Abstract
Prediction models for the identification of Lynch syndrome have been developed to quantify an individual’s risk of carrying a mismatch repair gene mutation and help clinicians decide for whom further risk assessment and genetic testing is necessary. There are diverse clinical settings in which a healthcare provider has the opportunity to assess an individual for Lynch syndrome. Prediction models offer a potentially feasible and useful strategy to systematically identify at-risk individuals, whether they are affected with colorectal cancer or not, and to help with management of the implications of molecular and germline test results. Given the complexity of diagnostic information currently available to clinicians involved in identifying and caring for patients with Lynch syndrome, prediction models provide a useful and complementary aid in medical decision-making. Systematic implementation of prediction models estimates should be considered in routine clinical care and at various stages of cancer risk assessment and prevention. In this manuscript, we review the main prediction models developed for Lynch syndrome, focus on their specific features and performance assessed in several validation studies, compare the models with other clinical and molecular strategies for the diagnosis of Lynch syndrome, and discuss their potential uses in clinical practice.
Keywords: Lynch syndrome; Prediction models; PREMM1,2,6; MMRPro; MMRPredict
Introduction
Lynch syndrome is the most common hereditary colorectal cancer (CRC) syndrome and is associated with underlying mutations in the mismatch repair (MMR) genes, mainly MLH1 and MSH2, but also MSH6, PMS2, and EPCAM. Individuals with Lynch syndrome have an increased risk of developing CRC, often at an early age, as well as other cancers, including those of the uterine tract lining, ovaries, stomach, small intestines, urinary tract, brain, and cutaneous sebaceous glands [1]. Early identification of individuals with MMR gene mutations is important as it allows for implementation of cancer prevention strategies such as intensified surveillance, prophylactic surgery or chemoprevention.
Several approaches are available to identify at-risk individuals and families with Lynch syndrome, including selecting individuals based on fulfillment of clinical criteria and molecular testing of tumors for evidence of microsatellite instability (MSI) through a universal (testing all CRCs) or targeted approach (based on age or family history criteria), followed by germline genetic testing for those deemed to be at increased risk. Prediction models in Lynch syndrome have been recently developed to quantify the risk of detecting a mutation based on personal and family history and help clinicians decide who should be referred for further risk assessment and/or genetic testing. In this manuscript, we will review the main prediction models developed for Lynch syndrome, focusing on their specific features and performance assessed in several validation studies, compare the models with other clinical and molecular strategies for the diagnosis of Lynch syndrome, and discuss their potential uses in clinical practice.
Identification of individuals with Lynch syndrome
Clinical criteria: Amsterdam criteria and Bethesda guidelines
Common strategies to identify individuals at risk for Lynch syndrome include fulfillment of clinical criteria such as the Amsterdam criteria or the revised Bethesda guidelines, which were developed by a consensus of experts [2, 3]. The Amsterdam criteria were originally developed for research purposes to distinguish families suspected of having hereditary non-polyposis colorectal cancer and determine the prevalence of MMR gene mutations [4]. The Bethesda guidelines were developed as a broader screening tool to identify patients whose tumors should be tested for MSI and were revised in 2004 to include both personal and family history features, including extracolonic malignancies associated with Lynch syndrome, age at diagnosis and pathologic characteristics of the tumor [2, 3]. However, the Amsterdam criteria and some components of the Bethesda guidelines are quite complex to apply and each aspect of personal and family history is weighed equally in both guidelines. Furthermore, these criteria encompass multiple diagnoses across generations and are not designed to determine the likelihood of carrying a genetic mutation for an individual patient. Several studies report that the Amsterdam criteria lack sensitivity and specificity for identification of individuals with Lynch syndrome and some have shown that the Bethesda guidelines may miss between 6 and 25 % of mutation carriers [5–8].
Molecular tumor screening: MSI and IHC testing
Germline mutations in the MMR genes result in the accumulation of mutations during DNA replication, particularly in repetitive sequences known as microsatellites. This microsatellite instability is a hallmark of tumors associated with Lynch syndrome. Mutations in the MMR genes also lead to loss of expression of the corresponding protein in the tumor. Therefore, immunohistochemical (IHC) analysis can be performed to assess which gene has been mutated. Lack of staining of MLH1 indicates either a germline mutation or methylation of the promoter, while lack of staining for MSH2 is a strong indicator for either an MSH2 or MSH6 mutation. From a clinical perspective, recognizing the heterodimeric partners MSH2–MSH6 and MLH1–PMS2 helps identify the causal mutations in Lynch syndrome by IHC. Nevertheless, approximately 15 % of sporadic cases also exhibit MSI, mainly due to somatic hypermethylation of the MLH1 promoter. Inclusion of BRAF mutation testing and MLH1 hypermethylation analyses on tumor tissue identifies sporadic microsatellite unstable CRC tumors and thereby may prevent unnecessary genetic evaluations [9].
Identification of Lynch syndrome patients
The identification of patients and families with Lynch syndrome remains a challenge. Some investigators have recommended identifying individuals based upon fulfillment of the revised Bethesda guidelines followed by MMR deficiency screening, while others recommend performing IHC on all CRC specimens [6, 10, 11]. Studies have consistently shown suboptimal assessment of family cancer history and genetics referrals among different health care professionals [12, 13]. In addition, a recent study carried out in the United States has shown that universal screening for Lynch syndrome among CRC patients based on IHC, with or without MSI testing, is feasible, but not well-adopted by cancer centers [14]. For all of the above reasons, different diagnostic strategies for identification of Lynch syndrome patients continue to be debated in the literature and clinical practice. Quantification of risk based on prediction models offers an additional means to identify those individuals at higher risk of being mutation carriers, whether affected or unaffected by cancer. The numerical estimates that the models provide can also be very helpful in communicating the results of the cancer risk assessment and its implications to patients and their family members.
Development of prediction models
Historical perspective: The Wijnen model and the Amsterdam plus model
In 1998, Wijnen and colleagues were the first to develop a multivariable model for the identification of MLH1 and MSH2 point mutations [15]. They identified three predictors in 184 unrelated kindreds at high risk of familial CRC: fulfillment of the Amsterdam criteria, mean age at diagnosis of CRC, and presence of endometrial cancer in the family. In 2004, a prediction model called Amsterdam plus, was developed from a familial cancer clinic population that added five variables to the Amsterdam criteria to improve its ability to predict MMR gene mutations: number of CRC and endometrial cancers in the family, number of individuals with two or more CRC or endometrial cancers, mean age at diagnosis, and number of individuals with five or more adenomas [16]. Both models include rather complex variables within the Amsterdam criteria, were developed using relatively small populations from dedicated high-risk clinics and have not been externally validated, potentially limiting their generalizability to other settings. These models also do not consider other important factors, such as the extracolonic tumors other than endometrial cancer that are part of Lynch syndrome, and have not become widely adopted in clinical practice.
Recent prediction models
In 2006, three prediction models were introduced to quantify an individual’s probability of carrying a MMR gene mutation most commonly associated with Lynch syndrome. These models include MMRPredict, MMRPro, and Prediction of Mismatch Repair Gene Mutations in MLH1 and MLH2 (PREMM1,2) [17–19]. The latter model has recently been extended to include prediction of MSH6 gene mutations and has been replaced by the PREMM1,2,6 model (Prediction of Mismatch Repair Gene Mutations in MLH1, MSH2, and MSH6) [20]. While the overall purpose of each model is the same, each has been developed very differently, Table 1 provides a comparison of the three prediction models.
Table 1.
Summary of the prediction models for the identification of mismatch repair gene mutation carriers
| Model | Ascertainment | # Individuals tested | Outcome | Method | Molecular testing included | Predictors | Strengths | Limitations | Software |
|---|---|---|---|---|---|---|---|---|---|
| Dutch model [15] | Clinic-based | 184 | Point mutations in MLH1, MSH2 | Logistic regression analysis | NO | Young age at CRC diagnosis in the family; fulfillment of Amsterdam criteria; presence of endometrial cancer in the family | 1st predictive model developed for Lynch syndrome; simple equation | Developed in a high risk population; Amsterdam criteria as a predictor; not validated; MSH6 and large rearrangement analysis not included | http://www5.utsouthwestern.edu |
| Amsterdam - plus [16] | Derivation and validation: clinic-based | 250 and 94 | Point mutations in MLH1, MSH2, MSH6 | Logistic regression analysis | NO | Fulfillment of Amsterdam criteria; number of relatives with CRC and endometrial cancer; number of relatives with multiple CRC or endometrial cancer; mean age at diagnosis of CRC and endometrial cancer; number of relatives with >5 colonic adenomas | Better accuracy compared to Amsterdam criteria | Complex variables; non-individualized risk prediction; software not available; large rearrangement analysis not included | N/A |
| MMRPredict [17] | Derivation: population-based CRC patients < 55 years Validation: population-based CRC patients < 45 years |
870 155 |
Overall estimate MLH1, MSH2, MSH6 | 2 stage: Multivariate logistic regression analysis. MSI/IHC data to refine prediction | YES | Proband: age, gender, tumor’s location, multiple tumors Family: CRC age (dichotomized at 50 years), endometrial cancer in 1st degree relative) |
Population-based cohort; gene-specific estimate and refinement of prediction with molecular data; individualized risk prediction; provides clinical applicability at different cut-offs | Developed and validated in a young onset CRC population; extracolonic Lynch-associated tumors other than endometrial not included |
http://www1.hgu.mrc.ac.uk Softdata/MMRpredict.php |
| MMRPro [18] | Derivation: population and clinic-based Validation: clinic-based |
N/A 279 |
Gene-specific estimate MLH1, MSH2, MSH6 | Mendelian and Bayesian analysis | YES | Proband and family: CRC and endometrial cancer status, age at diagnosis, relation to the proband, current age or at last follow- up, MSI/IHC, germline testing result | Offers pre- and post-sequencing gene-specific estimates; accounts for unaffected relatives and family size; considers molecular data; offers risk prediction of developing cancer in unaffected members | Time consuming process with pedigree drawing; extracolonic Lynch-associated neoplasms other than endometrial cancer not included |
http://www5.utsouthwestern.edu http://astor.som.jhmi.edu/BayesMendel/ |
| PREMM1,2,6 [20] | Derivation: clinic-based Validation: population and clinic-based |
4,538 1,827 |
Gene-specific estimate MLH1, MSH2, MSH6 | Logistic regression analysis | NO | Proband: age, gender, number of CRC, other Lynch-associated tumors Family: age and number of 1st and 2nd degree relatives with CRC and other Lynch-associated tumors |
Provides gene-specific estimates and individualized risk prediction; provides clinical applicability at different cut offs; easy-to-use | Molecular tumor data not incorporated; family size and unaffected individuals are not considered | http://www.dfci.org/premm |
CRC colorectal cancer, IHC immunohistochemistry, MSI microsatellite instability, N/A not available
MMRPredict
The MMRPredict model was developed using data from unselected patients with CRC diagnosed under the age of 55 years and applies logistic regression methodology [17]. The mutation prevalence in the development cohort of 875 subjects was 4 % and included mutations in MLH1 (n = 15), MSH2 (n = 16), and MSH6 (n = 7). The model gives an overall likelihood of carrying a mutation in MLH1, MSH2 or MSH6, but does not provide gene-specific risk estimates. MMRPredict calculates the overall risk estimate based on information obtained in two stages. Variables in the model’s first stage pertain to clinical information and include the patient’s age at CRC diagnosis, gender, location of tumor (proximal versus distal), multiple CRCs (synchronous or metachronous), and presence and age(s) of CRC and/or endometrial cancer diagnoses in first-degree relatives (FDR). The second stage incorporates tumor molecular diagnostic information (MSI and IHC testing) to refine the risk estimate provided in the first stage. MMRPredict does not include extracolonic cancer history in the proband. Family history is limited to FDRs, and does not take into account the number of relatives affected with CRC or endometrial cancer. The investigators also validated the model in an independent, retrospective series of patients diagnosed with CRC before the age of 45 years. Overall, the model would perform best in patients with young-onset CRC, as it was developed and validated in patients diagnosed before age 55 and 45 years respectively, and does not generate risk estimates for probands that are affected with extracolonic Lynch syndrome associated cancers but have not had CRC.
MMRPro
The MMRPro model was developed using published values for MMR mutation prevalence and penetrance for MLH1, MSH2, and MSH6 gene mutations and unlike MMRPredict and PREMM, estimates risk based on a Bayesian approach [18]. Independent, external validation was performed by the investigators involved in its development using data from 279 patients presenting to familial cancer registries where the mutation prevalence was 43 % with 51 MLH1, 63 MSH2, and 7 MSH6 gene mutation carriers. The model includes data for the proband and for each FDR and second-degree relative (SDR) on the presence of CRC and/or endometrial cancer, age at diagnosis, and current age or age at last follow-up for those unaffected by these cancers. The MMRPro model does not take into account the presence of multiple CRCs in the proband or any Lynch syndrome associated cancers other than endometrial cancer. The model incorporates molecular tumor testing results including MSI and IHC testing. It calculates the overall risk of carrying any one of the three gene mutations and provides separate risk estimates for each of the genes. The model’s unique features are that it can calculate the residual probability of finding a mutation if molecular tumor testing is negative and provide estimates of future colorectal and endometrial cancer risks for unaffected individuals (including known mutation carriers), untested individuals, and individuals in whom no mutation is found.
PREMM1,2,6
The PREMM1,2,6 model was developed using genotype and phenotypic data from 4,539 individuals who underwent genetic testing based on either personal or family history of cancer using multivariable polytomous logistic regression methodology [20]. The mutation prevalence was 12 % and the cohort represents the largest number of unrelated gene mutation carriers used for model development and validation (204 MLH1, 250 MSH2, and 71 MSH6 gene mutations). Proband specific variables include gender, the occurrence and age at CRC diagnosis (including multiple CRC diagnoses), endometrial cancer and other Lynch syndrome-associated cancers (including cancers of the ovary, stomach, kidney, ureter, bile duct, small bowel, brain (glioblastoma multiforme), pancreas, or sebaceous gland). Variables related to family members are limited to FDR and SDR cancer histories and include the number of relatives with CRC, endometrial cancer, or other Lynch syndrome-associated cancers as well as the minimum age at diagnosis of each cancer among relatives. The PREMM1,2,6 model replaces the original PREMM1,2 model which calculated risk of MLH1 and MSH2 gene mutations based on the same aforementioned variables except for gender and omits the presence of polyps in the proband. The model does not include molecular tumor data in risk prediction or data on unaffected family members. The PREMM1,2,6 model was externally validated by the investigators involved in its development among 1,827 population-based CRC cases and high-risk patients recruited through an international consortium.
Performance of prediction models compared to clinical criteria
With the introduction of Lynch syndrome prediction models, their ability to identify gene mutation carriers as compared to existing clinical criteria has also been studied. The performance of each model compared to Amsterdam and Bethesda guidelines has been assessed in each respective development cohort and in most cases, found to exceed the clinical criteria’s ability to discriminate carriers from noncarriers. MMRPredict performed similarly to the Bethesda guidelines at a cut-off of 0.005 with sensitivity of 95 % for both strategies, specificity of 14 and 38 %, and positive predictive value of 5 and 6 % respectively. MMRPredict performed better than the Amsterdam criteria alone and in combination with MSI testing [17]. The PREMM1,2 model selected 20 % more individuals than the revised Bethesda guidelines at a 5 % cut-off value, yielding higher sensitivity (94 vs 74 % respectively) but with 10 % lower specificity [19]. At this cut-off, the miss rate of mutation carriers was 6 % for PREMM1,2 compared with 26 % for Bethesda and 37 % for Amsterdam II criteria. In an independent, population-based study of 1,222 CRC cases that compared the PREMM1,2 and MMRPredict models for the identification of MLH1 and MSH2 gene mutation carriers, at a cut-off score of ≥5 %, the sensitivity of the revised Bethesda guidelines and PREMM1,2 was 100 % (95 % CI 71–100 %) compared to 87 % sensitivity (95 % CI 51–99 %) for MMRPredict [21, 22]. Use of the Amsterdam II criteria alone yielded 50 % sensitivity while specificity was highest at 98 %. Lastly, in the sample used to validate MMRPro’s performance as determined in the development cohort, MMRPro better identified carriers from noncarriers than the Bethesda guidelines, with and without MSI testing [18].
In a recent study by Green et al., the three models were compared among 725 unselected CRC cases diagnosed before age 75 years and 18 individuals (2.5 %) were identified with MMR gene mutations: eleven carriers met Amsterdam I criteria, one met Amsterdam II criteria, five others met only the Bethesda criteria, and one met none of these criteria [23]. The Bethesda guidelines yielded 94 % sensitivity and in comparing the models at cut-off points that would produce this same sensitivity, specificities were higher for the models, ranging from 61 to 88 % compared to 51 % for the Bethesda criteria (95 % CI 47.2–54.7 %).
Performance of prediction models compared to molecular tumor screening
One study has compared the performance of molecular tumor testing to prediction of MMR gene mutation status using the PREMM1,2,6 model [24]. The model’s ability to discern gene mutation carriers from noncarriers compared to molecular tumor testing was evaluated in 1,651 patients with CRC recruited through an international consortium of population and clinic-based family registries. There were 239/2,651 (14 %) mutation carriers and when assessed individually, the AUCs for PREMM1,2,6, IHC, and MSI were 0.90, 0.82, and 0.78 respectively. The strategy which most improved the PREMM1,2,6 model’s discriminative ability among both population and clinic-based cases was adding IHC tumor testing to PREMM1,2,6 prediction where AUC improved to 0.94 in the overall and population-based cohorts and 0.92 in the clinic based cohort. There was no additional value of MSI testing to the combination of PREMM1,2,6 + IHC. The impact of the proband’s age at CRC diagnosis on the strategies’ ability to discriminate mutation carriers from noncarriers was assessed and for every 10 year increase in the initial age of CRC diagnosis, IHC’s performance decreased with AUC of 0.85 for pro-bands diagnosed by 50 years, 0.84 for CRC diagnosed by 60 years, and 0.82 for CRC diagnoses by 70 years. In contrast, the performance of PREMM1,2,6 increased slightly with every 10 year increase in age at time of CRC diagnosis: AUC of 0.87 to 0.88 to 0.90 for CRC diagnoses made by 50, 60 and 70 years respectively. Similar to IHC, the discriminatory ability of MSI testing was 0.83 for CRC cases diagnosed by ages 50 and 60 years, which decreased to 0.81 for cases diagnosed by 70 years.
Validation of prediction models
A number of studies independently validate the performance and accuracy of the three recently developed prediction models, other than those aforementioned validation studies performed by the investigators involved in the development of each model (Table 2). Only one study provides external validation and comparison of the PREMM1,2,6 model to MMRPro and MMRPredict, as the PREMM1,2,6 model was most recently introduced in 2011 [25]. In a multicenter US-based study of 230 referred patients at high-risk of CRC, 113 gene mutation carriers were identified where the three models’ performance was similar: AUCs were 0.76 for MMRPredict (95 % CI 0.68–0.84), 0.78 for PREMM1,2,6 (95 % CI 0.72–0.84), and 0.82 for MMRPro (95 % CI 0.74–0.86). The performance of each model was assessed across a range of sensitivities: to obtain a sensitivity of 90 %, a threshold for mutation testing of>4 % for MMRPro would provide 29 % specificity; a cut-off of >6 % for PREMM1,2,6 would be 38 % specific, and for MMRPro, a cut-off score of >7 % would provide 36 % specificity.
Table 2.
Comparison of prediction models and clinical criteria for the identification of Lynch syndrome
| Study | Cohort | Subjects N | MMR Gene Mutation Prevalence N (%) | MMRPredict AUC (95 % CI) | MMRPro AUC (95 % CI) | PREMM AUC (95 % CI) | Revised Bethesda AUC (95 % CI) | Amsterdam Criteria AUC (95 % CI) |
|---|---|---|---|---|---|---|---|---|
| Khan et al. [25] | High-Risk | 230 | 113 (49) | 0.76 (0.68–0.84) | 0.82 (0.74–0.86) | 0.78 (0.72–0.84) | AUC: 0.52 (n/a) Sens: 99 % Spec: 10 % |
AUC: 0.68 (n/a) Sens: 81 % Spec: 52 % |
| Pouchet et al. [26] | High-Risk | 81 | 39 (48) | 0.73 (0.61–0.86) | 0.73 (0.62–0.85) | 0.77 (0.65–0.88) | n/a | n/a |
| Monzon et al. [27] | High-Risk | 72 | 25 (35) | 0.86 (0.76–0.96) | 0.90 (0.82–0.98) | 0.93 (0.86–0.99) | n/a | n/a Sens: 76 % Spec: 74 % |
| Balmaña et al. [22] | Population-based | 1,222 | 8 (0.7) | 0.92 (0.83–1.0) | n/a | 0.93 (0.86–0.99) | n/a Sens: 100 % Spec: 77 % |
n/a Sens: 50 % Spec: 90 % |
| Green et al. [23] | Population-based | 725 | 18 (2.5) | 0.96 (0.94–0.97) | 0.95 (0.93–0.96) | 0.91 (0.89–0.93) | n/a Sens: 94 % Spec: 51 % |
n/a Sens:66 % Spec: 96 % |
| Tresallet et al. [28] | Population-based | 214 | 8 (3.7) | 0.76 (0.65–0.88) | 0.73 (0.49–0.98) | 0.76 (0.59–0.93) | n/a Sens: 75 % Spec: 59 % |
n/a Sens: 37 % Spec: 99 % |
AUC area under the receiver operating characteristic curve, n/a not available, Sens sensitivity, Spec specificity
Previous validation studies comparing PREMM1,2 to MMRPredict and MMRPro have yielded similar results among selected patients at high-risk for CRC. In an early study comparing the models’ ability to discern gene mutation carriers from noncarriers identified through a cancer genetics clinic (n = 81), MMRPredict, PREMM1,2 and MMRPro performed similarly (AUCs of 0.73, 0.77 and 0.73, respectively) in identifying the 39 carriers [26]. However, predictions were not possible in seven individuals with extracolonic cancers using the MMRPredict model as it requires a CRC diagnosis. The investigators also compared predictions for each model among different risk categories to determine whether there was over- or under-prediction of mutation carriers: all individuals in the highest risk category (75–100 %) according to PREMM1,2 were carriers whereas nearly half of the individuals identified by MMRPro were carriers. Within the lowest high-risk category of <10 %, MMRPro and PREMM1,2 identified the majority as noncarriers whereas MMRPredict underestimated the carriers, missing eight carriers. MMRPredict overpredicted gene mutation carrier status between 10 and 75 % and had the most variability (dispersion) of probability estimates compared with PREMM1,2 and MMRPro. The authors concluded that the PREMM1,2 model was more accurate in detecting carriers and MMRPro was more accurate in detecting noncarriers.
A similar study of 25 gene mutation carriers detected among 72 cases evaluated through a high-risk genetics clinic found comparable performance characteristics between the three models: AUCs of 0.86 (95 % CI 0.7–0.96), 0.89 (95 % CI 0.81–0.98) and 0.93 (95 % CI 0.86–0.99) for the MMR-Predict, MMRPro and PREMM1,2 models, respectively where PREMM1,2 demonstrated the best performance for predicting carrier status based on the positive likelihood ratios at >10, >20 and >30 % probability thresholds [27].
The three studies that validate the models among unselected, population based CRC cases are limited by the few mutation carriers in each cohort: 8/214, 8/1,222 and 18/725 mutation carriers (Table 2) [21, 23, 28]. The latter study, which assessed patients with CRC diagnosed before age 75 years found that all the models overestimated MMR gene mutation carrier status by 2.1 to 4.3 fold. The investigators further corrected the prediction estimates for bias introduced by family size and age structure on the non-Bayesian derived models (MMRPredict and PREMM1,2) and improvement in discriminating of carriers from noncarriers was noted for both models. With this correction, the authors concluded that MMRPredict performed best in their dataset, which achieved a sensitivity of 94 % (95 % CI 73–99 %) and a specificity of 91 % (95 % CI 88–93 %), but recommend use of any of the models in identifying patients with CRC who would benefit from molecular tumor testing and further evaluation for Lynch syndrome.
Prediction models and cost-effective strategies for the identification of Lynch syndrome
To the best of our knowledge, only one cost-effectiveness analysis compares different strategies in identifying MMR gene mutation carriers among CRC patients which includes prediction model estimates [29]. Results from a Markov modeling analysis examine clinical criteria (Amsterdam and Bethesda guidelines), the three prediction models (MMRPro, MMRPredict, and PREMM1,2,6), tumor testing or up-front germline mutation testing followed by directed screening and risk-reducing surgery. When clinical criteria were met or prediction model scores were >5 % probability of carrying a gene mutation, IHC testing followed by germline testing was offered or germline testing was directly pursued. Under the assumption of perfect implementation of all strategies, tumor testing strategies were more costly than clinical criteria and prediction model strategies. However, with decreased implementation rates of clinical criteria strategies compared with tumor testing, the latter became more cost-effective. The most cost-effective strategy to evaluate for Lynch syndrome among newly diagnosed patients with CRC diagnosed before age 70 years was IHC testing (plus BRAF mutation testing for MLH1 protein loss), followed by targeted MMR gene sequencing; an incremental cost-effectiveness ratio of $36,200 per life year gained resulted from this strategy. However, the critical determinant of cost-effectiveness of any strategy was highly dependent on the number of relatives per proband who underwent germline testing and opportunities for cancer risk reduction.
A second cost-effective analysis examines screening approaches for Lynch syndrome among individuals without cancer in the general population [30]. In a simulation-based analysis using the PREMM1,2,6 model to detect individuals with MMR gene mutations and their at-risk relatives, integrated models of CRC and endometrial cancers with a 5-generation family history to predict health and economic outcomes of 20 primary screening strategies (with a wide range of compliance levels) were analyzed. These strategies were characterized by different screening ages for starting risk assessment and different risk thresholds above which to recommend genetic testing. For each strategy, 100,000 simulated individuals representative of the US population were followed from 20 years and the outcomes were compared with current practice. The results indicate that risk assessment starting at ages 25, 30, or 35 years, followed by genetic testing of those with mutation risks exceeding 5 % assessed by the PREMM1,2,6 model, reduced CRC and endometrial cancer incidence in MMR gene mutation carriers by approximately 12.4 and 8.8 %, respectively. For a population of 100,000 individuals with 392 mutation carriers, this strategy increased quality-adjusted life-years (QALY) by approximately 135 with an average cost-effectiveness ratio of $26,000 per QALY where the cost-effectiveness of screening for MMR gene mutations was comparable to currently well-accepted cancer screening activities for CRC, cervical and breast cancer. These results suggest that assessing unaffected individuals’ risk for Lynch syndrome based on their family cancer history starting between the ages of 25–35 years is appropriate and cost-effective.
Summary of comparison of models
The MMRPredict, MMRPro and PREMM1,2,6 models all provide a quantitative assessment of the risk of being a MMR gene mutation carrier. There is ample evidence that each of the models have superior performance characteristics in terms of sensitivity, specificity, positive and negative predictive values to support the use of the models over the existing clinical guidelines for the diagnosis and evaluation for Lynch syndrome.
Deciding which Lynch syndrome prediction model to chose in the risk assessment process depends on both the clinical setting in which it is applied and the patient population that is being evaluated. Validation studies show that the models have similar ability to discriminate gene mutation carriers from noncarriers. While the models were all developed for the same purpose, the populations in which they were validated brings to light each model’s specific characteristics that may impact the accuracy of the provided risk estimates and help healthcare providers determine which may be more appropriate to use in their respective clinical settings.
MMRPredict is best applicable in the assessment of patients with young onset CRC and its use could provide less accurate results when used to evaluate families with Lynch syndrome associated cancers and older individuals affected by CRC. MMRPro’s predictions account for family size and unaffected relatives, the possibility of including molecular tumor data in the risk analysis, and the option of predicting gene mutation carrier status following germline testing. The major limitation in the widespread use of MMRPro in routine practice outside the genetics setting is the need to input the entire pedigree which is relatively time consuming. Its best use at the current time is likely to be as a genetic counseling tool in a specialized high-risk clinic or research setting. PREMM’s major advantage is that it is easy to use and has been extensively validated. It includes risk prediction based on personal and family cancer history up to second-degree relatives for a broad spectrum of extracolonic cancers. However, the model does not take into account family size and may overestimate the likelihood of a mutation in a pedigree with includes multiple, elderly family members who are unaffected by CRC or endometrial cancer. Furthermore, the model does not incorporate tumor testing results or provide post hoc risk estimates based on gene sequencing results. Given the ease with which one can use the PREMM1,2,6 model (it has been deemed less time-consuming than MMRPro in validation studies) [25], it may be best considered by diverse healthcare providers whose aim is primarily to identify those patients who should be referred for genetic evaluation and is likely to be most useful in the pre-testing decision-making process. Further data on the performance of models in diverse populations as well as the accuracy of the gene-specific predictions of MMRPro and PREMM1,2,6 are needed.
How can prediction models help clinicians?
The best approach in identifying patients at risk for Lynch syndrome is one that is attainable by a majority of clinical practices. Given the diverse clinical settings in which a healthcare provider has the opportunity to assess an individual for Lynch syndrome, prediction models offer a potentially feasible and useful strategy to systematically identify at-risk individuals, whether they are affected with CRC or not, and to help with management of the implications of molecular and germline test results.
Prediction models and the evaluation of patients with CRC
For the initial assessment of Lynch syndrome among patients with CRC, it is important to determine whether the tumor specimen is available for additional molecular diagnostic testing.
For patients with CRC and available tumor block
Molecular tumor testing, including IHC and MSI testing of the CRC tumor, complemented by BRAF testing for cases with loss of MLH1 protein expression, is an important strategy in the assessment for Lynch syndrome. Two possible approaches seem to be the most reasonable given emerging data—universal testing of all CRCs regardless of age with IHC and/or MSI, or only testing patients diagnosed before age 70 years, as the latter is likely to be more cost-effective [29].
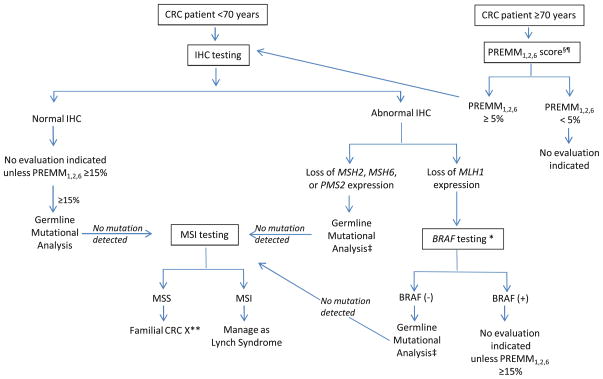
The models can complement molecular testing strategies in both of these scenarios (Fig. 1) [24]. To date, variable cutoff scores have been assessed for the PREMM1,2,6 model regarding its use compared to molecular tumor testing; comparable cut-off scores for the MMRPro and MMRPredict models have not yet been explored. A PREMM1,2,6 score of ≥5 % may be used as a first step in the assessment of Lynch syndrome in patients with CRC diagnosed above 70 years. Molecular tumor testing and genetic testing can likely not be pursued with PREMM1,2,6 scores<5 % in patients older than 70 years, as the likelihood of Lynch syndrome in the absence of significant family cancer history is unlikely and any abnormal tumor testing results are more likely to be age-related hypermethylation of the MLH1 gene promoter than a germline mutation. This approach can thereby limit the increased costs related to molecular diagnostic testing. For those patients with PREMM1,2,6 scores ≥5 %, IHC testing, along with BRAF testing for those tumors which display loss of MLH1, should be performed and results used to direct gene-specific DNA mutation analysis in order to ensure cost-efficiency. In cases were a BRAF mutation is detected, no germline testing would be necessary unless the PREMM1,2,6 score was >15 %. For patients with normal MSI or IHC, a PREMM1,2,6 cut-off of 15 % still necessitates DNA mutation analysis due to the possibility of false negative results. When no gene mutation is detected, MSI testing may be helpful for evaluating these patients as they are high risk for Lynch syndrome based on personal and family cancer history. MSI testing can stratify genetic testing results into either (1) indeterminate negative, where patients with MSI-high tumors should be treated as having as Lynch syndrome or (2) “Familial CRC X,” where patients with microsatellite-stable tumors do not need intensive cancer surveillance as those recommended for Lynch syndrome.
Fig. 1.
Proposed Algorithm for Systematic Evaluation for Lynch syndrome in Patients with Colorectal Cancer¥. CRC colorectal cancer, MSI microsatellite instability; MSS microsatellite stable, IHC immunohistochemistry.¥ In patients without available tumor for molecular diagnostic testing and in those individuals without colorectal cancer, a PREMM1,2,6 score ≥5 % should be followed by germline mutational analysis.§ PREMM1,2,6 score can be calculated at website http://www.dfci.org/premm; ¶ Other models (MMRpro, MMRpredict) may also be used with their own specified cut-off scores; * BRAF testing: + mutation present, − mutation absent/wild type; ** Surveillance recommendations based on personal and family history; ‡Gene-specific germline mutational analysis
For patients with CRC when a tumor block is unavailable
The opportunity to evaluate a patient for Lynch syndrome should not cease when their CRC tumor block is unavailable for molecular diagnostic testing. This may be the case when (1) the history of CRC was made years prior and tumor specimens can not be retrieved, (2) there are technical limitations to performing IHC or MSI testing, or (3) genetic evaluation and testing is necessary prior to any surgical decision-making. Risk assessment using the prediction models should be the strategy of choice when tumor testing is not feasible and is supported by the National Comprehensive Cancer Network where patients with a risk score of ≥5 % should be offered germline testing for Lynch syndrome [31].
Prediction models and the evaluation of individuals without CRC
Risk assessment based on family history is necessary in unaffected individuals to ensure appropriate surveillance for CRC and other associated cancers. When a family history of cancer is present, the quantifiable estimates provided by prediction models simplify the risk assessment process, making it easier for patients to understand risk and help them decide the next course of action regarding their genetic evaluation and/or surveillance strategies. Germline testing should be considered for those unaffected individuals with a risk estimate of ≥5 %, where no affected family member is available for testing, and is an approach that has been shown to be cost-effective [30].
Summary
Given the complexity of the diagnostic information available to clinicians involved in identifying and caring for patients with Lynch syndrome, prediction models provide a useful and complementary aid in medical decision-making. With growing evidence and consensus that the prediction models outperfom existing clinical criteria for Lynch syndrome, systematic implementation of prediction models estimates should be considered in routine clinical care and at various stages of cancer risk assessment and prevention.
Acknowledgments
This work was supported by the National Cancer Institute: K07 CA151769-02 (FK), R01 CA132829 (SS) and K24 CA113433 (SS). Additional support from the Louis V. Gerstner, Jr. Scholars Program (FK).
Contributor Information
Fay Kastrinos, Herbert Irving Comprehensive Cancer Center, Columbia University Medical Center, New York, NY, USA. Division of Digestive and Liver Diseases, Columbia University Medical Center, New York, NY, USA. Columbia University College of Physicians and Surgeons, New York, NY, USA.
Judith Balmaña, Department of Medical Oncology, Hospital Vall d’Hebron, Universitat Autònoma de Barcelona, Barcelona, Spain.
Sapna Syngal, Email: ssyngal@partners.org, Population Sciences Division, Dana-Farber Cancer Institute, 450 Brookline Avenue, Boston, MA 02215, USA. Division of Gastroenterology, Brigham and Women’s Hospital, Boston, MA, USA. Harvard Medical School, Boston, MA, USA.
References
- 1.Aarnio M, Mecklin JP, Aaltonen LA, Nyström-Lahti M, Järvinen HJ. Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer. 1995;64(6):430–433. doi: 10.1002/ijc.2910640613. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S. A National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89(23):1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 3.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JG, Vasen HF, Park KJ, Peltomaki P, de Leon MP, Rodriguez-Bigas MA, Lubinski J, Beck NE, Bisgaard ML, Miyaki M, Wijnen JT, Baba S, Lynch HT. Suspected hereditary nonpolyposis colorectal cancer: International Collaborative Group on hereditary non-polyposis colorectal cancer (ICG-HNPCC) criteria and results of genetic diagnosis. Dis Colon Rectum. 1999;42(6):710–715. doi: 10.1007/BF02236922. [DOI] [PubMed] [Google Scholar]
- 5.Syngal S, Fox EA, Eng C, Kolodner RD, Garber JE. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37(9):641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352(18):1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 7.Julié C, Trésallet C, Brouquet A, Vallot C, Zimmermann U, Mitry E, Radvanyi F, Rouleau E, Lidereau R, Coulet F, Olschwang S, Frébourg T, Rougier P, Nordlinger B, Laurent-Puig P, Penna C, Boileau C, Franc B, Muti C, Hofmann-Radvanyi H. Identification in daily practice of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer): revised Bethesda guidelines-based approach versus molecular screening. Am J Gastroenterol. 2008;103(11):2825–2835. doi: 10.1111/j.1572-0241.2008.02084.x. [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Carbonell L, Ruiz-Ponte C, Guarinos C, Alenda C, Payá A, Brea A, Egoavil CM, Castillejo A, Barberá VM, Bessa X, Xicola RM, Rodríguez-Soler M, Sánchez-Fortún C, Acame N, Castellví-Bel S, Piñol V, Balaguer F, Bujanda L, De-Castro ML, Llor X, Andreu M, Carracedo A, Soto JL, Castells A, Jover R. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012;61(6):865–872. doi: 10.1136/gutjnl-2011-300041. [DOI] [PubMed] [Google Scholar]
- 9.Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49(3):151–157. doi: 10.1136/jmedgenet-2011-100714. [DOI] [PubMed] [Google Scholar]
- 10.Piñol V, Castells A, Andreu M, Castellví-Bel S, Alenda C, Llor X, Xicola RM, Rodríguez-Moranta F, Payá A, Jover R, Bessa X. Gastrointestinal Oncology Group of the Spanish Gastro-enterological Association. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293(16):1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 11.Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, Hopper JL, Le Marchand L, Gallinger S, Newcomb PA, Haile R, Thibodeau SN, Gunawardena S, Jenkins MA, Buchanan DD, Potter JD, Baron JA, Ahnen DJ, Moreno V, Andreu M, de Leon MP, Rustgi AK, Castells A. EPI-COLON Consortium. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308(15):1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grover S, Stoffel EM, Bussone L, Tschoegl E, Syngal S. Physician assessment of family cancer history and referral for genetic evaluation in colorectal cancer patients. Clin Gastroenterol Hepatol. 2004;2(9):813–819. doi: 10.1016/s1542-3565(04)00352-0. [DOI] [PubMed] [Google Scholar]
- 13.Flynn BS, Wood ME, Ashikaga T, Stockdale A, Dana GS, Naud S. Primary care physicians’ use of family history for cancer risk assessment. BMC Fam Pract. 2010;11:45. doi: 10.1186/1471-2296-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beamer LC, Grant ML, Espenschied CR, Blazer KR, Hampel HL, Weitzel JN, MacDonald DJ. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol. 2012;30(10):1058–1063. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wijnen JT, Vasen HF, Khan PM, Zwinderman AH, van der Klift H, Mulder A, Tops C, Møller P, Fodde R. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med. 1998;339(8):511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 16.Lipton LR, Johnson V, Cummings C, Fisher S, Risby P, Eftekhar Sadat AT, Cranston T, Izatt L, Sasieni P, Hodgson SV, Thomas HJ, Tomlinson IP. Refining the Amsterdam Criteria and Bethesda Guidelines: testing algorithms for the prediction of mismatch repair mutation status in the familial cancer clinic. J Clin Oncol. 2004;22(24):4934–4943. doi: 10.1200/JCO.2004.11.084. [DOI] [PubMed] [Google Scholar]
- 17.Barnetson RA, Tenesa A, Farrington SM, Nicholl ID, Cetnarskyj R, Porteous ME, Campbell H, Dunlop MG. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, Watson P, Gruber SB, Euhus D, Kinzler KW, Jass J, Gallinger S, Lindor NM, Casey G, Ellis N, Giardiello FM, Offit K, Parmigiani G. Colon Cancer Family Registry. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balmaña J, Stockwell DH, Steyerberg EW, Stoffel EM, Deffenbaugh AM, Reid JE, Ward B, Scholl T, Hendrickson B, Tazelaar J, Burbidge LA, Syngal S. Prediction of MLH1 and MSH2 mutations in Lynch syndrome. JAMA. 2006;296:1469–1478. doi: 10.1001/jama.296.12.1469. [DOI] [PubMed] [Google Scholar]
- 20.Kastrinos F, Steyerberg EW, Mercado R, Balmaña J, Holter S, Gallinger S, Siegmund KD, Church JM, Jenkins MA, Lindor NM, Thibodeau SN, Burbidge LA, Wenstrup RJ, Syngal S. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140(1):73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balaguer F, Balmaña J, Castellví-Bel S, Steyerberg EW, Andreu M, Llor X, Jover R, Syngal S, Castells A. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Validation and extension of the PREMM1,2 model in a population-based cohort of colorectal cancer patients. Gastroenterology. 2008;134(1):39–46. doi: 10.1053/j.gastro.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balmaña J, Balaguer F, Castellví-Bel S, Steyerberg EW, Andreu M, Llor X, Jover R, Castells A, Syngal S. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Comparison of predictive models, clinical criteria and molecular tumor screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet. 2008;45:557–563. doi: 10.1136/jmg.2008.059311. [DOI] [PubMed] [Google Scholar]
- 23.Green RC, Parfrey PS, Woods MO, Younghusband HB. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst. 2009;101:331–340. doi: 10.1093/jnci/djn499. [DOI] [PubMed] [Google Scholar]
- 24.Kastrinos F, Steyerberg EW, Balmaña J, Mercado R, Gallinger S, Haile R, Casey G, Hopper JL, Lemarchand L, Lindor NM, Newcomb PA, Thibodeau SN, Syngal S The Colon Cancer Family Registry. Comparison of the clinical prediction model PREMM1,2,6 and molecular testing for the systematic identification of Lynch syndrome in colorectal cancer. Gut. 2012 doi: 10.1136/gutjnl-2011-301265. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan O, Blanco A, Conrad P, Gulden C, Moss TZ, Olopade OI, Kupfer SS, Terdiman J. Performance of Lynch syndrome predictive models in a multi-center US referral population. Am J Gastroenterol. 2011;106:1822–1827. doi: 10.1038/ajg.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouchet CJ, Wong N, Chong G, Sheehan MJ, Schneider G, Rosen-Sheidley B, Foulkes W, Tischkowitz M. A comparison of models used to predict MLH1, MSH2 and MSH6 mutation carriers. Ann Oncol. 2009;20:681–688. doi: 10.1093/annonc/mdn686. [DOI] [PubMed] [Google Scholar]
- 27.Monzon JG, Cremin C, Armstrong L, Nuk J, Young S, Horsman DE, Garbutt K, Bajdik CD, Gill S. Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer. 2010;126:930–939. doi: 10.1002/ijc.24808. [DOI] [PubMed] [Google Scholar]
- 28.Tresallet C, Brouquet A, Julié C, Beauchet A, Vallot C, Ménégaux F, Mitry E, Radvanyi F, Malafosse R, Rougier P, Nordlinger B, Laurent-Puig P, Boileau C, Emile JF, Muti C, Penna C, Hofmann-Radvanyi H. Evaluation of predictive models in daily practice for the identification of patients with Lynch syndrome. Int J Cancer. 2012;130(6):1367–1377. doi: 10.1002/ijc.26144. [DOI] [PubMed] [Google Scholar]
- 29.Ladabaum U, Wang G, Terdiman J, Blanco A, Kuppermann M, Boland CR, Ford J, Elkin E, Phillips KA. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med. 2011;155:69–79. doi: 10.7326/0003-4819-155-2-201107190-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinh TA, Rosner BI, Atwood JC, Boland CR, Syngal S, Vasen HF, Gruber SB, Burt RW. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res. 2011;4:9–22. doi: 10.1158/1940-6207.CAPR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Comprehensive Cancer Network. [Accessed November 2012];Guidelines Version 2.2012 Colorectal Cancer Screening. MS-15 Retrieved from http://www.nccn.org/clinical.asp. [Google Scholar]