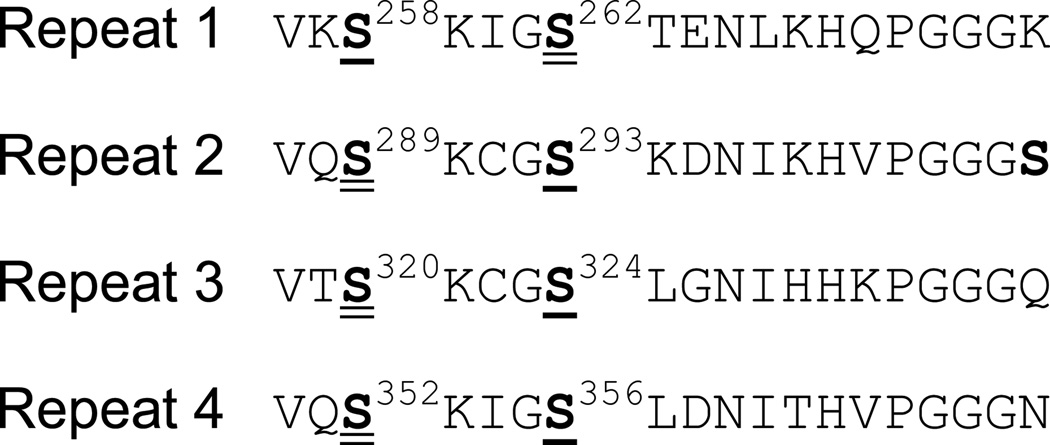Abstract
Hyperphosphorylation of microtubule-associated protein tau is thought to contribute to Alzheimer’s disease (AD) pathogenesis. We previously showed that DNA damage-activated cell cycle checkpoint kinases Chk1 and Chk2 phosphorylate tau at an AD-related site and enhance tau toxicity, suggesting potential roles of these kinases in AD. The purpose of this study is to systematically identify which sites in tau are directly phosphorylated by Chk1 and Chk2. Using recombinant human tau phosphorylated by Chk1 and Chk2 in vitro, we firstly analyzed tau phosphorylation at the AD-related sites by Western blot with phospho-tau-specific antibodies. Secondly, to globally identify phosphorylated sites in tau, liquid chromatography-tandem mass spectrometry (LC-MS/MS) was employed. These systematic analyses identified a total of 27 Ser/Thr residues as Chk1- or Chk2- target sites. None of them were proline-directed kinase targets. Many of these sites are located within the microtubule-binding domain and C-terminal domain, whose phosphorylation has been shown to reduce tau binding to microtubules and/or has been implicated in tau toxicity. Among these 27 sites, 13 sites have been identified to be phosphorylated in AD brains. Since DNA damage is accumulated in diseased brains, Chk1 and Chk2 may be involved in tau phosphorylation and toxicity in AD pathogenesis.
Keywords: Alzheimer’s disease, checkpoint kinase 1, checkpoint kinases 2, liquid chromatography, mass spectrometry, microtubule-associated protein tau, phosphorylation
Introduction
The checkpoint kinases Chk1 and Chk2 are Ser/Thr kinases that play critical roles in DNA damage-induced cell cycle checkpoint signaling pathways 1. In the presence of DNA damage or incomplete DNA replication, Chk1 and Chk2 are activated responding to the checkpoint signals emanating from the ATR (ataxia-telangiectasia and Rad-3 related) and ATM (ataxia-telangiectasia mutated), which leads to the activation of DNA repair, cell-cycle arrest, senescence or apoptosis 2. Chk1 and Chk2 have been reported to play important roles in a variety of processes including oogenesis, tissue growth, metabolic stress, tumorigenesis and neuronal survival 3–13.
Chk1 and Chk2 can phosphorylate a number of downstream effectors, such as BRCA1, Cdc25A, Cdc25C, E2F1, p53, and p73α, which play regulatory roles in DNA repair and cell cycle progression 14–20. More recently, in vitro studies using peptide library 21–23 or chemical genetics 24 have identified a number of potential substrates of Chk1 and/or Chk2. The list of targets produced from these studies revealed that Chk1 and/or Chk2 can phosphorylate proteins that play key roles in the cellular events such as RNA splicing, cell fate determination, and regulation of cytoskeleton, suggesting novel functions of Chk1 and Chk2 under physiological and pathophysiological conditions.
We have previously reported that the microtubule-associated protein tau is a novel substrate for Chk1 and Chk2 25. Under physiological conditions, tau is predominantly expressed in neurons and preferentially localizes to the axons, where it regulates microtubule dynamics 26. However, in Alzheimer’s disease (AD) brains, hyperphosphorylation of tau results in the formation of aggregates called paired helical filaments in neurofibrillary tangles, which is thought to contribute to AD pathogenesis 27. The longest isoform of tau has 85 potential Ser/Thr phosphorylation sites. To date, 45 sites have been identified to be phosphorylated in AD brains. Several Ser/Thr kinases including proline-directed protein kinases (CDK2, CDK5, GSK3α, GSK3β, MAPK, and SAPKs) and non-proline-directed protein kinases (CaMKII, Casein kinases 1, 1δ and 2, DYRK, MARK, the phosphorylase kinase, PKA, PKB/AKT, PKC, PKN and Tautubulin kinases 1 and 2) are known to phosphorylate tau 27–56. Also, protein phosphatase (PP) 1, PP2A, PP2B and PP5 are reported to dephosphorylate tau 57. However, what pathological events lead to dysregulation of these kinases and phosphatases and induce abnormal phosphorylation and toxicity of tau in AD still remains largely unknown.
Accumulation of DNA damage and activation of DNA repair have been observed in the brains of AD patients and animal models of AD 58–66. In our earlier work, we have reported that DNA damage-activated Chk1 and Chk2 phosphorylate tau at an AD-related site Ser262 in the microtubule-binding domain, whose phosphorylation is known to regulate tau binding to microtubules 25. Using a Drosophila model of human tau toxicity, we also showed that overexpression of Chk2 enhances tau-induced neurodegeneration, and tau phosphorylation at Ser262 plays an important role in this enhancement of tau toxicity 25. These observations suggest that aberrant activation of Chk1 and Chk2 may play a role in abnormal phosphorylation and toxicity of tau in AD pathogenesis. In addition to Ser262, there are a number of potential phosphorylation sites that are associated with AD and toxicity in tau. Whether Chk1 and/or Chk2 can phosphorylate tau at other AD-related sites has not been determined yet.
In this study, we systematically identified the Ser/Thr sites in tau that are directly phosphorylated by Chk1 and Chk2 in vitro. Western blot with phospho-tau specific antibodies and mass spectrometry (MS) analysis covered 98.5% of potential phosphorylation sites in tau-383 isoform (0N4R) and revealed that Chk1 and Chk2 phosphorylate tau at multiple AD-related sites. Interestingly, Chk1 and Chk2 phosphorylate many residues that are located within the microtubule-binding domain and C-terminal domain, whose phosphorylation has been reported to reduce tau binding to microtubules. These results suggest that Chk1 and Chk2 may contribute to abnormal phosphorylation and toxicity of tau in AD pathogenesis.
Methods
Phosphorylation of Tau by Chk1 and Chk2
Recombinant active human GST-tagged Chk1 (C0870, Sigma, St. Louis, MO, activity 169–229 nmol/min·mg) and Chk2 (C0995, Sigma, St. Louis, MO, activity 654–884 nmol/min·mg) were diluted to 1:2 and 1:5 respectively, mixed with 2 µg of recombinant human tau 0N4R (variant 3, NM_016834.3, T9825, Sigma, St. Louis, MO) in the reaction buffer containing 5 mM MOPS, pH 7.2, 2.5 mM glycerol 2-phosphate, 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, 0.05 mM DTT and 5µM ATP, and incubated at 30 °C for 3 h.
Western blot by Phospho-specific Antibodies
Recombinant human tau were incubated with recombinant active GST-tagged Chk1 (C0870, Sigma, St. Louis, MO) or Chk2 (C0995, Sigma, St. Louis, MO) with or without inhibitors (UCN-01 (Sigma, St. Louis, MO) or Chk2 inhibitor II (Sigma, St. Louis, MO)) as described above. Samples were mixed with SDS-Tris-Glycine sample buffer, separated by 10% Tris-Glycine gel and transferred to nitrocellulose membrane. The membranes were blocked with 5% milk (Nestle), blotted with antibodies described below, incubated with a secondary antibody (anti-mouse IgG, HRP-linked or anti-rabbit IgG, HRP-linked (GE Healthcare, Piscataway, NJ)) and developed using ECL plus Western Blot Detection Reagents (GE Healthcare, Piscataway, NJ). Phospho-Thr205, phospho-Ser214, phospho-Ser356, and phospho-Ser409 were purchased from Biosource (Life Technologies, Grand Island, NY). Anti-tau pSer320 (HIA3) was prepared in-house 53. All the experiments were carried out more than three times.
Chemicals and Reagents for Liquid Chromatography and Mass Spectrometry
Ammonium bicarbonate (NH4HCO3), iodoacetamide (IAM), calcium chloride (CaCl2) and formic acid were purchased from Sigma-Aldrich (St. Louis, MO). Trifluoroacetic acid (TFA) and tris(2-carboxyethyl)-phosphine (TCEP) were obtained from Pierce (Thermo Fisher Scientific, Waltham, MA). Porcine trypsin and Asp-N were acquired from Promega (Madison, WI) and Roche Applied Science (Indianapolis, IN), respectively. HPLC-grade water and acetonitrile (ACN) were from Thermo Fisher Scientific (Waltham, MA).
Reduction, Alkylation and Enzyme Digestion
The pH of tau protein samples with or without kinases were adjusted to pH 8.5 with 100 mM NH4HCO3. Proteins were reduced with 5 mM TCEP at 37 °C for 20 min and alkylated with 10 mM iodoacetamide for 30 min in the dark at room temperature. Two endopeptidases, trypsin and Asp-N were used to digest tau. In tryptic digestion, porcine trypsin was added at an enzyme to substrate ratio of 1:50 in the presence of 1 mM CaCl2. The sample mixture was incubated overnight at 37 °C in the dark and the digestion was quenched by adding trifluoroacetic acid (10%) to achieve a pH of 2–4. Samples were desalted with ZipTip C18 (Millipore, Billerica, MA) loaded with POROS R2 beads (Applied Biosystems, Foster City, CA) and eluted with 0.1% TFA in 50:50 ACN:H2O. The eluate was dried down and residue was reconstituted with 0.1% formic acid in 2:98 ACN:H2O for LC-MS3 analysis. The same procedures were followed when samples were digested with the endoproteinase Asp-N except CaCl2 was not added to the proteolysis.
Liquid Chromatography and Mass Spectrometry
A NanoAcquity UPLC system (Waters, Milford, MA) interfaced to an LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) equipped with a nanospray ionization source was employed for LC/MS3 analyses. Reversed-phase LC was performed on a Waters BEH130 C18 column (100 µm × 100 mm, 1.7 µm particle size). Samples were trapped and washed in a Waters Symmetry® C18 trap column (180 µm × 100 mm, 5 µm particle size) prior to separation in the capillary column. Gradient elution with 0.1% formic acid in water as solvent A and in ACN as solvent B, with solvent B raised from 1 to 50% in 30 minutes, then 50 to 85% in the next 10 min, was carried out. A flow rate of 500 nL/min was used.
The mass spectrometer was operated in positive mode with spray voltage at 2.1 kV, ion transfer tube voltage at 49 V and ion transfer tube temperature at 170 °C. No sheath and auxiliary gases were used. Ion signal threshold of 1,000 was used for MS/MS. A normalized collision energy of 35%, an activation of q = 0.25 and activation time of 30 ms were applied in MS/MS acquisitions. Data-dependent acquisition with automatic switching between MS and MS/MS modes was employed. A full scan was acquired at a target value of 1 × 106 ions with resolution (R) of 60,000 at m/z 400. The lock mass option, using the polydimethylcyclosiloxane ion (PCM; protonated (Si(CH3)2O))6) at m/z 445.120025, was enabled for the MS scan for accurate mass measurement. The top eight most intense ions were selected for fragmentation in the LTQ. Collision-induced dissociation (CID) at a target value of 10,000 ions was used for fragmentation. The following dynamic exclusion settings were applied to precursor ions chosen for MS/MS analysis: repeat count – 1; repeat duration – 30 s and exclusion duration – 120 s. A neutral loss experiment where data-dependent settings were chosen to trigger an MS3 scan when a neutral loss of 97.97, 48.99 or 32.66 m/z units (relative to the singly, doubly, or triply charged phosphorylated precursor ion, respectively), was detected among the 8 most intense product ions was performed to improve fragmentation of phosphopeptides. To ensure reliable mass spectrometric identification of phosphorylation sites, all of the experiments were repeated twice including phosphorylation reaction and mass spectrometric analysis.
Data Analysis
MS, MS/MS and MS3 spectra were searched against the human component of the NCBI non-redundant database (11/01/2010 version; 113,484 entries) using Sequest (Ver.27, Rev. 11) and Mascot (Ver. 2.3.01) algorithms. Searches were performed with full tryptic specificity (2 missed cleavages); carbamidomethylated cysteine residues (+57.0340 Da) as static modification and oxidized methionine, histidine and tryptophan (+15.9949 Da), deamidated asparagine and glutamine (+0.9840 Da), phosphorylated serine, threonine and tyrosine (+79.9663 Da) and dehydroalanine and dehydroaminobutyric acid (−18.0106 Da) as differential modifications. Precursor mass error tolerance of 10 ppm and default product ion mass error tolerance of above searching algorithms were used. Manual inspection of the tandem mass spectra and product ion lists was also conducted. Phosphorylation site assignment from database search was further evaluated with A-scores using Scaffold PTM (Proteome Software).67 The residue numbers used here correspond to the longest isoform of tau, 2N4R, which is commonly used to describe tau phosphorylation sites.
Results and Discussion
Phosphorylation of Tau at Major AD-related non-SP/TP Sites by Chk1 and Chk2 Revealed by Western Blot using Phospho-tau-specific Antibodies
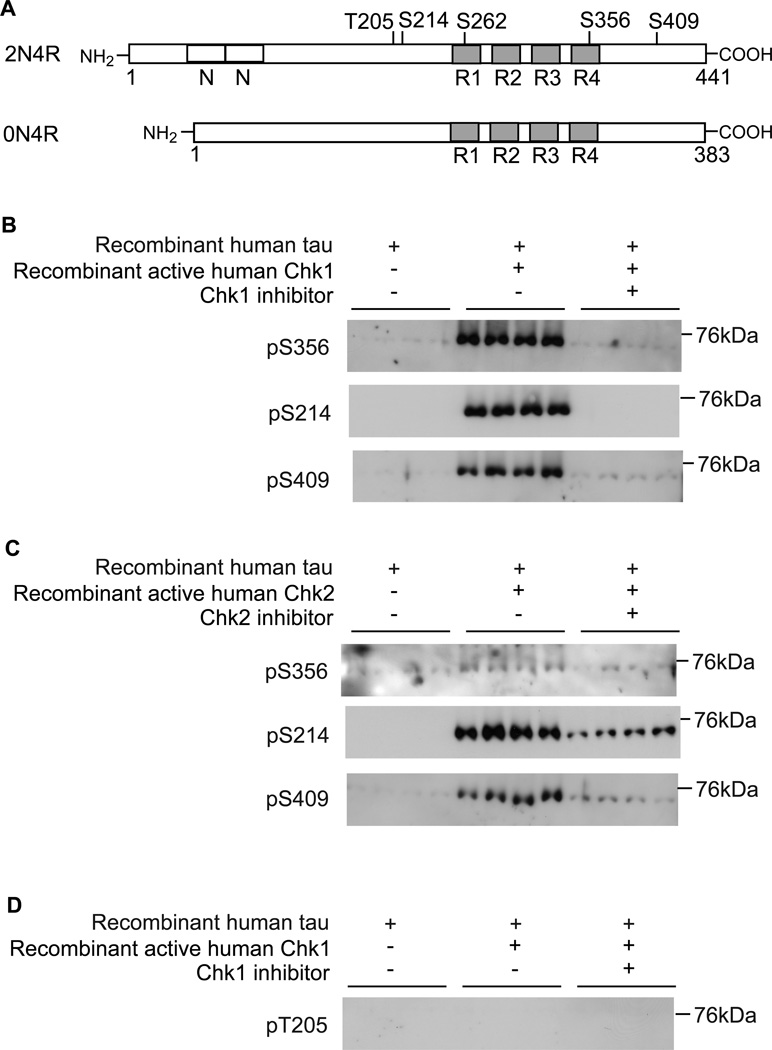
Well-characterized phospho-tau-specific antibodies are available for the sites in tau dominantly phosphorylated in AD brains. We utilized these antibodies to test whether Chk1 and Chk2 directly phosphorylate tau at AD-related sites. Recombinant wild-type human 0N4R tau, which has four microtubule-binding repeats (R) and no N-terminal insert (N) (Figure 1A), was phosphorylated by human Chk1 and Chk2 in vitro individually, and subjected to Western blot. Using this strategy, we have previously shown that Chk1 and Chk2 phosphorylate tau at an AD-related site Ser262 25, which is located in the KXGS motif in the first repeat region. Each microtubule-binding repeat region contains a KXGS motif, and phosphorylation at Ser356 in the fourth repeat region is also associated with AD. Using phospho-specific antibody for this site, we found that incubation of tau with recombinant active Chk1 substantially increased tau phosphorylation at Ser356. This phosphorylation was inhibited by Chk1-specific inhibitor (UCN-01) (Figure 1B). We carried out a similar experiment for Chk2. In contrast to Chk1, phosphorylation of tau at Ser356 by Chk2 was not detected clearly (Figure 1C).
Figure 1.
Phosphorylation of tau at AD-related non-SP/TP sites by Chk1 and Chk2. (A) Schematic representation of tau. Microtubule-binding repeats (R1-R4, gray boxes), N-terminal inserts (N, open boxes), and locations of Thr205, Ser214, Ser262, Ser356, and Ser409 are shown. (B) Chk1 directly phosphorylates tau at Ser356, Ser214 and Ser409. Recombinant human tau was incubated with recombinant active human Chk1 in the presence or absence of Chk1 inhibitor (UCN-01) and subjected to Western blot using anti-pSer356 tau, anti-pSer214 tau, and anti-pSer409 tau. (C) Chk2 phosphorylates tau at Ser214 and Ser409. Tau was incubated with recombinant active human Chk2 in the presence or absence of Chk2 inhibitor (Chk2 inhibitor II) and subjected to Western blot using anti-pSer356 tau, anti-pSer214 tau, and anti-pSer409 tau. Chk2 inhibitor diminished tau phosphorylation at these sites. (D) Chk1 does not phosphorylate tau at a proline-directed kinase target site Thr205. Tau was incubated with Chk1 in the presence or absence of inhibitors (UCN-01) and subjected to Western blot using anti-pThr205 tau.
In AD brains, tau is hyperphosphorylated at both proline-directed (SP/TP) sites and non-SP/TP sites. Since both Ser262 and Ser356 are non-SP/TP sites, and previous studies showed that Chk1 and Chk2 are non-SP/TP kinases 21–24, we further tested whether Chk1 and Chk2 can phosphorylate tau at major AD-related non-SP/TP sites, such as Ser214 and Ser409. Using phospho-tau specific antibodies, we found that Chk1 and Chk2 phosphorylated tau at these sites (Figure 1B and 1C). We also tested whether Chk1 can phosphorylate tau at an SP/TP site. As expected, Chk1 did not phosphorylate tau at Thr205, one of the AD-related SP/TP sites (Figure 1D). Taken together, these results demonstrate that Chk1 and Chk2 can phosphorylate tau at multiple major AD-related non-SP/TP sites.
Global Identification of Phosphorylation Sites of Tau Catalyzed by Chk1 and Chk2 Using LC-MS3
To systematically identify which sites in tau are directly phosphorylated by Chk1 and Chk2, we carried out liquid chromatography-tandem mass spectrometry (LC-MS3) analysis. Analysis of the tryptic digest from control tau yielded 33 peptides corresponding to 75% amino acid sequence coverage while the digest from Asp-N resulted to 22 peptides representing 41% amino acid sequence coverage. Combining the results from the two enzymatic digestions, the overall coverage for the control was found to be 90% (Figure 2A). A similar overall percent coverage was obtained from analysis of the longest isoform of tau.27
Figure 2.
Sequence coverage of (A) control, (B) Chk1-treated and (C) Chk2-treated tau proteins. Amino acid positions refer to 0N4R isoform. Peptides detected after separate digestion with trypsin and Asp-N are shown; those with solid underline are tryptic peptides while those with dashed underline are from Asp-N digestion. Serine and threonine residues in bold and italic are the phosphorylated sites detected. Amino acids in bold and lowercase are also found to be modified (i.e. oxidized (m,h), dehydrated (s, t), carbamidomethylated (c), deamidated (q, n)).
LC-MS3 analysis of tryptic digest of Chk1-treated tau identified 42 unique peptides that correspond to 76% amino acid sequence coverage of tau. Analysis of the Asp-N digest showed 24 unique peptides that correspond to 41% sequence coverage. An overall 90% amino acid sequence coverage was achieved when peptides from both trypsin and Asp-N were taken together (Figure 2B). Search results yielded 19 phosphorylation sites for the Chk1-treated sample (Table 1).
Table 1.
Sites on tau protein phosphorylated by Chk1 and Chk2 and identified by mass spectrometry. Thirteen novel phosphorylation sites are identified (bold and italicized). Residue numbers correspond to the longest isoform of tau, 2N4R.
| Phosphorylation Site |
Identified Peptide | Molecular Mass | Peptide ID Score | Ascore/Localization Probability | ||||
|---|---|---|---|---|---|---|---|---|
| Control | Chk1 | Chk2 | Control | Chk1 | Chk2 | |||
| T17 | DHAGTYGLGa | 889.3934 | 889.3922 | 969.3582 | 1.36* | 1.67* | 1.83* | 14/100 |
| T30 | DQGGYTMHQ | 1035.4073 | 1035.4065 | 1115.3727 | 45/1.4×10−3b | 39/6.5×10−3 | 40/3.6×10−3 | 29/100 |
| T123 | DEAAGHVTQARMVSKSK | 1813.9092 | 1893.8737 | 1813.9088 | 48/2.0×10−3 | 38/9.4×10−3 | 46/1.2×10−4 | 33/100 |
| T149 | GADGKTKIATPR | 1213.6782 | 1293.6452 | NDc | 2.99d | 2.39* | - | 50/100 |
| T169 | GQANATRIPAK | ND | 1205.5909 | ND | - | 2.39* | - | 1000/100 |
| S191 | TPPSSGEPPKSGDR | 1410.6750 | 1490.6391 | 1410.6728 | 3.25 | 2.52 | 3.27 | 21/93 |
| S208 | SGYSSPGSPGTPGSR | 1392.6265 | 1472.5971 | 1392.6283 | 4.25 | 3.39 | 4.02 | 26/99 |
| S214 | TPSLPTPPTREPK | 1419.7772 | 1499.7427 | 1499.7433 | 3.16 | 2.77 | 2.78 | 14/92; 20/98 |
| T245 | SRLQTAPVPMPDLK | 1551.8483 | 1631.8171 | 1631.8121 | 3.71 | 3.22 | 3.34 | 65/100; 65/100 |
| S258 | NVKSKIGSTENLK | 1416.7936 | 1576.7263 | 1416.7936 | ** | ** | ** | ** |
| S262 | IGSTENLKHQPGGGK | 1521.7895 | 1601.7592 | 1521.7956 | 44/1.8×10−3 | 42 (>45)*/1.2×10−3 | 42/5.5×10−3 | 10/99 |
| S285 | LDLSNVQSK | 1002.5345 | 1002.5341 | 1082.4988 | 3.04 | 2.97 | 2.73 | 38/100 |
| S289 | KLDLSNVQSK | 1130.6273 | 1210.5930 | 1210.5937 | 3.56 | 2.90 | 3.10 | 80/100; 80/100 |
| S293 | DLSNVQSKcGSKe | 1321.6295 | 1401.5947 | 1321.6275 | 47/1.4×10−3 | 53/9.6×10−5 | 43/3.1×10−3 | 12/100 |
| S305 | HVPGGGSVQIVYKPVDLSK | 1979.0915 | 2059.0540 | 1979.0912 | 3.57 | 3.58 | 3.89 | 64/100 |
| S324 | cGSLGNIHHKPGGGQVEVK | 1972.9821 | 2052.9453** | 1972.9821 | ** | ** | ** | ** |
| S352 | DRVQSKIGSL | 1101.6136 | 1181.5793 | 1181.5793 | 59/1.1×10−5 | 36/9.8×10−3 | 37/8.2×10−3 | 85/100; 108/100 |
| S356 | IGSLDNITHVPGGGNK | 1577.8178 | 1657.7840 | 1577.8183 | 73/1.8×10−6 | 74/1.1×10−6 | 30/8.3×10−3 | 49/100 |
| T377 | KIETHKLTFR | ND | 1351.6892 | 1351.6996 | - | 2.02* | 2.01* | 27/100; 49/100 |
| T386 | AKTDHGAEIVYK | 1330.6908 | 1410.6472 | 1410.6509 | 3.76 | 3.36 | 3.03 | 105/100; 112/100 |
| S400 | SPVVSGDTSPR | 1100.5473 | 1180.5152 | ND | 2.72 | 3.45 | - | 35/100 |
| T403 | SPVVSGDTSPR | 1100.5473 | 1100.5466 | 1180.5121 | 2.72 | 3.18 | 3.20 | 17/96 |
| T414 | DTSPRHLSNVSSTGSI | 1656.8058 | 1736.7672** | 1656.8058 | 3.81 | ** | 3.71 | ** |
| S433 | DEVSASLAKQGL | 1216.6292 | 1216.6281 | 1296.5943 | 3.91 | 3.21 | 2.94 | 18/100 |
| S435 | DEVSASLAKQGL | 1216.6292 | 1216.6281 | 1296.5954 | 3.91 | 3.21 | 2.71 | 22/100 |
Amino acid in boldface corresponds to phosphorylated residue
Ion score/expectation value from Mascot
ND: Phosphorylated or non-phosphorylated form not detected
XCorr value from Sequest
Amino acid in lower case corresponds to a modified residue
Identified with less stringent scores (For Mascot scores, number enclosed in parenthesis is the ion score needed to establish identity)
Phosphorylation was detected in MS/MS and MS3 scans after manual inspection.
Proteolysis of the Chk2-treated tau sample generated 37 tryptic peptides which make up 77% of the amino acid sequence. The use of Asp-N as the proteolytic enzyme yielded 19 unique tau peptides corresponding to 38% sequence coverage. Overall, 88% of the amino acid sequence was covered by both trypsin and Asp-N (Figure 2C). Twelve sites were found to be phosphorylated by Chk2 (Table 1). These 12 sites include 6 of Chk1-target sites leaving the 13 phosphosites unique to Chk1..
LC-MS3 analyses did not detect phosphorylation of tau at Ser262 by Chk2 or phosphorylation of tau at Ser409 by either Chk1 or Chk2. In contrast, phosphorylation at these sites were detected by Western blot using the phospho-specific antibodies (Figure 1 and previous publication 25), suggesting that phospho-specific antibodies are more sensitive to detect tau phosphorylation than LC-MS3. For example, our previous data indicated that tau phosphorylation at Ser262 by Chk1 was much efficient than that by Chk2 25. In LC-MS3 analysis, we detected phosphorylated form of Ser262 from in vitro Chk1 phosphorylation reactions but not from in vitro Chk2 phosphorylation reactions. These results suggest that tau phosphorylation at Ser262 by Chk2 is below the detection limit of LC-MS3. As for Ser409, in LC-MS3 analysis, we detected non-phosphorylated Ser409 (in the peptide where Thr414 phosphorylation was identified) in both in vitro phosphorylation reactions catalyzed by Chk1 and Chk2. In general, due to the negative charge introduced by phosphorylation, the ionization efficiency of peptides is greatly reduced in positive ion mass spectrometry analysis, which resulted to lower sensitivity of detecting phosphorylated peptides compared to the corresponding non-phosphorylated peptides. Therefore, some less efficient phosphorylations at certain sites in our study may not be identified by the LC-MS3 analysis.
Sequest and Mascot searches plus manual MS/MS and MS3 spectra analysis and interpretation yielded a total of 25 phosphorylation sites. Among these, 12 and 6 are uniquely phosphorylated by Chk1 and Chk2, respectively. There are 6 sites that were phosphorylated by both kinases, namely Ser214, Thr245, Ser289, Ser352, Thr377, and Thr386. As summarized in Table 1, molecular masses of the phosphopeptides differ by a mass of 80 daltons (Da) from the corresponding nonphosphopeptides. Evaluation of phosphorylation sites assigned from database search using Scaffold PTM resulted with A-scores for all the sites ≥ 10. Manual inspection of the MS/MS spectra and product ion lists was conducted to confirm for the phosphorylated sites identified.
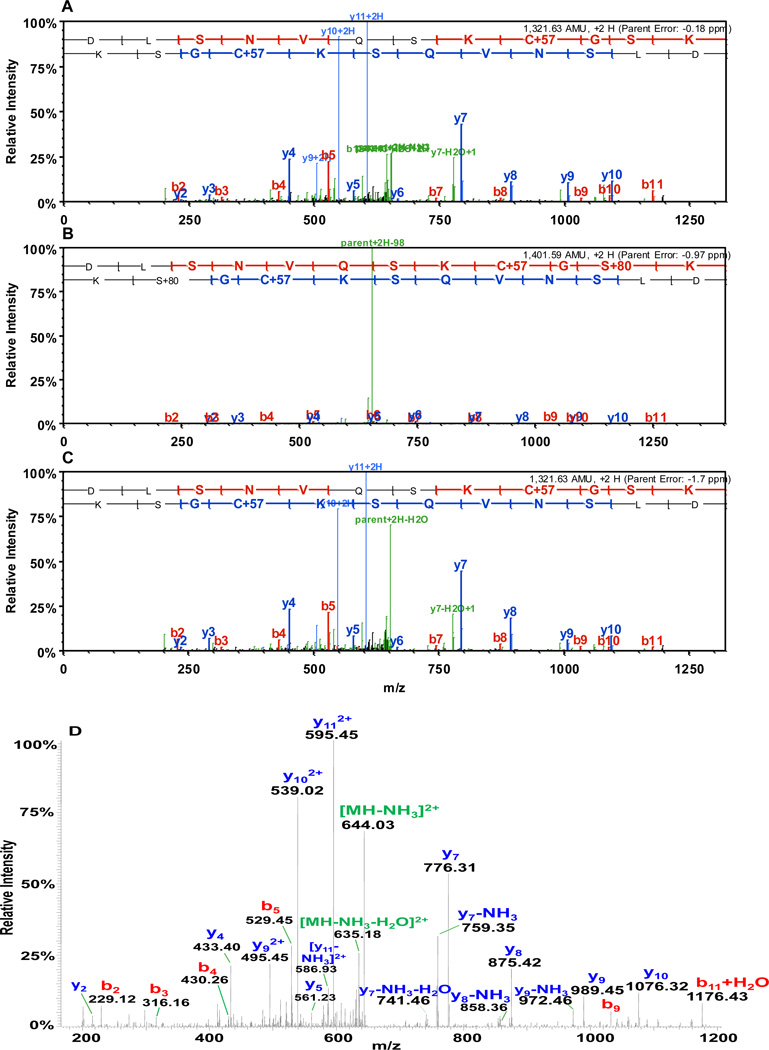
Representative MS/MS and MS3 spectra of a tau peptide phosphorylated by Chk1 only is shown in Figure 3. Detection of the corresponding nonphosphopeptide in the Chk2-treated sample suggests that phosphorylation at this particular site, Ser293, is unique to Chk1. The same is true for peptides phosphorylated by Chk2 only. MS/MS spectra of the other novel phosphopeptides identified are provided as supplementary data (Figure S1–S12).
Figure 3.
MS/MS spectra of a tau peptide from (A) control, (B) Chk1-treated, (C) Chk2-treated samples and (D) MS3 spectrum showing that Ser293 was phosphorylated by Chk1 only. The detection of the corresponding nonphosphopeptide in the sample treated with Chk2 suggests that phosphorylation at residue Ser293 is unique to the Chk1-treated sample.
Overlapping and Distinct Tau Phosphorylation Sites by Chk1 and Chk2
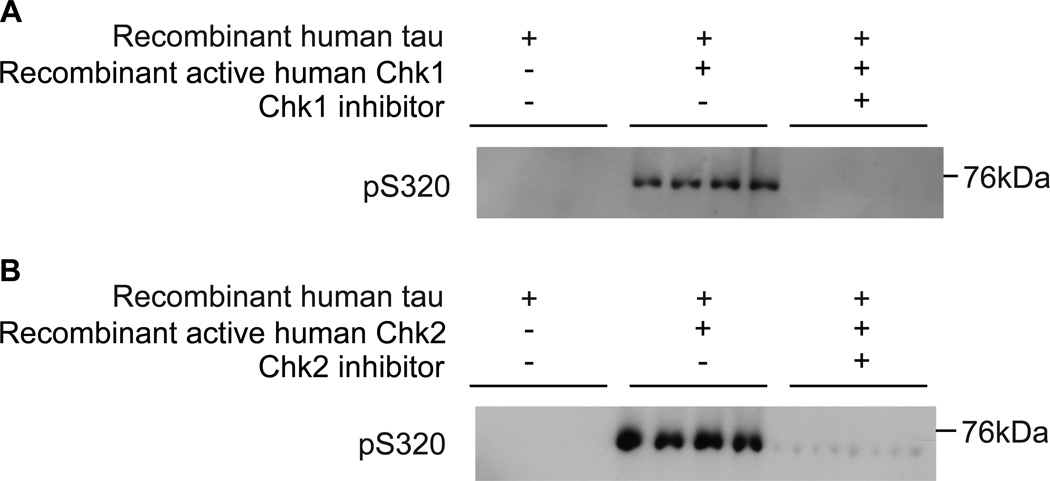
There are a total of 66 serine and threonine (40 Ser and 26 Thr) residues in 0N4R tau. Ninety seven percent (97%) of serine and threonine residues were covered in this LC-MS3 analysis, which left 1 Ser (Ser320) and 1 Thr (Thr319) not detected. With no miscleavage, the resulting tryptic peptide (VTSK) containing these two residues was too small to be detected with MS considering practical mass range settings. These residues were also not detected with Asp-N digestion possibly because the peptide was too big (31 amino acid residues). Between these two sites, the sequence surrounding Ser320 matches the published consensus for Chk2, K-X-X-S/T 21–23, and phospho-specific antibody for Ser320 is available 53. Western blot analysis using anti-phospho Ser320 tau antibody revealed that Chk1 and Chk2 phosphorylate tau at Ser320 (Figure 4).
Figure 4.
Phosphorylation of tau at Ser320 by Chk1 and Chk2. (A) Chk1 phosphorylates tau at Ser320. Recombinant human tau was incubated with recombinant active human Chk1 in the presence or absence of inhibitors (UCN-01) and subjected to Western blot using anti-pSer320 tau. (B) Chk2 phosphorylates tau at Ser320. Tau was incubated with recombinant active human Chk2 in the presence or absence of inhibitors (Chk2 inhibitor II) and subjected to Western blot using anti-pSer320 tau.
Taken together, our systematic analyses covered 98.5% of potential phosphorylation sites and identified 12 Chk1-target sites, 6 Chk2-target sites, and 9 sites that are phosphorylated by both Chk1 and Chk2 in tau (Table 2).
Table 2.
Sites on tau protein phosphorylated by Chk1 and Chk2 identified by mass spectrometry and Western blotting. Phosphorylated residues in AD brain are indicated. Other kinases that were reported to phosphorylate particular sites are also listed. Sites phosphorylated by both Chk1 and Chk2 are in boldface.
| Phosphorylation Site |
Method of Detection | Phosphorylated in AD brain? |
Other kinases that phosphorylate site | |
|---|---|---|---|---|
| Chk1 | Chk2 | |||
| T17 | LC-MS | CK1δ | ||
| T30 | LC-MS | |||
| T123 | LC-MS | |||
| T149 | LC-MS | CK1δ, GSK-3β | ||
| T169 | LC-MS | CK1δ | ||
| S191 | LC-MS | Yes | ||
| S208 | LC-MS | Yes | CK1δ, tau tubulin kinase1, tau tubulin kinase 2 |
|
| S214 | LC-MS, Western blot |
LC-MS, Western blot |
Yes | AKT, CDK2, CDK5, CK1δ, PKA |
| T245 | LC-MS | LC-MS | GSK-3β, p38, PKA | |
| S258 | LC-MS | Yes | CK1δ, GSK-3β, PKA, PKC, PKN | |
| S262 | LC-MS, Western blot25 |
Western blot25 | Yes | CaMKII, CK1δ, GSK-3α, GSK- 3β, MARK, phosphorylase kinase, PKA |
| S285 | LC-MS | CK1δ, GSK-3β, phosphorylase kinase |
||
| S289 | LC-MS | LC-MS | Yes | CK1δ, GSK-3β |
| S293 | LC-MS | MARK, PKC | ||
| S305 | LC-MS | CK1δ, GSK-3β, MARK, p38, phosphorylase kinase, PKA, PKC |
||
| S320 | Western blot | Western blot | MARK, PKN | |
| S324 | LC-MS | GSK-3α, GSK-3β, MARK, PKA, PKC, PKN |
||
| S352 | LC-MS | LC-MS | CK1δ, GSK-3β, phosphorylase kinase, PKA, PKC, PKN |
|
| S356 | LC-MS, Western blot |
Yes | CaMKII, CK1δ, GSK-3α, GSK- 3β, JNK, MARK, p38, PKA |
|
| T377 | LC-MS | LC-MS | ||
| T386 | LC-MS | LC-MS | CK2, CK1δ | |
| S400 | LC-MS | Yes | CK2, GSK-3β | |
| T403 | LC-MS | Yes | GSK-3β | |
| S409 | Western blot | Western blot | Yes | GSK-3β, PKA |
| T414 | LC-MS | Yes | CK1δ, CK2, GSK-3β, PKA | |
| S433 | LC-MS | Yes | CK1δ | |
| S435 | LC-MS | Yes | CK1δ, PKA | |
Consensus Sequences for Chk1 and Chk2 in Tau
Tables 3 and 4 show the amino acid sequence at the phosphorylation sites for Chk1 and Chk2, respectively. Peptide sequences were centered on each phosphorylation site and extended to 15 amino acids in length to determine the occurrence of a phosphorylation motif or substrate consensus type. None of the Chk1- or Chk2- target sites were followed by proline, indicating that neither Chk1 nor Chk2 phosphorylate tau at proline-directed kinase target sites. A commonly observed consensus sequence for tau peptides phosphorylated by Chk1, in this study, is R/K-X-X-S/T. This is in agreement with the findings of O’Neill and co-workers who have utilized peptide library analyses to determine optimal substrate motifs for Chk1 and Chk2. 21 Ten out of 21 phosphopeptides follow this published substrate consensus type (corresponding to phosphosites Ser214, Thr245, Ser262, Ser293, Ser320, Ser324, Ser352, Ser356, Thr386, and Ser409). Of these 10, Ser214 also has R in position −5 and L in position +1 (both residues favor Chk1 phosphorylation), Ser320 has L in position −5 (also Chk’s favored amino acid),21 Thr245 has A in position +1, Ser356 has R in position −7 and L in position +1 and Thr386 also has R in position −7. These amino acid residues are shown in boldface in Table 3. Four other peptides have leucine/hydrophobic amino acid/arginine in −5 position relative to the phosphorylation site (Thr123, Ser208, Ser258, Ser289, Ser305). Another phosphopeptide has phenylalanine at the +1 position (Thr377) where glutamine, methionine and other hydrophobic amino acids yield favorable preference values according to the peptide library analyses. No specific sequence pattern was observed for the remaining 5 phosphopeptides (Thr149, Thr169, Ser191, Ser400 and Thr414) and the amino acid residues at certain positions did not comply either with the results derived from the peptide library study.
Table 3.
Amino acid sequence at the phosphorylation site for Chk1. Residues in italic tested positive with Western blot.
| Phosphorylation Site | Amino Acid Residue | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −7 | −6 | −5 | −4 | −3 | −2 | −1 | Site | +1 | +2 | +3 | +4 | +5 | +6 | +7 | |
| T123 | D | E | A | A | G | H | V | T | Q | A | R | M | V | S | K |
| T149 | A | K | G | A | D | G | K | T | K | I | A | T | P | R | G |
| T169 | Q | K | G | Q | A | N | A | T | R | I | P | A | K | T | P |
| S191 | S | S | G | E | P | P | K | S | G | D | R | S | G | Y | S |
| S208 | G | S | P | G | T | P | G | S | R | S | R | T | P | S | L |
| S214 | G | S | R | S | R | T | P | S | L | P | T | P | P | T | R |
| T245 | S | A | K | S | R | L | Q | T | A | P | V | P | M | P | D |
| S258 | P | D | L | K | N | V | K | S | K | I | G | S | T | E | N |
| S262 | N | V | K | S | K | I | G | S | T | E | N | L | K | H | Q |
| S289 | L | D | L | S | N | V | Q | S | K | C | G | S | K | D | N |
| S293 | N | V | Q | S | K | C | G | S | K | D | N | I | K | H | V |
| S305 | K | H | V | P | G | G | G | S | V | Q | I | V | Y | K | P |
| S320 | V | D | L | S | K | V | T | S | K | C | G | S | L | G | N |
| S324 | K | V | T | S | K | C | G | S | L | G | N | I | H | H | K |
| S352 | D | F | K | D | R | V | Q | S | K | I | G | S | L | D | S |
| S356 | R | V | Q | S | K | I | G | S | L | D | N | I | T | H | V |
| T377 | K | I | E | T | H | K | L | T | F | R | E | N | A | K | A |
| T386 | R | E | N | A | K | A | K | T | D | H | G | A | E | I | V |
| S400 | V | Y | K | S | P | V | V | S | G | D | T | S | P | R | H |
| S409 | D | T | S | P | R | H | L | S | N | V | S | S | T | G | S |
| T414 | H | L | S | N | V | S | S | T | G | S | I | D | M | V | D |
Amino acids shaded in gray are not included in the phosphopeptide sequence detected.
Residues that favor Chk1 phosphorylation at −3, −5, −7 and +1 positions relative to phosphorylation site, according to peptide library analyses, are shown in boldface.
Table 4.
Amino acid sequence at the phosphorylation site for Chk2. Residues in italic tested positive with Western blot.
| Phosphorylation Site | Amino Acid Residue | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −7 | −6 | −5 | −4 | −3 | −2 | −1 | Site | +1 | +2 | +3 | +4 | +5 | +6 | +7 | |
| T17 | V | M | E | D | H | A | G | T | Y | G | L | G | D | R | K |
| T30 | R | K | D | Q | G | G | Y | T | M | H | Q | D | Q | E | G |
| S214 | G | S | R | S | R | T | P | S | L | P | T | P | P | T | R |
| T245 | S | A | K | S | R | L | Q | T | A | P | V | P | M | P | D |
| S262 | N | V | K | S | K | I | G | S | T | E | N | L | K | H | Q |
| S285 | I | N | K | K | L | D | L | S | N | V | Q | S | K | C | G |
| S289 | L | D | L | S | N | V | Q | S | K | C | G | S | K | D | N |
| S320 | V | D | L | S | K | V | T | S | K | C | G | S | L | G | N |
| S352 | D | F | K | D | R | V | Q | S | K | I | G | S | L | D | S |
| T377 | K | I | E | T | H | K | L | T | F | R | E | N | A | K | A |
| T386 | R | E | N | A | K | A | K | T | D | H | G | A | E | I | V |
| T403 | S | P | V | V | S | G | D | T | S | P | R | H | L | S | N |
| S409 | D | T | S | P | R | H | L | S | N | V | S | S | T | G | S |
| S433 | A | T | L | A | D | E | V | S | A | S | L | A | K | Q | G |
| S435 | L | A | D | E | V | S | A | S | L | A | K | Q | G | L | |
Amino acids shaded in gray are not included in the phosphopeptide sequence detected.
Residues that favor Chk2 phosphorylation at −3 to −7, +1 and +2 positions relative to phosphorylation site, according to peptide library analyses, are in boldface.
In the case of peptides phosphorylated by Chk2, all showed at least one amino acid residue conforming to the sequence motif predicted by the peptide library analyses. Three phosphopeptides showed the published consensus sequence for Chk2, K-X-X-S/T, (Ser262, Ser320, and Thr386) while Ser214, Thr245, Ser352 and Ser409 showed the R-X-X-S/T motif. The phosphopeptide containing Ser320 also has leucine at position −5 and that of Thr386 has R at position −7, both residues favoring phosphorylation by Chk2. Among the peptides that have the R-X-X-S/T motif, the one with Thr245 also has a hydrophobic amino acid, alanine, at position −6 while that of Ser352 has isoleucine at position +2. Another phosphopeptide has lysine at position −4 (Ser285) while two have leucine at the −5 position (Ser289, Ser433). All amino acids that exhibited high preference values for Chk2 phosphorylation in the peptide library analyses at various positions are indicated in boldface in Table 4. 21 Four other phosphopeptides have hydrophobic amino acids at position −6 (Thr17, Thr377, Thr403, Ser435). The phosphopeptide containing Thr377 also showed residues favorable for Chk2 phosphorylation at positions +1 (F) and +2 (R). The remaining peptide has R at position −7 (Thr30). Considering the 9 phosphopeptides common to both kinases, seven (Ser214, Thr245, Ser262, Ser320, Ser352, Thr386 and Ser409) conform to the published substrate consensus type, R/K-X-X-S/T while the other two (Ser289 and Thr377) have amino acid residues at certain positions that help facilitate phosphorylation by the checkpoint kinases.
Figure 5 shows the sequence logos of the amino acid residues adjacent to the phosphorylation sites for Chk1 and Chk2, which were generated using Weblogo (Ver. 2.8.2) 68. Using the height of the amino acid residues shown on the sequence logos as a measure of their relative frequency at a particular position, preferred substrate composition for Chk1 and Chk2 are suggested. For Chk1, the preferred substrate would consist of −7(Xxx) −6(Xxx) −5(Xxx) −4(Ser) −3(Arg/Lys) −2(hydrophobic) −1(Gly/Lys) Ser/Thr +1(Lys) +2(Xxx) +3(Asn/hydrophilic) +4(Ser) +5(Lys/hydrophobic) +6(Arg/Lys/His) +7(Xxx). The preferred substrate of Chk2 would consist of −7(Xxx) −6(Xxx) −5(Lys/Leu) −4(Ser) −3(Arg/Lys) −2(hydrophobic) −1(hydrophobic) Ser/Thr +1(Xxx) +2(hydrophobic) +3(Gly) +4(Ser) +5(Lys/Leu) +6(Xxx) +7(Asn). Xxx indicates no strong preference for an amino acid.
Figure 5.
Sequence logos of amino acid residues adjacent to the phosphorylation sites for (A) Chk1 and (B) Chk2. The height of each amino acid residue indicates its relative frequency at a particular position, in this case, centered on the phosphorylation site. The amino acids are grouped and colored according to their charges, hydrophobicity and polarity. Arginine and lysine at position −3 prove to be the most frequently occurring amino acid residues at this location for both Chk1 and Chk2.
The phosphorylation sites detected for each of the microtubule-binding repeat of tau are shown in Figure 6. Serines in the SKXGS motif were found to be phosphorylated in all repeats. Our data indicated that the first serine residue could be phosphorylated by both Chk1 and Chk2 except for S258. Phosphorylation of the second serine residue prefers Chk1 more.
Figure 6.
Phosphorylation within the microtubule-binding repeat region of tau by Chk1 or Chk2. Phosphorylation sites in the microtubule-binding repeat region are indicated in boldface type. The first Ser except for Ser258 can be phosphorylated by both Chk1 and Chk2 (double-underlined). The second Ser within the KXGS motif are preferably phosphorylated by Chk1 (single-underlined).
Potential Involvement of Chk1 and Chk2 in Tau Phosphorylation and Toxicity in AD
In this study, we identified a total of 27 non-SP/TP sites that are phosphorylated by Chk1 or Chk2 (12 sites for Chk1, 6 sites for Chk2, and 9 sites for both Chk1 and Chk2) in tau. Although most of these sites except for Thr30, Thr123, Ser191 and Thr377 have been identified as targets of other kinases such as AKT, CaMKII, CDK2, CDK5, CK1δ, CK2, GSK-3α, GSK-3β, JNK, MARK, tau tubulin kinases, p38, PKA, PKC and PKN (Table 2) 27, 29, 34, 37, 50–56, 69–71, the overall phosphorylation pattern of tau by Chk1 or Chk2 did not match to that by any tau kinase. Most of these sites (19 out of 27) are located within the microtubule-binding domain (residues 244–369) and C-terminal domain (residues 369–441), which have been reported to affect conformation of tau and/or microtubule organization (Figure 7). For example, tau phosphorylation at AD-related Ser214, Ser262, Ser356, or Ser409 diminishes its binding to microtubules 72–74, and is thought to trigger abnormal metabolism and toxicity of tau 75.
Figure 7.
Phosphorylation sites on tau by Chk1 and Chk2. The residues phosphorylated by Chk1 but not Chk2 are shown in open boxes, the residues phosphorylated by Chk2 but not Chk1 are shown in black boxes, and the residues phosphorylated by both Chk1 and Chk2 are shown in gray boxes. Stars indicate the residues phosphorylated in AD brains, and an open circle indicates the residues possibly phosphorylated in AD brains. The amino acid sequence of the longest isoform of human brain tau (2N4R) is shown. Residues corresponding to N-terminal inserts are italicized. The microtubule-binding domain is underlined.
Our results revealed that Chk1 and Chk2 directly phosphorylate tau at several AD-related non-SP/TP sites. In AD brains, 24 non-SP/TP sites in tau have been reported to be phosphorylated 76. Among these AD-related sites, 10 are phosphorylated by Chk1 (Ser191, Ser208, Ser214, Ser258, Ser262, Ser289, Ser356, Ser400, Ser409 and Thr414) and 7 are phosphorylated by Chk2 (Ser214, Ser262, Ser289, Thr403, Ser409, S433 and Ser435) (Table 2). In contrast, in normal human brain, 9 sites in tau (Ser46, Thr181, Ser199, Ser202, Thr231, Ser404, Ser416, and two out of three phosphorylatable residues in Ser412/Ser413/Thr414) have been reported to be phosphorylated 27, 77, and none of these sites except for Thr414 are phosphorylated by Chk1 or Chk2. These results suggest that Chk1 and Chk2 may contribute to tau phosphorylation at non-SP/TP sites in the brain under pathological conditions.
While Chk1 and Chk2 activities are well characterized in nucleus, tau is primarily described as a cytoplasmic protein. Thus, it seems that there is a special discrepancy as to how Chk1 and Chk2 can phosphorylate tau in neurons. However, several studies suggest that Chk1 or Chk2 and tau can colocalize in neurons. Active Chk1 has been detected in the cytoplasm 78. Chk2 is predominantly located in the nucleus in non-neuronal cells; however, in post-mitotic neurons, active Chk2 has been detected both in the cytoplasm and the nucleus 79. Moreover, tau has been detected in the nucleus in neuronal and non-neuronal cells 80–86, suggesting that phosphorylation of tau by Chk1 or Chk2 could occur in the nucleus. In addition, it is also possible that, under pathological conditions or upon aging, nuclear membrane may be disrupted, which may promote localization of Chk1 and Chk2 to the cytoplasm or localization of tau to the nucleus.
Conclusion
We have systematically identified the sites in tau phosphorylated by Chk1 and Chk2 in vitro by using Western blot with phospho-tau-specific antibodies and LC-MS3. We have shown that Chk1 and Chk2 phosphorylate tau at a total of 27 non- SP/TP sites. Many of these sites, 13 out of 24 to date, are associated with AD, suggesting that tau phosphorylation by Chk1 and Chk2 may be involved in the pathogenesis of AD.
Supplementary Material
Acknowledgement
We thank Behavioral and Medical Science Research Consortium (BMRC) for Anti-pSer320 tau (HIA3). We thank LiJuan Zhao and Georgia Dolios for technical help. This work was supported by funds from the Farber Institute for Neurosciences and Thomas Jefferson University to KMI and KA and by the Alzheimer’s Association [NIRG-10-173189] to KA, as well as, in part, by grants from the Gilbert Foundation/American Federation for Aging Research to KMI, the Alzheimer’s Association [NIRG-08-91985] to KMI, the National Institutes of Health [R01AG032279-A1] to KMI, P30 NS061777 and S10 RR022415 to RW, and the Uehara Memorial Foundation to MS.
Abbreviations
- ACN
acetonitrile
- AD
Alzheimer’s disease
- ATR
ataxia-telangiectasia and Rad-3 related
- ATM
ataxia-telangiectasia mutated
- Chk1
checkpoint kinases 1
- Chk2
checkpoint kinases 2
- IAM
iodoacetamide
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- TCEP
tris(2-carboxyethyl)-phosphine
- TFA
trifluoroacetic acid
Footnotes
Conflict of Interest Disclosure
The authors declare no competing financial interest.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21(2):245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye W, Blain SW. Chk1 has an essential role in the survival of differentiated cortical neurons in the absence of DNA damage. Apoptosis. 2011;16(5):449–459. doi: 10.1007/s10495-011-0579-z. [DOI] [PubMed] [Google Scholar]
- 4.Malzer E, Daly ML, Moloney A, Sendall TJ, Thomas SE, Ryder E, Ryoo HD, Crowther DC, Lomas DA, Marciniak SJ. Impaired tissue growth is mediated by checkpoint kinase 1 (CHK1) in the integrated stress response. J Cell Sci. 2010;123(Pt 17):2892–2900. doi: 10.1242/jcs.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, Finkel T. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336(6078):225–258. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kume K, Koyano T, Kanai M, Toda T, Hirata D. Calcineurin ensures a link between the DNA replication checkpoint and microtubule-dependent polarized growth. Nat Cell Biol. 2011;13(3):234–242. doi: 10.1038/ncb2166. [DOI] [PubMed] [Google Scholar]
- 7.Bakhrat A, Pritchett T, Peretz G, McCall K, Abdu U. Drosophila Chk2 and p53 proteins induce stage-specific cell death independently during oogenesis. Apoptosis. 2010;15(12):1425–1434. doi: 10.1007/s10495-010-0539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niida H, Murata K, Shimada M, Ogawa K, Ohta K, Suzuki K, Fujigaki H, Khaw AK, Banerjee B, Hande MP, Miyamoto T, Miyoshi I, Shirai T, Motoyama N, Delhase M, Appella E, Nakanishi M. Cooperative functions of Chk1 and Chk2 reduce tumour susceptibility in vivo. EMBO J. 2010;29(20):3558–3570. doi: 10.1038/emboj.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.North KE, Franceschini N, Avery CL, Baird L, Graff M, Leppert M, Chung JH, Zhang J, Hanis C, Boerwinkle E, Volcik KA, Grove ML, Mosley TH, Gu C, Heiss G, Pankow JS, Couper DJ, Ballantyne CM, Linda Kao WH, Weder AB, Cooper RS, Ehret GB, O'Connor AA, Chakravarti A, Hunt SC. Variation in the checkpoint kinase 2 gene is associated with type 2 diabetes in multiple populations. Acta Diabetol. 2009;47(Suppl 1):199–207. doi: 10.1007/s00592-009-0162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat Rev Cancer. 2007;7(12):925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 12.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 13.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, Anderson CW, Appella E, Nakanishi M, Suzuki H, Nagashima K, Sawa H, Ikeda K, Motoyama N. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21(19):5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282(5395):1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395(6701):507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 16.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14(3):289–300. [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404(6774):201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24(2):708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5(5):401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez S, Prives C, Cordon-Cardo C. p73alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23(22):8161–8171. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.O'Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277(18):16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- 22.Seo GJ, Kim SE, Lee YM, Lee JW, Lee JR, Hahn MJ, Kim ST. Determination of substrate specificity and putative substrates of Chk2 kinase. Biochem Biophys Res Commun. 2003;304(2):339–343. doi: 10.1016/s0006-291x(03)00589-8. [DOI] [PubMed] [Google Scholar]
- 23.Kim MA, Kim HJ, Brown AL, Lee MY, Bae YS, Park JI, Kwak JY, Chung JH, Yun J. Identification of novel substrates for human checkpoint kinase Chk1 and Chk2 through genome-wide screening using a consensus Chk phosphorylation motif. Exp Mol Med. 2007;39(2):205–212. doi: 10.1038/emm.2007.23. [DOI] [PubMed] [Google Scholar]
- 24.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. A phosphoproteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12(8):R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iijima-Ando K, Zhao L, Gatt A, Shenton C, Iijima K. A DNA damage-activated checkpoint kinase phosphorylates tau and enhances tau-induced neurodegeneration. Hum Mol Genet. 2010;19(10):1930–1938. doi: 10.1093/hmg/ddq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matenia D, Mandelkow EM. The tau of MARK: a polarized view of the cytoskeleton. Trends Biochem Sci. 2009;34(7):332–342. doi: 10.1016/j.tibs.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Hanger DP, Byers HL, Wray S, Leung KY, Saxton MJ, Seereeram A, Reynolds CH, Ward MA, Anderton BH. Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J Biol Chem. 2007;282(32):23645–23654. doi: 10.1074/jbc.M703269200. [DOI] [PubMed] [Google Scholar]
- 28.Drewes G, Lichtenberg-Kraag B, Doring F, Mandelkow EM, Biernat J, Goris J, Doree M, Mandelkow E. Mitogen activated protein (MAP) kinase transforms tau protein into an Alzheimer-like state. EMBO J. 1992;11(6):2131–2138. doi: 10.1002/j.1460-2075.1992.tb05272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann K, Mandelkow EM, Biernat J, Piwnica-Worms H, Mandelkow E. Abnormal Alzheimer-like phosphorylation of tau-protein by cyclin-dependent kinases cdk2 and cdk5. FEBS Lett. 1993;336(3):417–424. doi: 10.1016/0014-5793(93)80849-p. [DOI] [PubMed] [Google Scholar]
- 30.Goedert M, Hasegawa M, Jakes R, Lawler S, Cuenda A, Cohen P. Phosphorylation of microtubule-associated protein tau by stress-activated protein kinases. FEBS Lett. 1997;409(1):57–62. doi: 10.1016/s0014-5793(97)00483-3. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds CH, Utton MA, Gibb GM, Yates A, Anderton BH. Stress-activated protein kinase/c-jun N-terminal kinase phosphorylates tau protein. J Neurochem. 1997;68(4):1736–1744. doi: 10.1046/j.1471-4159.1997.68041736.x. [DOI] [PubMed] [Google Scholar]
- 32.Kitano-Takahashi M, Morita H, Kondo S, Tomizawa K, Kato R, Tanio M, Shirota Y, Takahashi H, Sugio S, Kohno T. Expression, purification and crystallization of a human tau-tubulin kinase 2 that phosphorylates tau protein. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63(Pt 7):602–604. doi: 10.1107/S1744309107028783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato S, Cerny RL, Buescher JL, Ikezu T. Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J Neurochem. 2006;98(5):1573–1584. doi: 10.1111/j.1471-4159.2006.04059.x. [DOI] [PubMed] [Google Scholar]
- 34.Tomizawa K, Omori A, Ohtake A, Sato K, Takahashi M. Tau-tubulin kinase phosphorylates tau at Ser-208 and Ser-210, sites found in paired helical filament-tau. FEBS Lett. 2001;492(3):221–227. doi: 10.1016/s0014-5793(01)02256-6. [DOI] [PubMed] [Google Scholar]
- 35.Greenwood JA, Scott CW, Spreen RC, Caputo CB, Johnson GV. Casein kinase II preferentially phosphorylates human tau isoforms containing an amino-terminal insert. Identification of threonine 39 as the primary phosphate acceptor. J Biol Chem. 1994;269(6):4373–4380. [PubMed] [Google Scholar]
- 36.Woods YL, Cohen P, Becker W, Jakes R, Goedert M, Wang X, Proud CG. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J. 2001;355(Pt 3):609–615. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89(2):297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 38.Paudel HK. The regulatory Ser262 of microtubule-associated protein tau is phosphorylated by phosphorylase kinase. J Biol Chem. 1997;272(3):1777–1785. [PubMed] [Google Scholar]
- 39.Kawamata T, Taniguchi T, Mukai H, Kitagawa M, Hashimoto T, Maeda K, Ono Y, Tanaka C. A protein kinase, PKN, accumulates in Alzheimer neurofibrillary tangles and associated endoplasmic reticulum-derived vesicles and phosphorylates tau protein. J Neurosci. 1998;18(18):7402–7410. doi: 10.1523/JNEUROSCI.18-18-07402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanger DP, Hughes K, Woodgett JR, Brion JP, Anderton BH. Glycogen synthase kinase-3 induces Alzheimer's disease-like phosphorylation of tau: generation of paired helical filament epitopes and neuronal localisation of the kinase. Neurosci Lett. 1992;147(1):58–62. doi: 10.1016/0304-3940(92)90774-2. [DOI] [PubMed] [Google Scholar]
- 41.Hong M, Lee VM. Insulin and insulin-like growth factor-1 regulate tau phosphorylation in cultured human neurons. J Biol Chem. 1997;272(31):19547–19553. doi: 10.1074/jbc.272.31.19547. [DOI] [PubMed] [Google Scholar]
- 42.Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33(1):95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 43.Moreno FJ, Munoz-Montano JR, Avila J. Glycogen synthase kinase 3 phosphorylation of different residues in the presence of different factors: analysis on tau protein. Mol Cell Biochem. 1996;165(1):47–54. doi: 10.1007/BF00229744. [DOI] [PubMed] [Google Scholar]
- 44.Godemann R, Biernat J, Mandelkow E, Mandelkow EM. Phosphorylation of tau protein by recombinant GSK-3beta: pronounced phosphorylation at select Ser/Thr-Pro motifs but no phosphorylation at Ser262 in the repeat domain. FEBS Lett. 1999;454(1–2):157–164. doi: 10.1016/s0014-5793(99)00741-3. [DOI] [PubMed] [Google Scholar]
- 45.Wang JZ, Wu Q, Smith A, Grundke-Iqbal I, Iqbal K. Tau is phosphorylated by GSK-3 at several sites found in Alzheimer disease and its biological activity markedly inhibited only after it is prephosphorylated by A-kinase. FEBS Lett. 1998;436(1):28–34. doi: 10.1016/s0014-5793(98)01090-4. [DOI] [PubMed] [Google Scholar]
- 46.Lund ET, McKenna R, Evans DB, Sharma SK, Mathews WR. Characterization of the in vitro phosphorylation of human tau by tau protein kinase II (cdk5/p20) using mass spectrometry. J Neurochem. 2001;76(4):1221–1232. doi: 10.1046/j.1471-4159.2001.00130.x. [DOI] [PubMed] [Google Scholar]
- 47.Scott CW, Spreen RC, Herman JL, Chow FP, Davison MD, Young J, Caputo CB. Phosphorylation of recombinant tau by cAMP-dependent protein kinase. Identification of phosphorylation sites and effect on microtubule assembly. J Biol Chem. 1993;268(2):1166–1173. [PubMed] [Google Scholar]
- 48.Robertson J, Loviny TL, Goedert M, Jakes R, Murray KJ, Anderton BH, Hanger DP. Phosphorylation of tau by cyclic-AMP-dependent protein kinase. Dementia. 1993;4(5):256–263. doi: 10.1159/000107331. [DOI] [PubMed] [Google Scholar]
- 49.Johnson GV. Differential phosphorylation of tau by cyclic AMP-dependent protein kinase and Ca2+/calmodulin-dependent protein kinase II: metabolic and functional consequences. J Neurochem. 1992;59(6):2056–2062. doi: 10.1111/j.1471-4159.1992.tb10094.x. [DOI] [PubMed] [Google Scholar]
- 50.Sironi JJ, Yen SH, Gondal JA, Wu Q, Grundke-Iqbal I, Iqbal K. Ser-262 in human recombinant tau protein is a markedly more favorable site for phosphorylation by CaMKII than PKA or PhK. FEBS Lett. 1998;436(3):471–475. doi: 10.1016/s0014-5793(98)01185-5. [DOI] [PubMed] [Google Scholar]
- 51.Ksiezak-Reding H, Pyo HK, Feinstein B, Pasinetti GM. Akt/PKB kinase phosphorylates separately Thr212 and Ser214 of tau protein in vitro. Biochim Biophys Acta. 2003;1639(3):159–168. doi: 10.1016/j.bbadis.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Yang SD, Yu JS, Shiah SG, Huang JJ. Protein kinase FA/glycogen synthase kinase-3 alpha after heparin potentiation phosphorylates tau on sites abnormally phosphorylated in Alzheimer's disease brain. J Neurochem. 1994;63(4):1416–1425. doi: 10.1046/j.1471-4159.1994.63041416.x. [DOI] [PubMed] [Google Scholar]
- 53.Taniguchi T, Kawamata T, Mukai H, Hasegawa H, Isagawa T, Yasuda M, Hashimoto T, Terashima A, Nakai M, Mori H, Ono Y, Tanaka C. Phosphorylation of tau is regulated by PKN. J Biol Chem. 2001;276(13):10025–10031. doi: 10.1074/jbc.M007427200. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto H, Yamauchi E, Taniguchi H, Ono T, Miyamoto E. Phosphorylation of microtubule-associated protein tau by Ca2+/calmodulin-dependent protein kinase II in its tubulin binding sites. Arch Biochem Biophys. 2002;408(2):255–262. doi: 10.1016/s0003-9861(02)00556-8. [DOI] [PubMed] [Google Scholar]
- 55.Tseng HC, Lu Q, Henderson E, Graves DJ. Phosphorylated tau can promote tubulin assembly. Proc Natl Acad Sci U S A. 1999;96(17):9503–9508. doi: 10.1073/pnas.96.17.9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynolds CH, Betts JC, Blackstock WP, Nebreda AR, Anderton BH. Phosphorylation sites on tau identified by nanoelectrospray mass spectrometry: differences in vitro between the mitogen-activated protein kinases ERK2, c-Jun N-terminal kinase and P38, and glycogen synthase kinase-3beta. J Neurochem. 2000;74(4):1587–1595. doi: 10.1046/j.1471-4159.2000.0741587.x. [DOI] [PubMed] [Google Scholar]
- 57.Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur J Neurosci. 2005;22(8):1942–1950. doi: 10.1111/j.1460-9568.2005.04391.x. [DOI] [PubMed] [Google Scholar]
- 58.Iijima K, Gatt A, Iijima-Ando K. Tau Ser262 phosphorylation is critical for Abeta42-induced tau toxicity in a transgenic Drosophila model of Alzheimer's disease. Hum Mol Genet. 2010;19(15):2947–2957. doi: 10.1093/hmg/ddq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smale G, Nichols NR, Brady DR, Finch CE, Horton WE., Jr Evidence for apoptotic cell death in Alzheimer's disease. Exp Neurol. 1995;133(2):225–230. doi: 10.1006/exnr.1995.1025. [DOI] [PubMed] [Google Scholar]
- 60.Adamec E, Vonsattel JP, Nixon RA. DNA strand breaks in Alzheimer's disease. Brain Research. 1999;849(1–2):67–77. doi: 10.1016/s0006-8993(99)02004-1. [DOI] [PubMed] [Google Scholar]
- 61.Lassmann H, Bancher C, Breitschopf H, Wegiel J, Bobinski M, Jellinger K, Wisniewski HM. Cell death in Alzheimer's disease evaluated by DNA fragmentation in situ. Acta Neuropathol. 1995;89(1):35–41. doi: 10.1007/BF00294257. [DOI] [PubMed] [Google Scholar]
- 62.Lucassen PJ, Chung WC, Kamphorst W, Swaab DF. DNA damage distribution in the human brain as shown by in situ end labeling; area-specific differences in aging and Alzheimer disease in the absence of apoptotic morphology. J Neuropathol Exp Neurol. 1997;56(8):887–900. doi: 10.1097/00005072-199708000-00007. [DOI] [PubMed] [Google Scholar]
- 63.Davydov V, Hansen LA, Shackelford DA. Is DNA repair compromised in Alzheimer's disease? Neurobiol Aging. 2003;24(7):953–968. doi: 10.1016/s0197-4580(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 64.Su JH, Anderson AJ, Cummings BJ, Cotman CW. Immunohistochemical evidence for apoptosis in Alzheimer's disease. Neuroreport. 1994;5(18):2529–2533. doi: 10.1097/00001756-199412000-00031. [DOI] [PubMed] [Google Scholar]
- 65.Stadelmann C, Bruck W, Bancher C, Jellinger K, Lassmann H. Alzheimer disease: DNA fragmentation indicates increased neuronal vulnerability, but not apoptosis. J Neuropathol Exp Neurol. 1998;57(5):456–464. doi: 10.1097/00005072-199805000-00009. [DOI] [PubMed] [Google Scholar]
- 66.Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat Neurosci. 2013 doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 68.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanger DP, Anderton BH, Noble W. Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol Med. 2009;15(3):112–119. doi: 10.1016/j.molmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Paudel HK, Lew J, Ali Z, Wang JH. Brain proline-directed protein kinase phosphorylates tau on sites that are abnormally phosphorylated in tau associated with Alzheimer's paired helical filaments. J Biol Chem. 1993;268(31):23512–23518. [PubMed] [Google Scholar]
- 71.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry. 1999;38(12):3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 72.Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11(1):153–163. doi: 10.1016/0896-6273(93)90279-z. [DOI] [PubMed] [Google Scholar]
- 73.Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE, Mandelkow EM, Mandelkow E. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem. 1995;270(13):7679–7688. doi: 10.1074/jbc.270.13.7679. [DOI] [PubMed] [Google Scholar]
- 74.Liu F, Li B, Tung EJ, Grundke-Iqbal I, Iqbal K, Gong CX. Site-specific effects of tau phosphorylation on its microtubule assembly activity and self-aggregation. Eur J Neurosci. 2007;26(12):3429–3436. doi: 10.1111/j.1460-9568.2007.05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8(9):663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 76.Sergeant N, Bretteville A, Hamdane M, Caillet-Boudin ML, Grognet P, Bombois S, Blum D, Delacourte A, Pasquier F, Vanmechelen E, Schraen-Maschke S, Buee L. Biochemistry of Tau in Alzheimer's disease and related neurological disorders. Expert Rev Proteomics. 2008;5(2):207–224. doi: 10.1586/14789450.5.2.207. [DOI] [PubMed] [Google Scholar]
- 77.Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Yoshida H, Titani K, Ihara Y. Proline-directed and non-proline-directed phosphorylation of PHF-tau. J Biol Chem. 1995;270(2):823–829. doi: 10.1074/jbc.270.2.823. [DOI] [PubMed] [Google Scholar]
- 78.Oe T, Nakajo N, Katsuragi Y, Okazaki K, Sagata N. Cytoplasmic occurrence of the Chk1/Cdc25 pathway and regulation of Chk1 in Xenopus oocytes. Dev Biol. 2001;229(1):250–261. doi: 10.1006/dbio.2000.9968. [DOI] [PubMed] [Google Scholar]
- 79.Lukas C, Bartkova J, Latella L, Falck J, Mailand N, Schroeder T, Sehested M, Lukas J, Bartek J. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res. 2001;61(13):4990–4993. [PubMed] [Google Scholar]
- 80.Sultan A, Nesslany F, Violet M, Begard S, Loyens A, Talahari S, Mansuroglu Z, Marzin D, Sergeant N, Humez S, Colin M, Bonnefoy E, Buee L, Galas MC. Nuclear tau, a key player in neuronal DNA protection. J Biol Chem. 2011;286(6):4566–4575. doi: 10.1074/jbc.M110.199976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loomis PA, Howard TH, Castleberry RP, Binder LI. Identification of nuclear tau isoforms in human neuroblastoma cells. Proc Natl Acad Sci U S A. 1990;87(21):8422–8426. doi: 10.1073/pnas.87.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Loomis PA, Zinkowski RP, Binder LI. A novel tau transcript in cultured human neuroblastoma cells expressing nuclear tau. J Cell Biol. 1993;121(2):257–267. doi: 10.1083/jcb.121.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sjoberg MK, Shestakova E, Mansuroglu Z, Maccioni RB, Bonnefoy E. Tau protein binds to pericentromeric DNA: a putative role for nuclear tau in nucleolar organization. J Cell Sci. 2006;119(Pt 10):2025–2034. doi: 10.1242/jcs.02907. [DOI] [PubMed] [Google Scholar]
- 84.Thurston VC, Zinkowski RP, Binder LI. Tau as a nucleolar protein in human nonneural cells in vitro and in vivo. Chromosoma. 1996;105(1):20–30. doi: 10.1007/BF02510035. [DOI] [PubMed] [Google Scholar]
- 85.Cross DC, Munoz JP, Hernandez P, Maccioni RB. Nuclear and cytoplasmic tau proteins from human nonneuronal cells share common structural and functional features with brain tau. J Cell Biochem. 2000;78(2):305–317. [PubMed] [Google Scholar]
- 86.Alonso AD, Di Clerico J, Li B, Corbo CP, Alaniz ME, Grundke-Iqbal I, Iqbal K. Phosphorylation of tau at Thr212, Thr231, and Ser262 combined causes neurodegeneration. J Biol Chem. 2010;285(40):30851–30860. doi: 10.1074/jbc.M110.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.