SUMMARY
Protein translation is an energetically demanding process that must be regulated in response to changes in nutrient availability. Herein, we report that the thiolation status of wobble-uridine (U34) nucleotides present on lysine, glutamine or glutamate tRNAs reflects intracellular methionine and cysteine availability, and regulates cellular translational capacity and metabolic homeostasis. tRNA thiolation is important for growth under nutritionally challenging environments and required for efficient translation of genes enriched in lysine, glutamine, and glutamate codons, which frequently encode proteins important for translation and growth-specific processes. tRNA thiolation is down-regulated during sulfur starvation in order to decrease sulfur consumption and growth, and its absence leads to a compensatory increase in enzymes involved in methionine, cysteine, and lysine biosynthesis. Thus, tRNA thiolation enables cells to modulate translational capacity according to the availability of sulfur amino acids, establishing a functional significance for this conserved tRNA nucleotide modification in cell growth control.
INTRODUCTION
The process of protein synthesis consumes enormous amounts of energy and must be carefully regulated in response to nutrient availability (Warner et al., 2001). The translational capacity and output of a cell is typically increased to promote growth and proliferation (Jorgensen and Tyers, 2004), or decreased during nutrient limitation or quiescence. In eukaryotes, much of this translational regulation in response to nutrients is controlled by the TORC1 and PKA signaling pathways, which regulate the translation machinery, rRNA, and tRNA biogenesis (Proud, 2002; Wullschleger et al., 2006; Zaman et al., 2008). While connections between these nutrient-sensitive signal transduction pathways and translation are increasingly well-studied, much remains unclear about how the regulation of protein translation is tied to the nutrients themselves.
Interestingly, many tRNAs contain unconventional, conserved nucleotide modifications (Gustilo et al., 2008; Phizicky and Hopper, 2010). When the genetic code was deciphered, it became apparent that the base at the “wobble position” on tRNA anticodons could pair with more than one base at the third codon position (Crick, 1966). Two sets of tRNA uridine modifications are present at the wobble position (U34) on tRNALys (UUU), tRNAGlu (UUC) and tRNAGln (UUG) (Gustilo et al., 2008; Phizicky and Hopper, 2010). These are an mcm5 modification, which denotes a methoxycarbonylmethyl functional group at the 5 position (termed uridine mcm5), which is often accompanied by thiolation where a sulfur atom replaces oxygen at the 2 position (termed uridine thiolation, or s2U) (Figure 1A). These modifications are often found together but can exist separately on their own (Chen et al., 2011b; Yarian et al., 2002) (Figure 1A). Although these conserved modifications have been known for a long time, an underlying logic for their biological purpose remains unclear.
Figure 1. tRNA uridine thiolation levels reflect sulfur amino acid availability.
(A) tRNA structure, wobble uridine modifications, and structures of methoxycarbonylmethyl (mcm5) and methoxycarbonylmethyl + thiolation (mcm5s2) uridine modifications.
(B) Experimental approach for measuring tRNA uridine modifications by LC-MS/MS.
(C) tRNA uridine thiolation amounts decrease in minimal media. Relative abundance of thiolated (mcm5s2) or mcm5-modified tRNA uridines in cells grown in rich (YPD, YPL) and minimal media (SD, SL) were measured. (Mean±SD, n=4, with experimental duplicates. *** indicates p< 0.001, ** indicates p<0.01, * indicates p< 0.05). See also Figure S1.
(D) tRNA uridine thiolation amounts are rescued by sulfur-containing amino acids. Cells were grown as described in (B), with or without supplementation of the indicated amino acids, ammonium sulfate (NH4)2SO4, or a non-sulfur amino acid mixture (non-S). Tyrosine was excluded due to poor solubility. (Mean±SD, n=3, with experimental duplicates). See also Figure S1.
(E) Simplified schematic of sulfur amino acid metabolism in budding yeast. Cysteine and methionine are derived from homocysteine, and can be interconverted.
(F) Intracellular sulfur amino acid levels decrease in SD minimal medium. Cells were grown in YPD, SD, or SD supplemented with methionine (150 μM) (see panel B), and sulfur metabolites were measured by LC-MS/MS. Note: relative decreases of both cysteine and methionine in SD mirror the decrease in uridine thiolation abundance. (Mean±SD, n=4, with experimental duplicates). See also Table S1 and Figure S1.
The proteins that modify these tRNA uridines are better understood biochemically. In yeast, the elongator complex protein Elp3p and the methyltransferase Trm9p are required for uridine mcm5 modifications (Begley et al., 2007; Chen et al., 2011a; Huang et al., 2005; Kalhor and Clarke, 2003). Uridine thiolation requires multiple proteins transferring sulfur derived from cysteine onto the uracil base (Goehring et al., 2003b; Leidel et al., 2009; Nakai et al., 2008; Nakai et al., 2004; Noma et al., 2009; Schlieker et al., 2008). This sulfur transfer proceeds through a mechanism shared with a protein ubiquitylation-like modification, called “urmylation”, where Uba4p functions as an E1-like enzyme to transfer sulfur to Urm1p. These tRNA uridine modifications can modulate translation. For example, tRNALys (UUU) uridine modifications enable the tRNA to bind both lysine cognate codons (AAA and AAG) at the A and P sites of the ribosome, aiding tRNA translocation (Murphy et al., 2004; Phelps et al., 2004; Yarian et al., 2002). Uridine modified tRNAs have an enhanced ability to “wobble” and read G-ending codons, forming a functionally redundant decoding system (Johansson et al., 2008). However, only a handful of biological roles for these modifications are known. Uridine mcm5 modifications allow the translation of AGA and AGG codons during DNA damage (Begley et al., 2007), influence specific telomeric gene silencing or DNA damage responses (Chen et al., 2011b), and function in exocytosis (Esberg et al., 2006). These roles cannot fully explain why these modifications are ubiquitous, or how they are advantageous to cells.
Interestingly, studies in yeast link these tRNA modifications to nutrient-dependent responses. Both modifications consume metabolites derived from sulfur metabolism, primarily S-adenosylmethionine (SAM) (Kalhor and Clarke, 2003; Nau, 1976), and cysteine (Leidel et al., 2009; Noma et al., 2009). These modifications appear to be downstream of the TORC1 pathway, as yeast lacking these modifications are hypersensitive to rapamycin (Fichtner et al., 2003; Goehring et al., 2003b; Leidel et al., 2009; Nakai et al., 2008), and interactions can be detected between Uba4p and Kog1/TORC1 (Laxman and Tu, 2011). These modification pathways also play critical roles in nutrient stress-dependent dimorphic foraging yeast behavior (Abdullah and Cullen, 2009; Goehring et al., 2003b; Laxman and Tu, 2011). We reasoned that deciphering the interplay between these modifications, nutrient availability and cellular metabolism would reveal a functional logic to their biological importance.
Herein, we show that tRNA uridine thiolation abundance reflects sulfur-containing amino acid availability, and functions to regulate translational capacity and amino acid homeostasis. Uridine thiolation represents a key mechanism by which translation and growth are regulated synchronously with metabolism. These findings have significant implications for our understanding of cellular amino acid-sensing mechanisms, and with the accompanying manuscript (Sutter et al., 2013), show how sulfur-containing amino acids serve as sentinel metabolites for cell growth control.
RESULTS
tRNA uridine thiolation amounts reflect intracellular sulfur amino acid availability
We were intrigued by connections between tRNA uridine modification pathways and nutrients, especially since mutants of tRNA uridine-modifying enzymes were hypersensitive to rapamycin (Figure S1A). We first tested whether tRNA uridine modification amounts changed in response to different nutrient environments. To qualitatively assay tRNA uridine thiolation, tRNAs were resolved on urea-PAGE gels containing the sulfur-coordinating mercury agent APM (Nakai et al., 2008) (Supplemental Information). We confirmed that the enzyme Uba4p is required for all tRNA thiolation (Figure S1B). While the majority of tRNALys (UUU), tRNAGlu (UUC) and tRNAGln (UUG) were thiolated in cells growing either in YPD (rich medium) or under continuous glucose-limitation, a fraction of these tRNAs remained unthiolated (Figure S1B), suggesting that this modification was not constitutive, and might change in abundance under specific conditions.
We then developed targeted LC-MS/MS methods to quantitatively measure amounts of thiolated, methoxycarbonylmethyl-modified (mcm5s2), or unthiolated, methoxycarbonylmethyl-modified (mcm5) tRNA uridines (Figure S1C). We grew cells under several nutrient conditions including rich (YP), or synthetic (S), minimal defined medium with either glucose (D) or lactate (L) as the carbon source (Figure 1B), and measured relative uridine modification amounts from purified tRNAs. We observed a significant decrease in relative amounts of thiolated uridine in cells grown in minimal media, particularly in non-fermentable SL medium compared to fermentable SD medium (Figure 1C). In all samples, amounts of unthiolated (mcm5) uridines always increased when thiolated (mcm5s2) uridines decreased, suggesting the mcm5 modification is more constitutive. Collectively, these data suggest the thiolation modification in particular is regulated by nutrient availability.
Both SD and SL minimal medium contain sufficient biosynthetic precursors for growth. However, a key difference compared to YP media is the absence of free amino acids. Therefore, we tested if specific amino acids were critical for tRNA uridine thiolation. We measured thiolated uridine amounts from tRNAs purified from cells grown in SD medium supplemented with individual amino acids. Thiolated uridine abundance was restored exclusively by sulfur-containing amino acids methionine and cysteine, but not other amino acids alone or in combination (Figure 1D, S1D). Excess ammonium sulfate also failed to restore thiolated uridine amounts (Figure 1D, S1D). These data reveal that tRNA uridine thiolation is responsive specifically to the availability of reduced sulfur equivalents in the cell.
Although cysteine is the sulfur donor for tRNA uridine thiolation, methionine and cysteine can be interconverted to one another in yeast (Figure 1E). We therefore asked if thiolated uridine amounts correlated with intracellular sulfur amino acid abundance. We determined intracellular methionine, cysteine, SAM and S-adenosylhomocysteine (SAH) abundance using targeted LC-MS/MS methods (Figure 1F). Compared to YPD medium, cells grown in SD medium showed substantially decreased methionine and cysteine abundance, which was restored upon methionine addition (Figure 1F). Such sulfur amino acid depletion was more considerable between non-fermentable YPL and SL media (Sutter et al., 2013). We estimated that cysteine was present at nM concentrations, while methionine and SAM were present at ~10–50 μM. Furthermore, the ratio of SAM:SAH decreased substantially upon switching to SD or SL from rich media (Table S1). These data suggest that tRNA uridine thiolation amounts are tuned to reflect intracellular sulfur amino acid availability.
tRNA uridine thiolation is important under challenging growth conditions
Why might cells modulate tRNA uridine thiolation levels depending on sulfur amino acid abundance? Mutant strains lacking these modifications do not exhibit significant growth phenotypes under standard nutrient-rich growth conditions (Figure S1A) unless exposed to rapamycin, caffeine, or oxidative stress (Leidel et al., 2009; Nakai et al., 2008). We hypothesized that stronger phenotypes resulting from a lack of these tRNA modifications might emerge under more challenging growth environments.
During continuous nutrient-limited growth, prototrophic strains of budding yeast exhibit robust oscillations in oxygen consumption in a phenomenon termed the yeast metabolic cycle (YMC) ((Tu et al., 2005) and Figure 2A). During the YMC, synchronized cells shift between three metabolic states, OX (oxidative) where genes specific to growth (e.g., ribosome biogenesis, translation machinery) increase in expression, RB (reductive-building) where genes specific to DNA replication and the cell cycle peak, and RC (reductive-charging) where cells are quiescent-like with increased expression of stress and survival genes (Figure 2A). Sulfur metabolism is not only tightly regulated during the YMC but is also critical for maintaining such cycles (Murray et al., 2003; Tu et al., 2005; Tu et al., 2007). Thus, we turned to the YMC to provide insights into the specific biological roles of tRNA uridine modifications.
Figure 2. tRNA uridine thiolation is required for normal metabolic cycles.
(A) Schematic describing the yeast metabolic cycle (YMC) and its three phases, OX, RB, and RC. Numbers indicate time points of typical sample collections for Figure S2.
(B) Thiolation-deficient strains cannot undergo normal metabolic cycles. Metabolic cycles of WT, uba4Δ and urm1Δ (uridine thiolation-deficient), and elp3Δ and trm9Δ (uridine mcm5- deficient) cells are shown across a 20 h time period. See also Figure S2.
(C) Schematic of the tRNA uridine thiolation/sulfur transfer pathway and urmylation pathway. The tRNA uridine thiolation pathway shares components with the protein urmylation pathway. Uba4p/Urm1p are required for both pathways, while Ncs2p/Ncs6p are only required for tRNA uridine thiolation. Ahp1p is the only known urmylated yeast protein. See also Figure S2.
(D) Uba4p catalytic activity is required for normal metabolic cycles. The metabolic cycles of catalytically dead uba4 mutant (C225A and C397A) compared to WT are shown, along with the location of the catalytic cysteine residues within Uba4p.
(E) Protein urmylation is not required for metabolic cycles. ahp1Δ strains show normal metabolic cycles. ncs2Δ and ncs6Δ strains are incapable of tRNA uridine thiolation, but are not involved in protein urmylation. Both ncs2Δ and ncs6Δ strains exhibit cycles comparable to uba4Δ and urm1Δ strains. See also Figure S2.
Transcript levels of genes encoding uridine-modifying enzymes (URM1, ELP3 and TRM9, but not UBA4) are periodic in the YMC (Tu et al., 2005), peaking during the OX/growth phase (Figure S2A). Genes induced during this phase typically have important roles in growth (Brauer et al., 2008; Cai et al., 2011; Tu et al., 2005). Accordingly, the abundance of the thiolation-specific and mcm5-specific enzymes increased during the OX/growth phase as well (Figure S2B), suggesting growth-specific roles for these modifications. Total amounts of tRNAs harboring these modifications (e.g. tRNAGlu (UUC)) also increased specifically during the growth phase (Figure S2C). We also compared the relative amounts of these tRNA uridine modifications (in proportion to all other tRNA nucleotides present at that time) across the YMC (Figure S2D and Experimental Procedures), and found that they remained constant across the different phases.
Mutants of key metabolic regulators of cell growth or division often display strong metabolic cycle phenotypes (Cai et al., 2011; Chen et al., 2007). tRNA thiolation-deficient cells (uba4Δ and urm1Δ) were unable to maintain normal metabolic cycles, showing weak, unstable oscillations with short periodicity (Figure 2B). This observed phenotype in thiolation-deficient cells is pronounced, since mutants of many non-essential genes show no cycling phenotype at all. In contrast, strains deficient in mcm5-modified uridines (elp3Δ or trm9Δ) had near-normal metabolic cycles (Figure 2B), while mutants lacking both tRNA uridine modifications did not cycle (Figure S2E). These data suggest critical roles for tRNA uridine thiolation, and more permissive roles for mcm5-modified uridines, during continuous nutrient-limited growth. Overexpressing mcm5-modified tRNALys (UUU), tRNAGlu (UUC) and tRNAGln (UUG) was insufficient to rescue the aberrant YMC phenotype of the uba4Δ mutant (Figure S2F). These data suggest essential roles for tRNA thiolation under challenging growth environments.
tRNA uridine thiolation requires proteins shared by the protein urmylation pathway (Figure 2C) (Goehring et al., 2003b; Schlieker et al., 2008). The observed phenotypes could alternatively be due to non-catalytic functions of Uba4p, protein urmylation, or other unknown functions of these proteins. To test these possibilities, we first mutated key catalytic residues required for the sulfur transfer activity of Uba4p (C225A and C397A) (Schmitz et al., 2008). Strains with these mutations behaved identically to uba4Δ and urm1Δ strains (Figure 2D), showing that Uba4p catalytic activity is required for normal cycling. Next, we tested roles for protein urmylation. Only one yeast protein not part of the urmylation pathway, Ahp1p, has been identified to be urmylated, which occurs during oxidative stress (Goehring et al., 2003a; Van der Veen et al., 2011) (Figure 2A). However, ahp1Δ strains showed normal metabolic cycles (Figure 2E). We measured global protein urmylation under different nutrient conditions by Western blot. Urmylation of unidentified target proteins was low or barely detectable (Figure S2G), particularly in SL medium and chemostat cultures. Finally, cells lacking Ncs2p or Ncs6p, which are required for tRNA uridine thiolation, but not protein urmylation (Noma et al., 2009) (Figure 2C), exhibited disrupted metabolic cycles identical to uba4Δ or urm1Δ strains (Figure 2E). Collectively, these data demonstrate that tRNA thiolation, and not protein urmylation, is important for the coordination of growth and metabolic cycling under challenging nutrient environments.
tRNA uridine thiolation regulates carbohydrate metabolism and amino acid synthesis
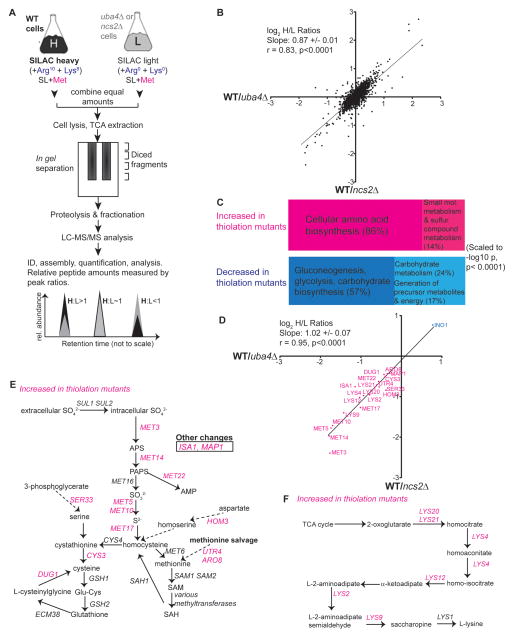
To investigate which cellular proteins are affected by tRNA thiolation, we performed an unbiased analysis of protein abundance in WT and thiolation-deficient cells using a stable isotope labeling with amino acids in culture (SILAC) experiment (Figure 3A). To rule out contributions from protein urmylation, we independently compared WT to either the uba4Δ mutant (lacking both uridine thiolation and protein urmylation) or the ncs2Δ mutant (lacking only uridine thiolation). Experiments were performed in SL medium, where tRNA thiolation is regulated (Figure 1C, 4A). Cells were grown in SL supplemented with methionine (to promote maximal tRNA thiolation in WT), and either heavy or light arginine and lysine (Figure 3A). Approximately ~1900 proteins, or one-third of the yeast proteome, were unambiguously measured in both samples (Table S2).
Figure 3. tRNA uridine thiolation regulates amino acid biosynthesis and sugar metabolism.
(A) Schematic of the SILAC quantitative proteomic approach used to assess global changes in protein levels in thiolation-deficient cells (Supplemental Information).
(B) tRNA uridine thiolation is responsible for most uba4Δ phenotypes. Correlation plot comparing normalized log2 H/L ratios from WT/ubaΔ (X-axis) and WT/ncs2Δ (Y-axis) samples, for all proteins detected in both samples (~1900 total). ncs2Δ and uba4Δ strains showed similar profiles (slope=0.87, r=0.83). See also Table S2 for complete data, Table S3 and S4 for GO analysis data.
(C) tRNA uridine thiolation regulates sugar and carbohydrate metabolism as well as sulfur and lysine amino acid metabolism. Proteins increased (top panel) or decreased (bottom panel) by at least 1.4 fold in thiolation-deficient strains (compared to WT cells) were analyzed by GO term grouping, to find significantly enriched terms (significance threshold p<0.0001). Consolidated GO-term groups were visualized using the descriptive, scaled TreeMaps shown (Supplemental Information).
(D) Proteins related to sulfur amino acid metabolism or lysine metabolism that increase or decrease in uba4Δ or ncs2Δ mutant cells were compared using log2 H/L ratio plots. WT/uba4Δ (X-axis) ratios for these proteins are compared against WT/ncs2Δ (Y-axis). Both samples showed a nearly 1:1 correlation (r=0.95, slope=1).
(E) Sulfur amino acid metabolism-related proteins that increase in tRNA uridine thiolation-deficient mutants. Proteins involved in methionine or cysteine biosynthesis or salvage that specifically increased in both uba4Δ and ncs2Δ mutants are indicated in magenta. See also Figure S3.
(F) Lysine metabolism-related proteins that increased in tRNA uridine thiolation-deficient mutants (uba4Δ, ncs2Δ) compared to WT are indicated in magenta.
Figure 4. Uba4p is negatively regulated during sulfur amino acid starvation.
(A) npr2Δ and npr3Δ strains exhibit excessive tRNA thiolation during sulfur starvation. Relative tRNA thiolation amounts were measured in WT, npr2Δ and npr3Δ cells grown in YPL or SL medium. Thiolation increased in npr2Δ and npr3Δ strains compared to WT cells only in SL (Mean±SD, n=3, with experimental duplicates. ** indicates p<0.01). See also Figure S4.
(B) Thiolation-deficient npr2Δ/uba4Δ strains exhibit slower growth compared to npr2Δ in SL medium. Growth curves of the indicated strains following switch from YPL to SL medium were measured over ~50 hours in batch cultures starting at an OD600 of ~0.1. (Mean±SD, n=3, *** indicates p<0.001). See also Figure S4
(C) npr2Δ cells have abnormally higher levels of sulfur-containing metabolites. Relative amounts of cysteine, methionine and SAM were measured in WT and npr2Δ cells growing in YPL or SL by LC-MS/MS
(D) Uba4p abundance decreases upon switch to SL medium. Western blot shows amounts of Myc-UBA4 present over time in cells growing in YPL, SL or SL+0.5mM methionine (Mean±SD, n=3)
(E) UBA4 and URM1 transcript levels remain relatively stable upon sulfur amino acid starvation. Relative amounts of UBA4 and URM1 transcripts in cells growing in the indicated conditions were measured by RT-qPCR (normalized to snR14). See also Figure S4.
The two sets of experiments (WT vs. uba4Δ or WT vs. ncs2Δ), showed exceptional correlation (Pearson’s coefficient r=0.83, p<0.0001), and a ~1:1 ratio for all proteins detected (slope = 0.87) (Figure 3A), indicating that the extent of changes in protein levels in either uba4Δ or ncs2Δ cells (each compared to WT) was nearly identical. This further suggests that tRNA thiolation defects, and not protein urmylation defects, recapitulate the phenotypes observed with the uba4Δ strains under the conditions tested. Next, we selected proteins that either decreased or increased in both uba4Δ cells and ncs2Δ cells compared to WT cells, by > 1.4 fold. Only a small fraction of the proteins detected (<5% for each set) met these criteria, with the majority of the detected proteins remaining relatively unchanged in abundance (Table S2).
These proteins were analyzed using Gene Ontology (GO) for significantly enriched GO terms, using stringent exclusion criteria (p<0.0001). All detected proteins that decreased in thiolation-deficient strains grouped to GO pathways related to sugar and carbohydrate metabolism (Figure 3C and Table S3). These include enzymes involved in glycolysis and inositol synthesis, suggesting that reduced tRNA thiolation signals cells to down-regulate carbon metabolism. We similarly analyzed proteins that increased in thiolation-deficient mutants compared to WT, which broadly grouped to cellular amino acid biosynthesis (86%), small molecule metabolism and sulfur compound metabolism (Figure 3C and Table S4). In both uba4Δ and ncs2Δ mutants, all these proteins increased to a comparable extent relative to WT cells (Figure 3D, slope =1, Pearson’s coefficient r=0.95, p<0.0001), and did not appear to be due to increased transcription (Figure S3).
We further examined the functional roles of the proteins related to amino acid metabolism that increased in abundance in thiolation-deficient mutants, and observed that nearly all of them are involved in the synthesis of methionine, cysteine (Figure 3E) or lysine (Figure 3F), and not their degradation. In addition, methionine salvage enzymes including Map1p, Utr4p, and Aro8p also increased in the mutants (Figure 3E). All enzymes in the lysine biosynthetic pathway, as well as twelve enzymes in the extensive sulfur amino acid metabolism pathway increased in abundance in mutants lacking tRNA thiolation (Figure 3E, F). Intriguingly, lysine codons are recognized and translated by a uridine thiolated tRNA. Thus, despite the presence of excess methionine and lysine, cells deficient in tRNA uridine thiolation cannot accurately gauge availability of these amino acids, and upregulate pathways promoting their accumulation. Collectively, these data reveal that thiolated tRNA levels reciprocally regulate amino acid and carbohydrate metabolism to help achieve metabolic homeostasis.
tRNA thiolation and Uba4p protein levels are actively down-regulated during sulfur amino acid limitation
Upon switch from YPL to SL medium where tRNA thiolation is decreased, yeast cells also induce autophagy that is dependent on a protein complex containing Iml1p, Npr2p, and Npr3p (Wu and Tu, 2011). Since this complex regulates cellular responses to sulfur amino acid limitation (Sutter et al., 2013), we tested if tRNA thiolation, a sulfur-consuming process, might also be regulated by this complex. We compared the relative abundance of thiolated tRNA uridines in WT, npr2Δ or npr3Δ strains growing in YPL or SL medium. In both npr2Δ and npr3Δ strains, thiolated uridine abundance was significantly higher than in WT strains only after switch to SL (Figure 4A and S4A). Furthermore, both npr2Δ and npr3Δ mutant strains grew faster than WT cells in these conditions (Figure 4B, S4B and described in detail in (Sutter et al., 2013)). Eliminating tRNA thiolation by deleting uba4Δ reduced the amount of unchecked growth in the npr2Δ mutant, suggesting that tRNA thiolation is typically reduced to decrease growth rates upon switch to sulfur amino acid-limited growth conditions (Figure 4B). Direct biochemical associations between epitope tagged-versions of Uba4p and the Iml1p/Npr2p/Npr3p complex could not be reliably assessed since most deletions of Uba4p at the N- or C-terminus resulted in complete inactivation of Uba4p (Figure S4C). However, we observed that amounts of cysteine, methionine, and in particular SAM, were abnormally high in npr2Δ mutant cells in SL (Figure 4C), which likely contributes to excessive tRNA thiolation under these conditions. These data suggest that the Iml1p/Npr2p/Npr3p complex negatively regulates tRNA thiolation partly by altering sulfur amino acid availability.
To further address how tRNA uridine thiolation might be down-regulated during sulfur amino acid starvation, we measured protein abundance of components of the tRNA thiolation machinery in cells grown in rich or minimal medium. We observed a decrease in amounts of Uba4p, as well as the sulfur carrier Urm1p, upon switch to SL medium, which was attenuated by the presence of methionine (Figure 4D, Figure S4D). However, amounts of the other tRNA thiolation proteins (Ncs2p and Ncs6p) did not decrease to a similar extent under these conditions (Figure S4D). These data strongly suggest that Uba4p and Urm1p abundance are regulated by sulfur amino acid availability, and that tRNA thiolation amounts also decrease in part due to reduced levels of these proteins. The decrease in Uba4p and Urm1p appeared to be occurring post-transcriptionally (Figure 4E), and was not dependent on Npr2p (Figure S4E). Furthermore, inhibiting protein synthesis by cycloheximide treatment increased the degradation rate of Uba4p only slightly (Figure S4F). Thus, when sulfur amino acids become limiting, cells actively down-regulate tRNA uridine thiolation by reducing abundance of Uba4p and Urm1p, along with reduced sulfur substrate availability.
Genes with functions associated with translation and growth are especially dependent on thiolated tRNAs for translation
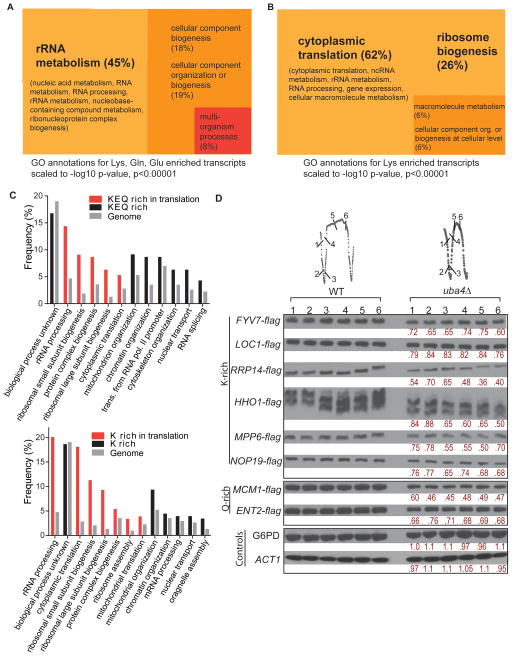
tRNA uridine modifications improve reading of A or G ending codons by facilitating wobble base-pairing (Chen et al., 2011b; Johansson et al., 2008; Murphy et al., 2004). However, a logic for why these modifications are tailored specifically to Lys (K), Glu (E), and Gln (Q) tRNAs remains unclear. In particular, our SILAC experiments revealed that cells deficient in tRNA thiolation upregulate enzymes involved in lysine biosynthesis (Figure 3C, 3F). To understand the distinctiveness of these codons, we performed an unbiased, genome-wide analysis of codon usage in yeast to assess classes of transcripts enriched in K (as well as E and Q) codons (Table S5). For our analysis, we noted that (a) K, E and Q have two codons each, but the yeast genome is biased towards codons requiring cognate uridine-modified tRNAs for translation (AAA 58%, GAA 70% and CAA 69%) and (b) the uridine modifications enable tRNAs to recognize and translate both cognate codons for each amino acid (Johansson et al., 2008). We therefore grouped both codons together for analysis. We selected genes clustered at over two standard deviations over the mean (Z≥2) for the frequency of occurrence of K, E or Q, or all three codons, and identified highly significant shared Gene Ontology (GO) terms, using an exceptional p-value cutoff <0.00001 (Table S6). We found that genes highly enriched for all three (K, E, Q) codons are substantially overrepresented in rRNA processing, ribosomal subunit biogenesis and other translation/growth-specific biological processes (Figure 5A and Table S6) (p<10−7). Secondly, K codon rich genes are especially overrepresented in processes related to rRNA formation, translation factors, ribosomal subunit biogenesis, and mitochondrial organization (Table S6 and Figure 5B) (p<10−10), while E and Q rich codons are broadly overrepresented in growth-specific processes (Figure S5A, B). Collectively, transcripts enriched in codons recognized by thiolated tRNAs, particularly lysine, are highly overrepresented in processes involved in ribosome, rRNA function, and translation. We also GO Slim mapped frequencies of these GO clusters (by biological process) in K, E, Q-enriched, or K-enriched genes with their corresponding genome-wide frequencies (Figure 5C). Genes involved in protein translation and ribosome biogenesis again were overrepresented in the list of K or K, E, Q enriched codons.
Figure 5. Lys, Gln, Glu codon-enriched genes are overrepresented in translation and growth-related functions.
(A) From a genome-wide analysis of codon usage frequency (Table S5), the genes with the highest frequencies of Lys, Gln and Glu (K, Q, E) (Z≥2) were selected and analyzed by Gene Ontology (GO) for highly significant GO terms (209 genes, p<0.00001), and visualized using scaled TreeMaps (See also Supplemental Information, Figure S5 and Table S6)
(B) Genes with the highest frequencies of Lys (K) codons (Z≥2) were selected and analyzed by Gene Ontology (GO) for highly significant GO terms (204 genes, p<0.00001), and visualized using scaled TreeMaps. (See also Table S6 and Figure S5 for genes overrepresented for Gln and Glu codons)
(C) Granular GO annotations for K, Q, E or K codon enriched genes to GO terms by biological processes, shown in comparison to genome-wide frequencies of the same biological process. Biological processes related to translation are shown in red
(D) Decreased levels of K and Q-rich proteins in tRNA thiolation-deficient cells. Several genes enriched for K or Q codons were arbitrarily selected, flag epitope tagged at their C-termini, and the encoded proteins were detected by Western blot in WT and uba4Δ mutant cells grown under continuous glucose-limitation. Quantifications of relative protein amounts comparing uba4Δ with WT are shown in red. Note: the 6 independent time points from each strain cycle can be compared since cells were maintained simultaneously at exactly the same cell density and fed the same medium. See also Figure S5.
To test if tRNA thiolation might be important for the translation of transcripts enriched in these codons, we measured the protein levels of several lysine or glutamine codon-rich genes picked from this dataset arbitrarily in WT and thiolation-deficient uba4Δ strains grown continuously under glucose-limitation (Figure 5D). Notably, the abundance of each K or Q-rich protein tested was reproducibly decreased in uba4Δ mutants across each surveyed time point (Figure 5D), with decreases ranging from ~15% to ~40% (Figure 5D). These decreases in protein levels were unlikely due to changes in transcript levels of these genes (Figure S5C). Such decreases in protein levels were less apparent when the cells were grown in YPD rich medium (Figure S5D). Thus, tRNA uridine thiolation appears to be required for optimal translation of transcripts enriched in these codons, especially under more challenging growth environments. Since genes enriched in these codons function predominantly in the translation process, these data suggest that tRNA uridine thiolation functions to regulate the overall translational capacity of the cell in tune with sulfur amino acid availability.
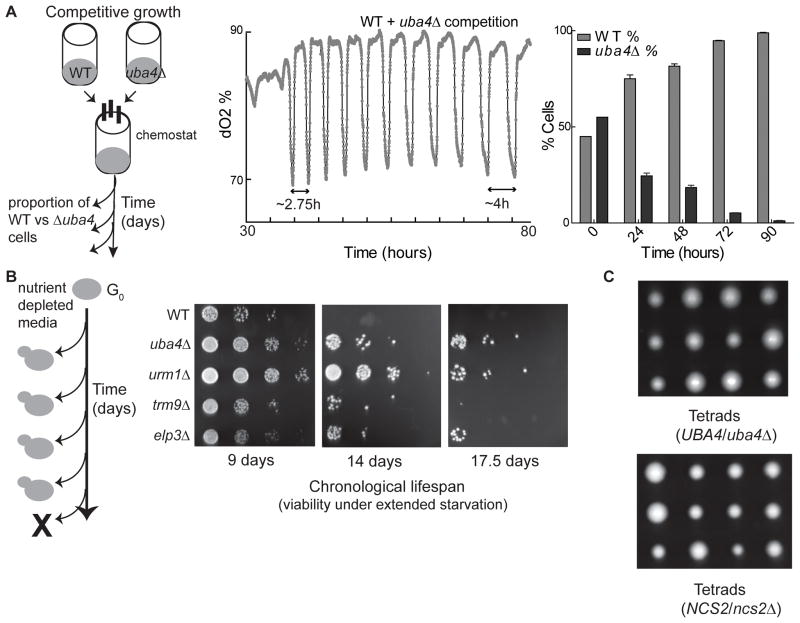
tRNA uridine modifications promote growth at the cost of survivability
For microorganisms, competitive growth advantages during nutrient limitation can be critical for their successful propagation. Competitive growth assays between WT and thiolation-deficient cells under glucose-limited conditions revealed that tRNA thiolation provided cells a strong growth advantage, allowing them to take over a population in rapid fashion (Figure 6A and Figure S6A). In contrast, accumulating evidence suggests that an overall slowing of metabolism during fasting, when cells have decreased growth and translation, functions to increase longevity or survival (Blagosklonny and Hall, 2009). One measure of survivability is a test of chronological lifespan, where yeast remain in exhausted batch cultures over time and are tested for their ability to produce colonies upon transfer to fresh medium (Figure 6B). We compared chronological lifespans between WT, thiolation-deficient (uba4Δ, urm1Δ) and mcm5-deficient (elp3Δ, trm9Δ) mutants. The absence of either modification increased chronological lifespan, and the trend correlated with the severity of metabolic cycle defects. tRNA thiolation-deficient strains survived the longest, while mcm5-deficient strains survived longer than WT strains, but less than thiolation-deficient strains (Figure 6B).
Figure 6. tRNA uridine modifications promote growth at the cost of survivability.
(A) Competitive growth between WT and tRNA thiolation-deficient cells during nutrient limited growth in chemostats was measured as depicted on the left. The percentage of each strain within the population was measured for several days. Thiolation-deficient cells were out-competed by WT cells in less than 3 days. See also Figure S6
(B) Chronological lifespan of mutants with disrupted tRNA uridine modifications. WT or mutant cells were grown in batch cultures in SD medium as depicted on the left. Cell survivability was assessed after the indicated number of days.
(C) Tetrads of sporulated diploid cells carrying a single copy uba4Δ or ncs2Δ deletion germinated on YPD medium. All the slower-growing spores were verified to be uba4Δ or ncs2Δ.
Finally, mutants lacking tRNA thiolation showed very minor growth defects in YPD glucose-rich medium (Figure S1). We hypothesized that phenotypes due to thiolation-deficiency could be masked due to compensation arising from metabolic adaptations (e.g., Figure 3) as well as the accumulation of mcm5-modified uridines. Indeed, we observed that mcm5-uridine abundance increased in thiolation-deficient cells (Figure S6). To minimize chances for compensation and adaptation in mutants, we deleted a single copy of either UBA4 or NCS2 in diploid cells, and examined the growth of newly-germinating uba4Δ or ncs2Δ haploid cells produced from sporulation (Figure 6C). These haploid mutants lacking tRNA thiolation now exhibited pronounced growth defects even on YPD rich medium (Figure 6C), indicating that the absence of tRNA thiolation acutely compromises growth.
DISCUSSION
Our findings reveal that cells co-opt tRNAs to link growth and translational capacity to the availability of a key nutrient, via a post-transcriptional nucleotide modification on the tRNA itself (Figure 7). We show that uridine thiolation on tRNAs decreases with reduced availability of the sulfur-containing amino acids cysteine and methionine. This serves as a cue to increase cysteine and methionine synthesis and salvage, signifying the importance of these sulfur amino acids. Furthermore, mRNA transcripts biased for Gln and Glu and in particular Lys codons, which are read by thiolated tRNAs, predominantly encode components of the translational machinery and other growth-related processes. Therefore, decreased levels of tRNA thiolation may be sensed by the translational machinery to modulate translational capacity. Thiolation-deficient cells in particular upregulate lysine biosynthetic enzymes, presumably to compensate for defects in translating lysine-specific codons. Thus, yeast cells utilize tRNA thiolation levels to gauge their metabolic state and translational capacity in order to achieve metabolic homeostasis (Figure 7).
Figure 7. tRNA uridine thiolation gauges sulfur amino acid availability to regulate cell growth and translation.
In sulfur amino acid-replete conditions (left), cells have high amounts of tRNA uridine thiolation, which increases translational capacity and cell growth. Sulfur amino acid starvation (right) results in decreased tRNA uridine thiolation, due to a decrease in Uba4p amounts as well as reduced sulfur equivalents. A reduction in tRNA thiolation functions to down-regulate translation and cell growth. This results in a feedback mechanism which serves to increase methionine, cysteine, and lysine synthesis and salvage. Decreased tRNA uridine thiolation may also limit translation of components of the translation machinery, which often harbor a large number of Lys, Gln and Glu codons. Thus, the abundance of thiolated tRNAs enables cells to integrate sulfur amino acid availability with supportable rates of translation and cell growth.
The uridine thiolation modification appears to be more critical than the mcm5-modification during nutrient-limited growth. This is consistent with previous observations (Murphy et al., 2004; Phelps et al., 2004) describing how tRNAlys (UUU) uridine thiolation enhances ribosomal binding and translocation of recognized codons nearly as much as multiple modifications (mcm5U34+t6A37) on tRNALys together. This is in addition to the enhanced ability of tRNAs with concurrent mcm5 and s2 modified uridines to read A and G (wobble) ending codons (Chen et al., 2011b; Esberg et al., 2006; Johansson et al., 2008). Moreover, recent studies suggest that cells finely regulate ribosome speed, and thus protein synthesis efficiency, using patterns of gene codon usage (Tuller et al., 2010). In particular, the translation of the first ~30–50 codons is slow, due to a bias for codons translated by more limiting tRNAs, leading to a “ramping” process of translation (Tuller et al., 2010). Positively charged residues such as lysines have specifically been suggested to be major determinants of ribosomal velocity and translation rate (Charneski and Hurst, 2013) and protein quality control (Brandman et al., 2012). It is possible that cells use similar modes of modulating translation capacity through specific nutrient-sensitive tRNA modifications targeted towards specific residues, particularly lysine.
How many intracellular sulfur equivalents could be consumed for tRNA uridine thiolation? Rapidly growing yeast cells contain an estimated ~3 million copies of total tRNA molecules (Phizicky and Hopper, 2010). Of 274 yeast tRNA genes, 30 (10.5%) encode just the three tRNAs with thiolated uridines (UUU, UUC and UUG anticodons), out of 61 anticodon tRNAs. The tRNA gene copy number correlates with tRNA expression levels in respiratory and fermentative growth conditions (Percudani et al., 1997; Tuller et al., 2010). Using this as a baseline, ~300,000 tRNA molecules in a single yeast cell could be thiolated, resulting in ~20 μM of uridine thiolated tRNAs during sulfur and carbon replete conditions in a ~30 fl yeast cell (Jorgensen et al., 2002), comparable to total intracellular methionine concentrations (Table S1). Changes in thiolated uridine abundance therefore reflect substantial changes in the availability of reduced sulfur. In the accompanying manuscript, we describe how autophagy is induced when cells are switched to conditions that make it hard to synthesize sufficient levels of methionine (Sutter et al., 2013). Upon switch to the same sulfur-limited conditions, tRNA thiolation is down-regulated as means to spare the consumption of sulfur during a time when cells must decrease translation rates. Preventing such sulfur “wasting” by reducing tRNA thiolation appears to be a key aspect of translational regulation. Such regulation of tRNA thiolation appears to occur downstream of TORC1 as well as the Iml1p/Npr2p/Npr3p complex. How these pathways modulate tRNA thiolation will be an important area of future research.
Integrating amino acid homeostasis with a single tRNA modification also allows cells to directly regulate the balance between growth and survival. During times of unpredictable nutrient availability, translation needs to be carefully regulated. Using a tRNA modification to sense sulfur amino acid availability and integrate it with translational capacity may provide cells with significant growth advantages under challenging nutrient environments, enabling cells to maximize translation rates when methionine and cysteine are plentiful. Conversely, when sulfur resources become limiting, this process is down-regulated perhaps to conserve sulfur for other processes important for cell survivability.
In closing, our findings reveal how tRNA thiolation is involved in regulating cell growth, translation, sulfur metabolism, and metabolic homeostasis. Through use of this ancient, conserved tRNA nucleotide modification, we show how cells have evolved a means to judiciously regulate translation and growth in response to availability of sulfur as a sentinel nutrient. As such, the ability of specific tRNAs to wobble appears to be directly linked to cellular metabolism and the availability of reduced sulfur equivalents. Although there are particular differences in the regulation of sulfur metabolism in other species compared to yeast, the tRNA thiolation pathway is conserved in all eukaryotes, and the modification conserved throughout all kingdoms of life. Therefore, it is likely that certain aspects of amino acid sensing and growth regulation through the tRNA thiolation modification may occur with a similar logic in other organisms including mammals.
EXPERIMENTAL PROCEDURES
Yeast strains and method
The prototrophic CEN.PK strain background was used in all experiments. Strains are listed in Table S7. Additional details as well as cell collection, protein extraction, immunopurifications, urmylation assays and protein detection methods are described in detail in the Supplemental Information.
RNA purifications
Small RNA species (primarily all tRNAs) were isolated from yeast cells as described in the Supplemental Information.
LC-MS/MS based detection and quantification of tRNA modifications
Targeted LC-MS/MS methods to detect and quantify tRNA uridine modifications were developed and described in the Supplemental Information.
APM polyacrylamide gel electrophoresis and northern blotting
tRNAs containing thiolated uridine were detected by Northern blotting, using polyacrylamide gels containing (N-acryloylamino)phenylmercuric chloride (APM), as described in the Supplemental Information.
Metabolic cycles
Chemostat growth and production of metabolic cycles was performed as described previously (Tu et al., 2005).
Sulfur metabolite analysis
Metabolites were extracted and sulfur-containing metabolites were measured using targeted LC-MS/MS methods described previously (Tu et al., 2007).
Genome-wide codon usage analysis
The complete ORF yeast genome was analyzed for codon composition, sorted, scored and significant enrichments analyzed using Gene Ontology as described in the Supplemental Information.
mRNA isolation and RT-qPCR
Total RNA from yeast cells grown in different media was isolated using a MasterPure Yeast RNA isolation kit (Epicentre), DNAse treated, RNA reverse transcribed into cDNA, and transcript levels measured by qPCR as described in the Supplemental Information.
Cell protein labeling and SILAC analysis
WT, uba4Δ and ncs2Δ cells were grown in SL medium supplemented with 20 μg/ml methionine, 50 μg/ml heavy or light lysine and 50 μg/ml heavy or light arginine, harvested, proteins extracted and analyzed by mass spectrometry as described in the Supplemental Information.
Chronological lifespan assays
Chronological lifespan assays comparing WT with mutant strains were performed using cultures grown in SD medium (without additional amino acids), and allowed to persist in stationary phase over a period of ~20 days (Figure 6B).
Supplementary Material
HIGHLIGHTS.
Sulfur amino acid availability is reflected by tRNA thiolation amounts
tRNA thiolation is actively down-regulated when sulfur amino acids are limiting
Sulfur amino acids modulate translational capacity using thiolated tRNAs
tRNA thiolation is important for amino acid homeostasis
Acknowledgments
We thank Uttam Tambar for suggestions during the synthesis of APM, Randal Halfmann, Paul Dutchak and other members of the Tu Lab for extensive discussions and critical input. This work was supported by the UTSW Endowed Scholars Program, award R01GM094314 from NIGMS, the Burroughs Wellcome Fund, the David and Lucile Packard Foundation, and the Damon Runyon Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdullah U, Cullen PJ. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot Cell. 2009;8:1362–1372. doi: 10.1128/EC.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging (Albany NY) 2009;1:357–362. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS biology. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Anderson JT, Bystrom AS. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PLoS One. 2011a;6:e20783. doi: 10.1371/journal.pone.0020783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Eliasson M, Ryden P, Bystrom AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011b;7:e1002258. doi: 10.1371/journal.pgen.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- Crick FH. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJ, Schaffrath R. Elongator’s toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol. 2003;49:1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003a;2:930–936. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring AS, Rivers DM, Sprague GF., Jr Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Molecular biology of the cell. 2003b;14:4329–4341. doi: 10.1091/mbc.E03-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustilo EM, Vendeix FA, Agris PF. tRNA’s modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Current biology: CB. 2004;14:R1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Tu BP. Multiple TORC1-associated proteins regulate nitrogen starvation-dependent cellular differentiation in Saccharomyces cerevisiae. PloS one. 2011;6:e26081. doi: 10.1371/journal.pone.0026081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- Murphy FVt, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- Murray DB, Klevecz RR, Lloyd D. Generation and maintenance of synchrony in Saccharomyces cerevisiae continuous culture. Exp Cell Res. 2003;287:10–15. doi: 10.1016/s0014-4827(03)00068-5. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- Nau F. The methylation of tRNA. Biochimie. 1976;58:629–645. doi: 10.1016/s0300-9084(76)80387-2. [DOI] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic acids research. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- Phelps SS, Malkiewicz A, Agris PF, Joseph S. Modified nucleotides in tRNA(Lys) and tRNA(Val) are important for translocation. J Mol Biol. 2004;338:439–444. doi: 10.1016/j.jmb.2004.02.070. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Regulation of mammalian translation factors by nutrients. Eur J Biochem. 2002;269:5338–5349. doi: 10.1046/j.1432-1033.2002.03292.x. [DOI] [PubMed] [Google Scholar]
- Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008;105:18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Chowdhury MM, Hanzelmann P, Nimtz M, Lee EY, Schindelin H, Leimkuhler S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- Sutter BM, Wu X, Laxman S, Kim SH, Tu BP. Regulation of cell growth and autophagy by methionine and the SAM-responsive methylation of Protein Phosphatase 2A (PP2A) 2013 doi: 10.1016/j.cell.2013.06.041. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci U S A. 2011;108:1763–1770. doi: 10.1073/pnas.1014402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harbor symposia on quantitative biology. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Molecular biology of the cell. 2011;22:4124–4133. doi: 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277:16391–16395. doi: 10.1074/jbc.M200253200. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.