 12], and for HIV-1, the ability for the virus to integrate into the host genome [13].
In the 20th century, one major technological advance that enabled vaccine development was the use of cell culture that permitted the growth of viruses in vitro [6, 14]. Although HIV-1 is readily grown in cell culture, a vaccine for HIV-1 has remained elusive. The establishment of a latent pool of infected cells provides a persistent reservoir that is resistant to antiviral immune responses and to antiretroviral drugs [15]; the integration event that establishes this latent pool is thought to occur within hours to days after HIV-1 transmission [16–18]. This implies that a successful preventative HIV-1 vaccine will need to provide sterilizing immunity that is present at the time of exposure [17, 18]. All other vaccines rely upon secondary T and B cell anti-pathogen responses to prevent disease; thus, an effective HIV-1 vaccine may have to provide something achieved by no other vaccine to date. One response that may be able to provide protective immunity is anti-HIV-1 envelope antibodies. Recently, new techniques have probed the B cell repertoire of humans in the settings of infection [19–21] and vaccination [22], providing new insights into immune mechanisms that have prevented vaccine-induced protective HIV-1 antibody responses [23, 24]. In this review, we discuss how analysis of infection and vaccine candidate-induced antibodies and their genes may guide vaccine design.
The nature of HIV-1 protective antibody responses
The HIV-1 genome has extraordinary variability [10]. This feature combined with strategies for immune evasion exploited by the virus poses unprecedented challenges for inducing neutralizing antibodies with breadth of activity against most of the circulating strains of HIV-1. bnAbs against most clades and circulating recombinant forms can be spontaneously produced by rare subjects infected with HIV-1, but such antibodies only appear several years after infection [25]. During acute HIV-1 infection (AHI), the initial anti-HIV-1 antibody response is directed toward non-neutralizing epitopes on the gp41 envelope glycoprotein and does not appear to exert an anti-HIV-1 effect, as indicated by AHI gp41 antibody failure to select for virus escape mutants [26–29]. The first antibody response that can select virus escape mutants and neutralize transmitted/founder viruses does not appear until ~12–16 weeks after transmission, targeting the gp120 envelope glycoprotein, and has very limited breadth [30, 31]. The variability of the HIV-1 envelope glycoprotein effectively permits escape from immune control and quickly renders strain-specific neutralizing antibodies ineffective [10]. However, several years after HIV-1 transmission, approximately 20% of chronically HIV-1-infected subjects develop antibodies that neutralize multiple HIV-1 strains, with 2–4% of subjects developing serum antibodies that broadly neutralize most of the tested HIV-1 strains [25, 32, 33]. When they are made, bnAbs generally do not control viremia [25]; this lack of clinical impact might be a consequence of the appearance of bnAbs long after virus integration. Nonetheless, bnAbs can select for virus escape mutants, indicating promise for the prevention of HIV-1 transmission if bnAbs are present prior to HIV-1 exposure [34]. That anti-HIV-1 bnAbs may be effective in preventing infection if present at the time of exposure to the virus is also supported by results from non-human primate passive protection trials in which anti-HIV-1 envelope glycoprotein bnAbs at concentrations predicted to be achievable by immunization were able to block infection with chimeric simian-human immunodeficiency virus challenge [35–38]. Thus, to be effective, a preventive HIV-1 vaccine will need to induce broadly protective antibodies that are present at mucosal surfaces at the time of HIV-1 exposure.
Envelope targets of potentially protective antibodies
During the first two decades of the HIV-1 pandemic, only five bnAbs capable of neutralizing multiple primary HIV-1 isolates were identified (reviewed in [38–40]). These antibodies identified three epitope targets on the HIV-1 envelope glycoprotein: a post-translational glycan epitope on gp120 recognized by 2G12 [41, 42]; the CD4 binding site (CD4bs) recognized by b12 [43, 44]; and the membrane proximal external region (MPER) of gp41 recognized by 2F5 [42, 45, 46], 4E10 [42, 47], and Z13 [48]. Each of these antibodies display one or more unusual characteristics [24]: polyreactivity with human and/or non-human antigens, unusually long heavy chain complementarity determining region 3 (HCDR3) loops, and high levels of somatic mutation [23, 24, 49]. These characteristics suggested that bnAbs of these types mightbe limited by immune tolerance controls and/or be the result of tortuous and/or unfavored antibody maturation pathways [23, 24]. Attempts to consistently elicit antibodies with the activity of bnAbs through vaccination using conventional approaches have been unsuccessful [8, 23, 50]. The recent use of novel immunogens has been able to elicit antibodies that bind to the core epitope of bnAb 2F5, but such induced antibodies do not display broad neutralization, probably because they lack the lipid binding activity of the bnAb [51, 52]. The tolerance deletion faced by B cells expressing 2F5-like bnAbs was confirmed by experiments in mice that had 2F5 bnAb variable heavy (VH) chain and variable light (VL) chain genes knocked in, where bnAb-expressing B cells were limited by both central and peripheral tolerance mechanisms [53, 54].
In 2009, the ALVAC-prime AIDSVAX-boost RV144 trial demonstrated a modest degree of short-lived protection [55]. This trial had an estimated vaccine efficacy of 31% without inducing high levels of classical neutralizing antibodies [56], suggesting that vaccines targeting epitopes that induce non-neutralizing envelope antibodies might also be able to provide a measure of protection from HIV-1 transmission [57].
A recent immune correlates analysis of samples collected during the RV144 efficacy trial found that antibodies to the V1/V2 region of the HIV-1 envelope may have correlated inversely with infection risk [57]. This analysis raised the hypothesis that non-neutralizing V1/V2 antibodies may have played a role in protection, perhaps via antibody-dependent cellular cytotoxicity or other as yet unidentified mechanisms [57, 58]. Thus, IgG antibodies directed against the V1/V2 region may have non-neutralizing modes of virus inhibition that could be an important component of an effective vaccine strategy. At present, it remains unknown if this type of antibody response, as well as improvements to the magnitude and durability of the response, could be optimized by rational vaccine designs (Box 1). These new findings have provided new directions for investigation of the immune correlates of protection from HIV-1 transmission.
Box 1. Outstanding questions.
Can non-neutralizing antibodies exert a protective function through mechanisms other than classic neutralization as measured by conventional neutralizing assays (e.g., ADCC)?
Can immunogens be found that drive memory B cell maturation toward the production of bnAbs by targeting immunogens to bind to bnAb unmutated ancestors?
Can a vaccination regimen induce bnAbs in vaccinated subjects with diverse genetic backgrounds?
Can immunogens be optimized to drive the maturation of B cell clonal lineages toward the generation of bnAbs as opposed to toward easier to induce non- or poorly neutralizing HIV-1 envelope reactive antibodies?
The application of new technologies has resulted in the isolation of a large number of anti-HIV-1 bnAbs (summarized in Table 1). The first of the new bnAb epitopes was defined by PG9 and PG16, a pair of clonally related antibodies with greater neutralization potency than previously described bnAbs [20]. These bnAbs bind to an epitope on the gp120 envelope glycoprotein that is dependent on an asparagine in position 160 and that is usually conferred or stabilized by trimer formation [20, 21]. Subsequently, two additional groups of bnAbs that recognize this epitope were found; these were clonally related sets of four (CH01–CH04 [21]) and five (PGT141–PGT145 [59]) bnAbs. These new quaternary structure preferring conformational epitope-specific V1/V2 bnAbs share characteristics with previously described bnAbs in that they are all highly mutated, with VH nucleotide mutation frequencies ranging from 11.5% to 16% [20, 21, 59] (the mean frequency of human VH mutations found in secondary immune responses is approximately 5% [12, 26, 60]). This group of bnAbs also has exceptionally long, anionic HCDR3 sequences that are rich in tyrosines that are frequently sulfated (Figure 1) [20, 21, 61–64]. Structure/function analyses of the crystal structures of PG16 and CH04 antigen-binding fragments (Fabs) and the V1/V2 domain in complex with PG9 Fab revealed that the unusual conformation of the HCDR3 loop in these bnAbs is a critical feature necessary for neutralization breadth and potency [62–64]. Interestingly, these groups of bnAbs neutralize viruses with different patterns of breadth and potency, display different degrees of sensitivity to amino acid substitution, and different orientations of the HCDR3 loop in complex with the V1/V2 domain [21, 63]. Few of these bnAbs have been found to be polyreactive [20, 21, 59], suggesting that most quaternary structure preferring V1/V2 conformational epitope-specific bnAbs may not be subjected to deletion by tolerance control during their maturation. In fact, this specificity of bnAbs is now known to be relatively frequent among chronically infected broad neutralizers [20, 65]. These findings make the quaternary structure preferring V1/V2 conformational epitopes an attractive vaccine target.
Table 1.
Broadly neutralizing and crossreactive antibodies isolated since 2009
| Technology used | Antibody name | Antibody specificity | Refs |
|---|---|---|---|
| Broadly neutralizing antibodies (bnAbs) | |||
| Neutralization screening of cultured, unselected IgG+ memory B cells | PG9, PG16 | V1/V2 conformational epitope | [20] |
| CH01–CH04 | V1/V2 conformational epitope | [21] | |
| PGT141–PGT145 | V1/V2 conformational epitope | [59] | |
| PGT121–PGT123 | V3 epitope involving carbohydrates | [59] | |
| PGT125–PGT128, PGT130, PGT131 | V3 epitope involving carbohydrates | [59] | |
| PGT135–PGT135 | V3 epitope involving carbohydrates | [59] | |
| Fluorescence-activated cell sorting (FACS) using a resurfaced core gp120 molecule (RSC3) | VRC01, VRC02 | CD4 binding site | [19] |
| CH30–CH34 | CD4 binding site | [69] | |
| VRC-PG4, VRC-PG4b | CD4 binding site | [69] | |
| FACS and 454 deep sequencing | 3BNC117, 3BNC55, 12A12, 8ANC195, NIH45–46a | CD4 binding site | [70] |
| Crossreactive neutralizing antibodies | |||
| Neutralization screening of cultured, unselected IgG+ memory B cells | HJ16 | Near CD4 binding site | [66] |
| FACS of colostrum-derived B cells | CH08 | CD4-inducible epitope | [86] |
| FACS using MPER tetramer reagent | CAP206-CH12 | gp41 membrane proximal external region | [77] |
| Phage displayed immunoglobulin library | m66.6 | gp41 membrane proximal external region | [79] |
Representative antibodies of clonally related sequences.
Figure 1.
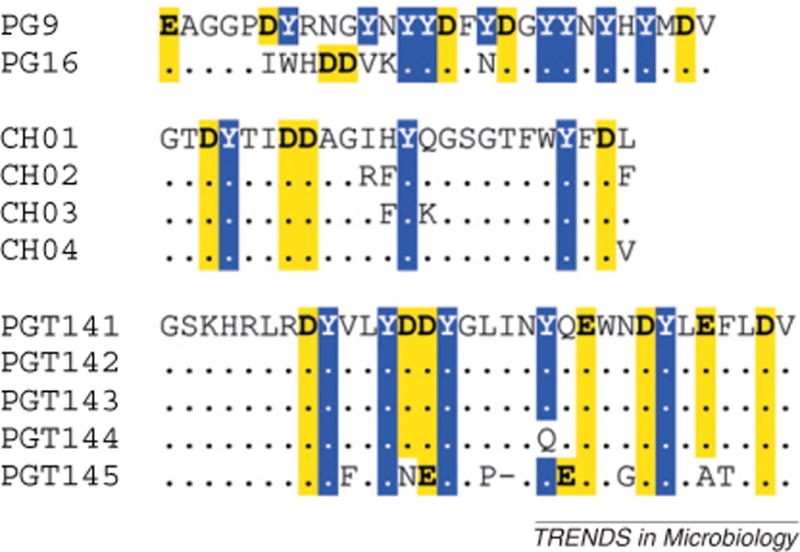
Heavy complementarity determining region 3 (HCDR3) amino acid sequences of the new quaternary structure preferring conformational epitope-specific V1/V2 broadly neutralizing antibodies. The HCDR3 amino acid sequences of each of the PG9/PG16, CH01–CH04 and PGT141–PGT145 clonal lineages were obtained from GenBank deposited sequences and manually aligned. Conserved amino acids within each clonal lineage are indicated by a dot (.); deletions are indicated by a dash (–). Anionic residues (aspartic acid and glutamic acid) are bold and highlighted in yellow; tyrosine residues are inverted and highlighted in blue.
The new CD4bs bnAb group includes antibodies that bind to epitopes near the CD4bs (e.g., HJ16 [66]) and those with footprints overlapping that of the CD4-gp120 contact region (i.e., VRC01–VRC03 [19, 67], PGV04, also known as VRC-PG04, [68], CH30–CH34 [69], and the HAAD motif antibodies [70]). Although these bnAbs recognize a similar epitope at the CD4bs as the previously described antibody b12, the new antibodies display greater neutralization breadth and potency. The crystal structures of Fabs of PGV04, VRC01, and VRC03 in complex with a gp120 core protein, combined with the analysis of large sets of heavy chain sequences obtained by 454 pyrosequencing, suggested a convergent mode of epitope recognition among CD4bs-specific bnAbs from different individuals [69]. The similarities in epitope recognition occurred despite only ~50% amino acid similarity in heavy chain sequence and the acquisition of divergent amino acid changes during somatic hypermutation [69]. These divergent changes maintained amino acids with similar chemical characteristics in the antibody binding domains, suggesting that epitope recognition may have been a driving force during affinity maturation [69]. Similarly, the CD4bs-specific HAAD motif antibodies were shown to have a core of amino acid similarities despite being derived from different subjects [70].
Unlike the quaternary structure preferring V1/V2 conformational epitope-specific bnAbs, the recently described CD4bs-specific bnAbs appear to use a restricted set of VH gene families. The majority, although not all, of these bnAbs use VH1-2 or VH1-46 gene segments despite being derived from different subjects [19, 68–70]. In addition, each of these CD4bs bnAbs has an extraordinarily high number of somatic mutations (30–32% for the VH gene, 17–20% for light chain variable gene nucleotides) and this level of mutation may be required for these antibodies to mediate broad neutralization [19, 69, 70]. Thus, CD4bs-specific bnAbs may be particularly difficult to induce by vaccines, given the level of chronic antigen stimulation needed to induce such profound levels of heavy and light chain mutation.
A third group of recently described bnAbs bind to carbohydrate epitopes in the gp120 V3 region (PGT121–PGT123 and PGT135–PGT137) or to gp120 protein carbohydrate-dependent V3 epitopes (PGT125–PGT128, PGT130 and PGT131) [59]. This group of antibodies appears to be more diverse than the V1/V2 bnAbs or the CD4bs bnAbs, with recognition of multiple carbohydrate-dependent epitopes. PGT130 and PGT131 show sensitivity to mutations in both the V3 and C4 regions of gp120, whereas PGT125–PGT128 show sensitivity to mutations in multiple locations in the V3 loop [59]. Antibodies PGT121–PGT123 and PGT135–PGT137 are sensitive to mutation of an asparagine at position 332 (N332) of the V3 loop, a position known to be critical for the recognition of the well-described bnAb 2G12 [71]. This group of bnAbs also competes with the binding of 2G12 to gp120, indicating that bnAbs with this pattern of reactivity can be found in different HIV-1-infected subjects. The high mannose glycans recognized by these bnAbs are similar to host glycans, suggesting that they may also be regulated by tolerance mechanisms.
The isolation of many new bnAbs and the efficacy seen in the RV144 trial boosted optimism for the possibility that a preventive HIV-1 vaccine can be formulated to induce protective antibody-mediated responses. The isolation of multiple specificities of bnAbs from chronically HIV-1-infected subjects suggests that the immune system has the capability to generate bnAb responses when properly stimulated. However, many challenges remain that necessitate the dissection of bnAb responses by integrating functional, genomic, and computational approaches to elucidate bnAb clonal lineage trees and to use them as templates for immunogen design.
B cell lineage vaccine design
For the design of future vaccine candidates, it may be useful to use clues provided by the newly isolated bnAbs, many of which have been found as clonal families of related antibodies [19–21, 59, 69, 70], and to use a newly proposed strategy, termed B cell lineage immunogen design [23], for construction of new vaccine candidates. B cell lineage design builds upon the observation that immunogens with the highest affinity for naïve B cell receptors are the most potent immunogens [72, 73]. Using the nucleotide sequence information from recovered clonally related bnAbs, unmutated ancestor (UA) and intermediate ancestor (IA) antibodies can be inferred using a combination of maximum likelihood methods and Bayesian inference [23]. The probability that a given UA or IA is correct can be calculated, and those UAs and IAs that are most likely to be correct can then be synthesized. Using these UA and IA antibodies, HIV-1 envelope constructs can be screened to select those with the highest binding affinity; the resulting series of envelopes can then be used as a set of experimental immunogens that should be the best priming and boosting immunogens for Env antibody induction [23]. Thus, study of clonal lineages of bnAbs and application of B cell lineage immunogen design can provide a theoretical map of the maturational pathway used by HIV-1-infected subjects, and thus provide us with a possible blueprint for their induction by vaccines [23, 74]. It is hoped that by using the information derived from clonal lineages along with computational biology approaches, it is possible to use the inferred sequence of the original B cell receptor that began the bnAb maturational process as a template to design immunogens that would drive B cells to make bnAbs in vaccinees (Figure 2).
Figure 2.
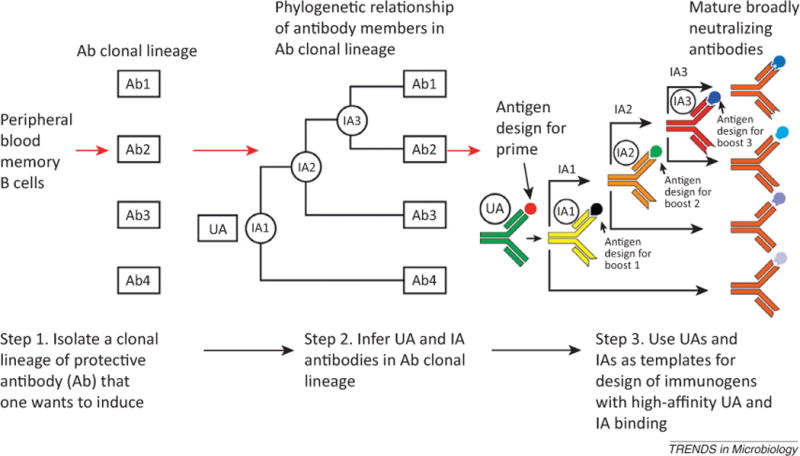
Steps of a B cell lineage-based approach to vaccine design. Step 1 is to isolate variable heavy (VH) and variable light (VL) chain members from the peripheral blood or tissues of patients containing broadly neutralizing antibodies (bnAbs) and to express these native Ig chain pairs as whole antibodies. Step 2 is to infer intermediate ancestor (IA) antibodies (labeled 1, 2, and 3) and the unmutated ancestor (UA) antibody. Step 3 requires producing the unmutated and intermediate ancestors as recombinant monoclonal antibodies (mAbs) and using structure-based alterations in the antigen [changes in envelope (Env) constructs predicted to enhance binding to the unmutated or intermediate ancestors] or deriving altered antigens using a suitably designed selection strategy. Vaccine administration might prime with the antigen that binds the unmutated ancestor most tightly, and this is then followed by sequential boosts with antigens optimized for binding to each intermediate ancestor. Shown here is an actual clonal lineage of the V1/V2-directed bnAbs CH01–CH04 [21]. Targeting the unmutated ancestor with an immunogen that has enhanced binding may induce higher antibody responses. If high-affinity ligands for unmutated ancestors cannot be found, then high-affinity ligands targeting the intermediate ancestors may be equally useful for triggering a response. Modified from [23] and reprinted with permission.
The bnAb response differs considerably from the initial antibody response in AHI [25–27, 75]; how the bnAb response evolves from the AHI response is not known. The B cell response during AHI is primarily directed against gp41 [27] and analysis of antibody clonal families during this response shows that such antibodies are often polyreactive [26]. In addition, inferred UAs of these clonal families often do not react with HIV-1 antigens, suggesting that they may have been originally directed against non-HIV-1 targets [26]. A similar pattern has been seen for some bnAbs; inferred UA of CD4bs-specific bnAbs show weak affinity, in the high micromolar range, for HIV-1 envelope glycoproteins [67, 69, 70]. By contrast, the inferred ancestor of the quaternary structure preferring V1/V2 conformational epitope-specific bnAb lineage CH01–CH04 bound to the AE.A244 gp120 Env at an affinity thought to be biologically relevant for naïve B cell triggering [21, 73, 76] and this inferred ancestor was able to neutralize four HIV-1 strains, including C.ZM233, astrain also neutralized by an inferred ancestor of PG9 [21, 62]. These data suggest that there are common motifs capable of binding UAs of disparate bnAb clonal lineages. It will be essential to map antibody maturation from AHI through the 3–4 years of infection required for the development of bnAbs in order to define the maturational pathways taken by Bcells. The sequence and structural changes that bnAb B cell receptors undergo, driven by Env antigens, should provide a blueprint for future vaccine candidates (Box 1) [23].
Another unanswered question is whether all vaccinated subjects will be able to make bnAbs with typical vaccination regimens (i.e., two to three immunizations) (Box 1). Although the mechanism of B cell gene rearrangement ensures a very broad initial antibody repertoire, person-to-person variation in this initial repertoire may preclude the existence of a B cell with the same specificity as the inferred ancestor of a given bnAb family. In this regard, the observed restriction of VH gene segments usage that has been described for CD4bs bnAbs may be a benefit or hindrance for vaccine design. The isolation of this class of bnAbs from multiple subjects indicates that some degree of convergent evolution among bnAbs of this specificity may occur [69, 70]. Conversely, if such restrictions are absolute, people with a limited use of the necessary gene segments may not be able to make such bnAbs, regardless of the immunogen used. Whether a similar situation holds for antibodies directed against the gp41 MPER is not yet known; the antibody CAP206-CH12 directed against the MPER uses the same VH1-69 gene segment as bnAb 4E10 [77]. An analysis of inferred ancestors of bnAb 2F5 showed that allelic variants of VH2-5 that encoded an aspartic acid at position 54 were capable of binding the 2F5 epitope, whereas those encoding an asparagine at that position bound much more weakly with an order of magnitude lower affinity; these data suggest that allelic variation could determine whether a vaccine recipient could make 2F5-like antibodies [78]. However, although bnAb 2F5 uses VH2-5, a new 2F5-like crossreactive neutralizing antibody (m66.6) uses VH5-51 [79], implying that there may be alternative pathways to 2F5-like bnAbs. Convergent antibody evolution may also apply to the quaternary structure preferring V1/V2 conformational epitope-specific bnAbs, which display HCDR3s with similar features despite using a diverse pool of VH families (Figure 1), thus suggesting that people with diverse genetic backgrounds may be able to make antibodies with similar reactivity against the same antigen [61, 62, 64]. These data suggest that there is no one single maturational pathway that every individual must follow in order to generate bnAbs of a particular specificity. Rather, it suggests that there may be many evolutionary possibilities and maturational pathways that share common features that can achieve the production of bnAbs. To maximize the success of lineage vaccine design strategies, immunogens should also be optimized to better bind to the desired Env bnAb paratopes in comparison to Env paratopic variants predicted to result in non-protective antibodies. Thus, immunogens may need tobeselected for their ability to steer the immune response away from non-protective responses at critical checkpoints during clonal evolution and induce dominant responses that contain signatures of broadly neutralizing antibodies (Box 1).
Manipulation of the naïve B cell pool may also provide a way to improve vaccine response. Knock-in mice expressing 2F5 VH and VL genes demonstrated that both central and peripheral tolerance mechanisms inhibit bnAb expressing B cells [53, 54]. Recent work suggests that manipulation of the B cell pool with the cytokine B lymphocyte stimulator (BLyS) can partially relieve peripheral tolerance in mice, thereby improving anti-HIV-1 responses [80]. Whether similar manipulation of human B cells combined with novel immunogen designs will result in vaccine-induced bnAbs is not known.
Finally, the design of immunogens should also take into account the variability in human leukocyte class II antigen (HLA class II) gene and allele usage, as differences in the HLA class II genetic make-up restrict the pool of peptides presented on the surface of antigen-presenting cells to CD4+ T helper cells: in the RV144 HIV ALVAC-prime AIDSVAX-boost efficacy trial, usage of the DRB1*11 and DRB1*16:02 HLA class II alleles was associated with lack of HIV-1 clade-specific neutralizing antibody responses, and vaccine recipients with HLA-DQ heterodimers encoded by DQA1*05:01 and DQB1*03:01 alleles were less likely to produce neutralizing antibodies [81].
Using new tools to assess vaccine-induced antibody responses
Plasma neutralization breadth can be mediated by either many [82] or only one or a few [19, 65] antibody specificities. Thus, the ability to detect precursors of bnAbs induced by vaccine candidates would be helpful. Several recent studies [65, 75, 82, 83] have shown that more than one specificity of antibodies contributing to plasma neutralization breadth can be generated in a single individual, suggesting it is unlikely that immunological barriers prevent concurrent development and maturation of multiple specificities of bnAbs. Concurrent production of both CD4bs (CH30–CH34) and quaternary structure preferring V1/V2 conformational epitope-specific (CH01–CH04) bnAbs was recently demonstrated in a chronically HIV-1-infected subject, showing that there is no absolute block on the development of more than one bnAb specificity [83]. It has been shown that the combination of two bnAbs can achieve almost pan-HIV-1 neutralization in vitro, both for CH01 and CH31 [83] and for PG9 and VRC01 [84]. Thus, determining if a candidate vaccine can initiate the maturation of multiple specificities of bnAbs is of critical importance.
The high degree of somatic mutation of isolated bnAbs suggests that they may require prolonged or multiple rounds of antigen-driven maturation to achieve breadth [85]. For this reason, a current critical question is whether bnAb or vaccine-driven maturation results in the persistence of antibody clonal families over time. This question has only recently been addressed, and, at present, the data from vaccination studies are mixed. The most studied model is influenza vaccination, where multiple antibody lineages are induced by repeated exposure to hemagglutinin antigens either through infection or vaccination [12, 60]. The mean VH mutation frequency of antibodies induced by repeated vaccination is ~5% [12, 60], although some antibodies with exceptionally high mutation frequencies (>25%) can be detected [12]. Interestingly, in the case of influenza vaccination, these highly mutated antibodies are not broadly crossreactive, suggesting that high degrees of somatic mutation by itself is not predictive of broad activity [12].
The techniques used to isolate bnAbs have recently been extended to the assessment of HIV-1 vaccine trials. Repeated immunization with gp120W6.1D resulted in increasing levels of mutation over the course of four immunizations, although the final mutation level (3.8%) was still rather modest compared with bnAbs [22]. In this same study, evidence was found for the persistence of clonal lineages over time, suggesting that candidate vaccines may be able to overcome one hurdle to bnAb development [22]. Furthermore, examination of the ALVAC-prime AIDSVAX-boost RV144 trial found that vaccine-induced monoclonal antibodies with modest degrees of mutation were able to mediate Tier 1 virus neutralization and antibody-dependent cellular cytotoxicity [56, 58]. Unfortunately, neither ALVAC-AIDSVAX nor gp120W6.1D vaccine induced high levels of bnAbs, and so new vaccine designs and formulations will be required to determine if a HIV-1 vaccine that induces bnAbs can be created [23].
Concluding remarks
The application of several new technologies has led to the isolation of a large number of new anti-HIV-1 bnAbs and crossreactive neutralizing antibodies. These new groups of HIV-1 envelope antibodies as well as the recent immune correlates analysis of the RV144 HIV efficacy trial have provided clues for strategies of rational vaccine design based on understanding of bnAb clonal lineage pathways.
By using the same recombinant antibody techniques to study human vaccine trials, we are now able to gauge the effectiveness of candidate vaccines by comparing maturation and clonal families of vaccine-induced antibodies with those of bnAbs arising in HIV-1-infected subjects. Current HIV-1 vaccine strategies appear to elicit antibodies with diverse specificities similar to those seen in chronic infection, but with somatic mutation profiles more comparable with those seen in AHI. This suggests that to elicit bnAbs by HIV-1 vaccines, we must harness our knowledge of the maturational pathways taken by those antibodies to guide maturation toward bnAb specificities and around tolerance deletion checkpoints.
Thus, identifying new clonal lineages of bnAbs, acquiring information on the reactivities of UA and intermediate antibodies, and identifying signatures that differentiate bnAbs from non-protective antibodies targeting the same epitopes are critical steps to facilitate the design of optimized immunogens capable of selectively eliciting broadly neutralizing responses.
References
- 1.World Health Organization. Global HIV/AIDS Response: Epidemic Update and Health Sector Progress Towards Universal Access. WHO Press; 2011. Progress Report 2011. [Google Scholar]
- 2.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JH, et al. HIV vaccines: lessons learned and the way forward. Curr Opin HIV AIDS. 2010;5:428–434. doi: 10.1097/COH.0b013e32833d17ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 5.Amanna IJ, et al. Protective immunity following vaccination: how is it defined? Hum Vaccin. 2008;4:316–319. doi: 10.4161/hv.4.4.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker JP, Katz SL. Childhood vaccine development: an overview. Pediatr Res. 2004;55:347–356. doi: 10.1203/01.PDR.0000106317.36875.6A. [DOI] [PubMed] [Google Scholar]
- 7.Plotkin SA. Six revolutions in vaccinology. Pediatr Infect Dis J. 2005;24:1–9. doi: 10.1097/01.inf.0000148933.08301.02. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson Hedestam GB, et al. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 9.Burton DR, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 10.Korber B, et al. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 11.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody MA, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS ONE. 2011;6:e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindmarsh P, Leis J. Retroviral DNA integration. Microbiol Mol Biol Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plotkin SA, Plotkin SL. The development of vaccines: how the past led to the future. Nat Rev Microbiol. 2011;9:889–893. doi: 10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 15.Margolis DM. Eradication therapies for HIV infection: time to begin again. AIDS Res Hum Retroviruses. 2011;27:347–353. doi: 10.1089/aid.2011.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen MS, et al. Acute HIV-1 infection. N Engl J Med. 2011;364:1943–1954. doi: 10.1056/NEJMra1011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMichael AJ, et al. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moody MA, et al. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J Virol. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes BF, et al. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verkoczy L, et al. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray ES, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao HX, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomaras GD, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goonetilleke N, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, et al. Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. J Virol. 2011;85:11196–11207. doi: 10.1128/JVI.05601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richman DD, et al. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 32.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen X, et al. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol. 2009;83:3617–3625. doi: 10.1128/JVI.02631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montefiori DC, Mascola JR. Neutralizing antibodies against HIV-1: can we elicit them with vaccines and how much do we need? Curr Opin HIV AIDS. 2009;4:347–351. doi: 10.1097/COH.0b013e32832f4a4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mascola JR. Passive transfer studies to elucidate the role of antibody-mediated protection against HIV-1. Vaccine. 2002;20:1922–1925. doi: 10.1016/s0264-410x(02)00068-3. [DOI] [PubMed] [Google Scholar]
- 38.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 39.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 41.Calarese DA, et al. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci USA. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchacher A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein–Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 43.McInerney TL, et al. A human IgG1 (b12) specific for the CD4 binding site of HIV-1 neutralizes by inhibiting the virus fusion entry process, but b12 Fab neutralizes by inhibiting a postfusion event. Virology. 1997;233:313–326. doi: 10.1006/viro.1997.8547. [DOI] [PubMed] [Google Scholar]
- 44.Roben P, et al. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conley AJ, et al. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc Natl Acad Sci USA. 1994;91:3348–3352. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stiegler G, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 48.Zwick MB, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 50.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity. 2010;33:542–554. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guenaga J, et al. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS ONE. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dennison SM, et al. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS ONE. 2011;6:e27824. doi: 10.1371/journal.pone.0027824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verkoczy L, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci USA. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verkoczy L, et al. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH × VL knockin mice reveals multiple tolerance controls. J Immunol. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 56.Montefiori DC, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonsignori M, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Changela A, et al. Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J Virol. 2011;85:2524–2535. doi: 10.1128/JVI.02335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pancera M, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pejchal R, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker LM, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falkowska E, et al. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J Virol. 2012;86:4394–4403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scanlan CN, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of a1→2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shih TAY, et al. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002;3:399–406. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- 73.Dal Porto JM, et al. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med. 2002;195:1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao X, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomaras GD, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shih TAY, et al. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- 77.Morris L, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS ONE. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alam SM, et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J Virol. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Z, et al. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J Virol. 2011;85:11401–11408. doi: 10.1128/JVI.05312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dosenovic P, et al. BLyS-mediated modulation of naive B cell subsets impacts HIV Env-induced antibody responses. J Immunol. 2012;188:6018–6026. doi: 10.4049/jimmunol.1200466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paris R, et al. HLA class II restriction of HIV-1 clade-specific neutralizing antibody responses in ethnic Thai recipients of the RV144 prime-boost vaccine combination of ALVAC-HIV and AIDSVAX® B/E. Vaccine. 2012;30:832–836. doi: 10.1016/j.vaccine.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 83.Bonsignori M, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Doria-Rose NA, et al. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol. 2012;86:3393–3397. doi: 10.1128/JVI.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haynes BF, et al. B cell responses to HIV-1 infection and vaccination: pathways to preventing infection. Trends Mol Med. 2011;17:108–116. doi: 10.1016/j.molmed.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Friedman J, et al. Isolation of HIV-1-neutralizing mucosal monoclonal antibodies from human colostrum. PLoS ONE. 2012;7:e37648. doi: 10.1371/journal.pone.0037648. [DOI] [PMC free article] [PubMed] [Google Scholar]


