Abstract
Objectives:
The biochemical effects of thiamine pyrophosphate on ischemia-reperfusion (IR) induced oxidative damage and DNA mutation in rat kidney tissue were investigated, and compared to thiamine.
Materials and Methods:
Rats were divided into four groups: Renal ischemia-reperfusion (RIR); thiamine pyrophosphate + RIR (TPRIR); thiamine + RIR (TRIR); and sham group (SG).
Results:
The results of biochemical experiments have shown that malondialdehyde (MDA) levels in rat kidney tissue after TRIR and TPRIR treatment were 7.2 ± 0.5 (P > 0.05) and 3.3 ± 0.3 (P < 0.0001) μmol/g protein, respectively. The MDA levels in the SG rat kidney tissue and in RIR group were 3.6 ± 0.2 (P < 0.0001) and 7.6 ± 0.6 μmol/g protein, respectively. Total glutathione (tGSH) levels in TRIR, TPRIR, SG, and RIR animal groups were 2.2 ± 0.3 (P > 0.05), 5.8 ± 0.4 (P < 0.0001), 6.2 ± 0.2 (P < 0.0001), and 1.7 ± 0.2 nmol/g protein, respectively. In the TRIR, TPRIR, SG, and RIR animal groups; 8-hydroxyguanine (8-OHGua)/Gua levels, which indicate mutagenic DNA, were 1.75 ± 0.12 (P > 0.05), 0.93 ± 0.1 (P < 0.0001), 0.85 ± 0.08 (P < 0.0001), and 1.93 ± 0.24 pmol/L, respectively.
Conclusions:
It has been shown that thiamine pyrophosphate prevents increase in mutagenic DNA in IR induced oxidative damage, whereas thiamine does not have this effect.
KEY WORDS: DNA mutation, ischemia-reperfusion, oxidative damage, rat, thiamine pyrophosphate
Introduction
Ischemia-reperfusion (IR) process, which is performed during kidney transplants and various urological procedures, leads to kidney damage.[1] IR injury is a complex condition and is the main cause of acute renal failure. As soon as reoxygenation is achieved by reperfusion, the enzyme xanthine oxidase, which is formed during ischemia[2] converts accumulated hypoxanthine to xanthine, resulting in a large amount of free oxygen radicals in the environment.[3] The free oxygen radicals, which are called reperfusion mediators, oxidate the cell membrane lipids and cause formation of toxic products, such as aldehydes and malondialdehyde (MDA).[4] Furthermore, it has been reported that excessive free oxygen radicals react with DNA, causing oxidative damage of DNA. This oxidative damage is characterized by DNA breaks in base chains and ribose-phosphate bonds, as well as double strand breaks. The initial damage is caused by formation of pyrimidine dimers due to breaks in thymine-adenine bonds. After reaction with free oxygen radicals, base changes in nucleic acids and chain breaks in DNA are encountered. If this damage cannot be repaired, DNA mutation will follow. 8-hydroxyguanine (8-OHGua) is a mutagenic form of DNA.[5] GSH and other antioxidants prevent harmful effects of free oxygen radicals on cellular structures, but when the antioxidant defense mechanisms fail, serious tissue damage occurs.[6] This suggests that treatment with antioxidants may be beneficial in IR injury.
Thiamine pyrophosphate, which we have used in our study for treatment of IR injury, is an active metabolite of thiamine. Thiamine pyrophosphate is formed by phosphatizing of thiamine in the liver. Thiamine increases the formation of antioxidants and nicotineamide adenine dinucleotide phosphate (NADPH) levels via pentose phosphate pathway.[7] Because IR damage is a rapidly developing process, thiamine pyrophosphate use, that its active metabolite, thought to be more effective. The antioxidant properties of thiamine pyrophosphate is not yet fully known. In recent years, studies have shown the antioxidant activity of thiamine pyrophosphate. In a study, it was found that IR induced oxidative damage in rat ovary was not prevented by thiamine, but it was prevented by thiamine pyrophosphate.[8] In addition, unlike thiamine, thiamine pyrophosphate is a cofactor for both pyruvate-2-oxoglutarate dehydrogenase complex that is required for mitochondrial adenosine triphosphate (ATP) synthesis and transketolase enzyme plays an important role in maintaining cellular redox status.[9] Therefore, thiamine pyrophosphate is important for cellular energy production and preservation. In literature, we have not found any information about protective effect of thiamine pyrophosphate in IR induced kidney injury in rats. The aim of our study was to investigate the biochemical effects of thiamine pyrophosphate on IR induced oxidative damage and DNA mutation in rat kidney tissue, and to compare it to that of thiamine.
Materials and Methods
Animals
In this study, 24 albino Wistar male rats of 230–245 g were used, which were supplied from Atatürk University Medical Application and Research Center. The animals were kept in room temperature (22°C) in groups and were fed. In document B.30.2.ATA.0.23.85-57 dated 5 April 2012, Atatürk University Local Ethical Committee of Experimental Animals (AUDHADYEK) approved that all the steps of this study were compliant with ethical rules.
Chemicals
Thiopental sodium was provided by IE Ulagay, Turkey. Thiamine and thiamine pyrophosphate were obtained from Biopharma, Russia.
General Procedure
The rats were divided into four groups as follows: Renal ischemia-reperfusion group (RIR); thiamine pyrophosphate + RIR group (TPRIR); thiamine + RIR group (TRIR), and sham group (SG). The surgical interventions on rats were performed in laboratories, under sterile conditions, using thiopental sodium (25 mg/kg intraperitoneal (i.p.)) anesthesia.
Performing Study
An hour before the thiopental sodium anesthesia, TPRIR group (n = 6) was given thiamine pyrophosphate 25 mg/kg i.p. and TRIR group was given thiamine 25 mg/kg i.p. In the RIR and SG groups, distilled water was injected via the same way. After thiopental sodium injection, rats were kept until appropriate moment for surgical intervention. The moment that animals remain motionless in supine position is considered the appropriate time to perform surgery. The kidneys of rats were reached through a unilateral dorsal incision on the left side. Subsequently, except for the SG group, a vascular clip was placed on lower part of renal artery and vein, and ischemia was maintained for 1 h. After this period, in the RIR, TPRIR, and TRIR groups; vascular clip was removed in order to provide reperfusion for 6 h. Afterwards, all the animals were terminated by high-dose anesthesia, both kidneys were removed and biochemical studies were performed. The biochemical results of TPRIR and TRIR groups were compared with results of the RIR and SG groups.
Biochemical Analysis of Renal Tissue
From each kidney, a sample of renal tissue of 0.2 g was obtained. The samples were homogenized in ice with 2 ml of 1.15% potassium chloride buffer for MDA analysis and phosphate buffer with a pH of 7.5 for other analyses. Then, they were centrifuged at 4°C, 10,000 rpm for 15 min. The supernatant was used as the analysis sample. For all the measurements, the tissue-protein estimation was performed using Bradford's method.[10]
MDA Analysis
The concentrations of renal lipid peroxidation were determined by estimating MDA using thiobarbituric acid test.[11] The rat renal tissues were rinsed with cold saline. The corpus mucosa was scraped, weighed, and homogenized in 10 ml of 100 g/L KCl. The homogenate (0.5 ml) was added to a solution containing 0.2 ml of 80 g/L sodium lauryl sulfate, 1.5 mL of 200 g/L acetic acid, 1.5 mL of 8 g/L 2-thiobarbiturate, and 0.3 mL distilled water. The mixture was incubated at 98°C for 1 h. Upon cooling, 5 ml of n-butanol:pyridine (15:l) was added. The mixture was vortexed for 1 min and centrifuged for 30 min at 4,000 rpm. The absorbance of supernatant was measured at 532 nm. The standard curve was obtained by using 1,1,3,3-tetramethoxypropane.
Total Glutathione (tGSH) Analysis
The amount of GSH in the total homogenate was measured according to the method of Sedlak and Lindsay with some modifications.[12] The sample was weighed and homogenized in 2 mL of 50 mM tris-HCl buffer containing 20 mM ethylenediamineteraacetic acid (EDTA) and 0.2 mM sucrose at pH 7.5. The homogenate was immediately precipitated with 0.1 mL of 25% trichloroacetic acid, and the precipitate was removed after centrifugation at 4,200 rpm for 40 min at 4°C and the supernatant was used to determine GSH levels. A total of 1500 μL of measurement buffer (200 mM tris-HCl buffer containing 0.2 mM EDTA at pH 7.5), 500 μL supernatant, 100 μL 5,5-dithiobis (2-nitrobenzoic acid) (DTNB) (10 mM), and 7900 μl methanol were added to a tube and vortexed and incubated for 30 min in 37°C. DTNB was used as a chromogen and it formed a yellow-colored complex with sulfhydryl (SH) groups. The absorbance was measured at 412 nm using a spectrophotometer. The standard curve was obtained by using reduced GSH.
Isolation of DNA from Renal Tissue
DNA was isolated from renal tissue using Shigenaga et al.'s, modified method.[13] The samples (for renal tissue 50 mg) were homogenized at 4°C in 1 mL of homogenization buffer (0.1 M NaCl, 30 mM Tris, pH 8.0, 10 mM EDTA, 10 mM 2-mercaptoethanol, 0.5% (v/v) Triton X-100) with six passes of a teflon glass homogenizer at 200 rpm. The samples were centrifuged at 4°C for 10 min at 1,000 g to pellet nuclei. The supernatant was discarded, and the crude nuclear pellet resuspended and rehomogenized in 1 mL of extraction buffer (0.1 M Tris, pH 8.0, 0.1 M NaCl, 20 mM EDTA) and recentrifuged as above for 2 min. The washed pellet was resuspended in 300 μl of extraction buffer with a wide orifice 200 μl Pipetman tip. The resuspended pellet was subsequently incubated at 65°C for 1 h with presence of 0.1 ml of 10% sodium dodecyl sulfate (SDS), 40 μL proteinase K, and 1.9 mL leukocyte lysis buffer. Then, ammonium acetate was added to crude DNA sample to give a final concentration of 2.5 mol/L and centrifuged in a microcentrifuge for 5 min. The supernatant was removed and mixed with two volumes of ethanol to precipitate DNA fraction. After centrifugation, the pellet was dried under reduced pressure and dissolved in sterile water. The absorbance (A) of this fraction was measured at 260 and 280 nm. Purification of DNA was determined as A 260/280 ratio 1.8.
DNA Hydrolysis with Formic Acid
Approximately 50 mg of DNA was hydrolyzed with 0.5 mL of formic acid (60%, v/v) for 45 min at 150°C. The tubes were allowed to cool. The contents were then transferred to Pierce microvials, covered with Kleenex tissues cut to size (secured in place using a rubber band), and cooled in liquid nitrogen. Formic acid was then removed by freeze drying. Before analysis by high performance liquid chromatography (HPLC), they were redissolved in the eluent (final volume 200 μL).
Measurement of 8-hydroxy-2 deoxyguanine (8-OH Gua)
The amount of 8-OH Gua and Gua were measured by using a HPLC system equipped with an electrochemical detector (HP Agilent 1100 module series, E.C.D. HP 1049 A), as described previously.[14] The amount of 8-OH Gua and Gua were analyzed on a 250 4.6 mm Supelco LC-18-S reverse-phase column. The mobile phase was 50 mM potassium phosphate, pH 5.5, with acetonitrile (97 volumes of acetonitrile and 3 volumes of potassium phosphate), and the flow rate was 1.0 mL/min. The detector potential was set at 0.80 V for measuring oxidized base. Gua and 8-OH Gua (25 pmol) were used as standards. The 8-OH Gua levels were expressed as number of 8-OH Gua molecules/105 Gua molecules.[15]
Statistical Analysis
All data were subjected to one-way analyze of variance (ANOVA) using Statistical Package for the Social Sciences (SPSS) 18.0 software. The differences among groups were analyzed using the least significant difference (LSD) option and a P-value < 0.05 was considered to be statistically significant. The results are presented as the mean ± standard error of mean (SEM).
Results
MDA, GSH and 8-OHGua levels in SG, TRIR, TPRIR and RIR groups were seen in Table 1.
Table 1.
Effect of thiamine pyrophosphate and thiamine on amount of 8-hydroxyguanine (8-OHGua), malondialdehyde (MDA), and total glutathione (tGSH) levels in kidney tissue after ischemia-reperfusion
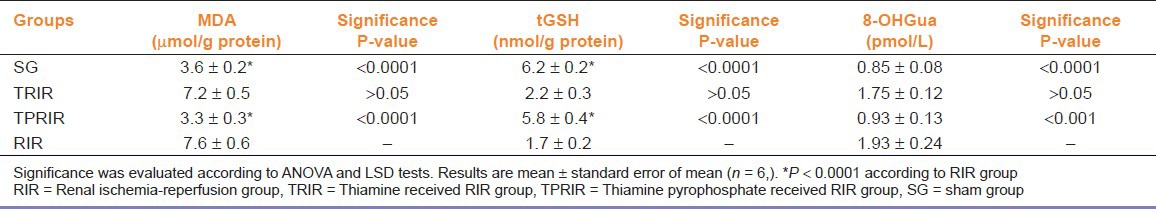
Effect of thiamine pyrophosphate and thiamine on MDA levels in kidney tissue after IR
As seen in Figure 1, MDA levels in TPRIR and TRIR groups were 3.3 ± 0.3 and 7.2 ± 0.5 μmol/g protein, while 3.6 ± 0.2 and 7.6 ± 0.6 μmol/g protein in the SG and RIR groups; respectively. MDA level is lower in the SG (P < 0.0001) and TPRIR groups than in the RIR group (P < 0.0001) It does not show any statistically significant difference of MDA levels between the TRIR and RIR groups (P > 0.05) [Figure 1].
Figure 1.
Effect of thiamine pyrophosphate and thiamine on malondialdehyde levels in kidney tissue after ischemia-reperfusion (IR)
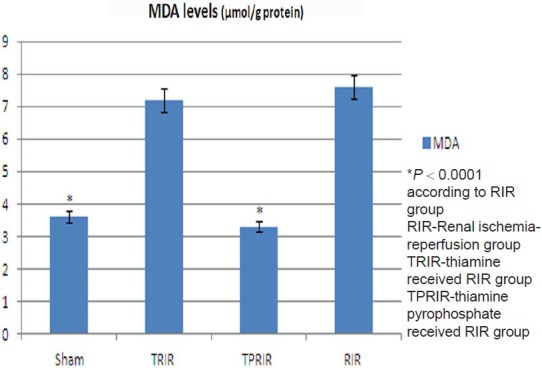
Effect of thiamine pyrophosphate and thiamine on tGSH levels in kidney tissue after IR
The tGSH levels in TPRIR, TRIR, SG, and RIR groups were 5.8 ± 0.4, 2.2 ± 0.3, 6.2 ± 0.2, and 1.7 ± 0.2 nmol/g protein, respectively. tGSH level is higher in SG (P < 0.0001) and TPRIR groups than in RIR group (P < 0.0001). tGSH levels of TPRIR and RIR groups were statistically not different (P > 0.05) [Figure 2].
Figure 2.
Effect of thiamine pyrophosphate and thiamine on glutathione levels in kidney tissue after ischemia-reperfusion
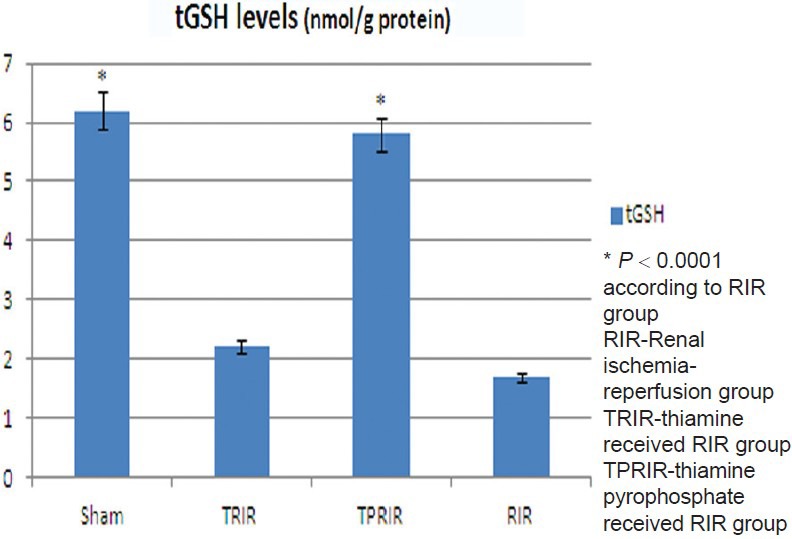
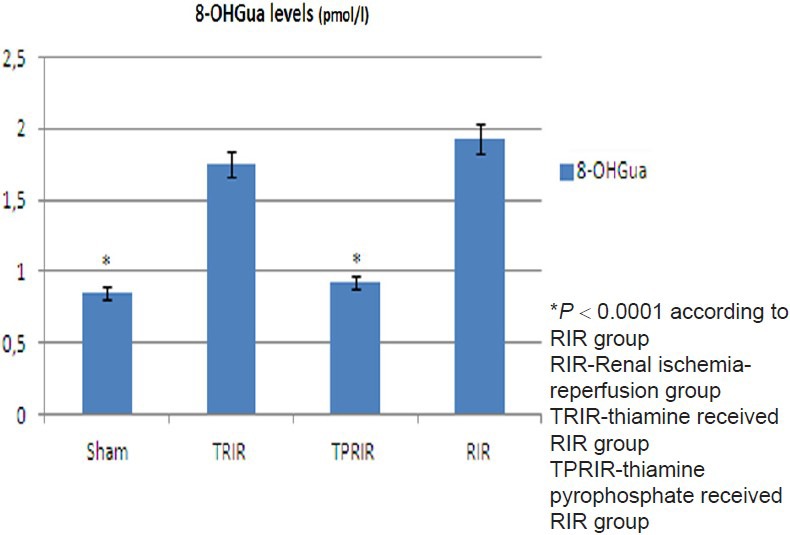
Effect of thiamine pyrophosphate and thiamine on 8-OHGua/Gua levels in kidney tissue after IR
8-OHGua/Gua levels in TPRIR, TRIR, SG, and RIR groups were 0.93 ± 0.1, 1.75 ± 0.12 pmol/L, 0.85 ± 0.08 pmol/L, and 1.93 ± 0.24 pmol/L, respectively. 8-OHGua/Gua level was lower in SG and TPRIR groups than in RIR group (P < 0.0001). 8-OH Gua/Gua levels of TPRIR and RIR groups did not differ statistically (P > 0.05) [Figure 3].
Figure 3.
Effect of thiamine pyrophosphate and thiamine on 8-hydroxyguanine/Guanine levels in kidney tissue after ischemia-reperfusion (IR)
Discussion
In this study, the biochemical effects of thiamine pyrophosphate on the IR induced oxidative stress and DNA mutation in rat kidney were investigated, and were compared to that of thiamine. It is known that tissue damage of various degrees occur after IR of the kidneys. The oxygen radicals such as superoxide anion (O2–), hydrogen peroxide (H2O2), hydroxyl radical (OH), and peroxynitrite anion (ONOO–) are thought to be responsible for tissue damage associated with IR.[16] During IR, the excessive free oxygen radicals increased the lipid peroxidation, impairing integrity of cellular structure.[17] In our study, MDA levels in kidney tissue in RIR group were significantly higher than the SG group. This indicates the presence of oxidative stress in rat kidney tissue in our experiment.
It is known that serious imbalance between free radical formation and antioxidant defense mechanisms during oxidative stress leads to tissue damage.[18] There are studies reporting an increase in MDA and a decrease in GSH levels in damaged kidney tissue after IR.[19] In tissues, there are enzymatic and nonenzymatic antioxidant mechanisms against oxidative damage. Reduced GSH is the most important endogenous nonenzymatic antioxidant. Reduced GSH plays a role in protection of cells from superoxide anion (O2–), hydrogen peroxide (H2O2), hydroxyl (OH), and peroxynitrite anion (ONOO–) radicals.[20] In some experimental kidney failure models, it was reported that reduced GSH decreases in case of kidney damage.[21] In our study, there was no statistically significant difference in MDA and GSH levels in kidney tissue between TPRIR and the SG group. However, the difference in MDA and GSH levels in kidney tissue was statistically significant between TRIR and RIR groups. These results indicate that thiamine pyrophosphate prevents oxidative stress induced by IR in kidney tissue.
The excessive free oxygen radicals do not only cause damage to the lipids, but also to DNA. The damage to DNA can be in the form of single and double strain breaks, abasic areas, base modifications, or formation of crosslinks between the proteins.[22] OH, which is a free radical, can lead to DNA damage. OH has to be very close to DNA in order to be effective and cause damage. Nevertheless, OH radicals cannot penetrate the cell nucleus. However, OH radicals, which come free as a result of the reaction of H2O2 with Fe-Cu ions in the nucleus, do induce DNA damage. Furthermore, because DNA consists of a large number of phosphate groups with a negative load, it is bounded to positive metal ions such as Fe2+/3+ and Cu1+/2+.[23] These DNA bonded metal ions react with H2O2 in the nucleus (on the DNA), and form toxic radicals such as OH leading to oxidative damage of DNA.[22] It was shown in tissue cultures, by increasing Fe3+ and Cu2+ concentrations, oxidative DNA base damage increases; and in the cells exposed to H2O2, the use of copper and/or iron chelators prevents oxidative damage to DNA.[24] This indicates the importance of use of antioxidants together with iron chelators in prevention of DNA damage. In our study, we have shown that thiamine pyrophosphate prevents increase in mutagenic DNA (8-OHGua) levels in case of oxidative damage induced by IR in rat kidney. Although Lukienko et al., demonstrated the antioxidant activity of thiamine,[25] in our study, thiamine could not prevent oxidative damage and associated increase in mutagenic DNA levels after IR in kidney tissue.
As a result, we have found an increase in mutagenic DNA levels in rat kidney tissue, associated with oxidative stress, after IR injury. It was found that thiamine pyrophosphate prevents increase in mutagenic DNA levels in case of oxidative damage. Thiamine, on the other hand, does not have this effect. The protective effect of thiamine pyrophosphate on IR damage of kidney can be due to its antioxidant activity.
Footnotes
Source of Support: Nil
Conflict Interest: None declared
References
- 1.Křen V, Walterová D. Silybin and silymarin—new effects and applications. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2005;149:29–41. doi: 10.5507/bp.2005.002. [DOI] [PubMed] [Google Scholar]
- 2.Lindsay TF, Liauw S, Romaschin AD, Walker PM. The effect of ischemia/reperfusion on adenine nucleotide metabolism and xanthine oxidase production in skeletal muscle. J Vasc Surg. 1990;12:8–15. doi: 10.1067/mva.1990.19946. [DOI] [PubMed] [Google Scholar]
- 3.McCord JM. Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med. 1985;312:159–63. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- 4.Del Maestro RF. An approach to free radicals in medicine and biology. Acta Physiol Scand Suppl. 1980;492:153–68. [PubMed] [Google Scholar]
- 5.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–70. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 6.Young IS, Woodside JV. Antioxidants in health and disease. J Clin Pathol. 2001;54:176–86. doi: 10.1136/jcp.54.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bomser J, Madhavi DL, Singletary K, Smith MA. In vitro anticancer activity of fruit extracts from Vaccinium species. Planta Med. 1996;62:212–6. doi: 10.1055/s-2006-957862. [DOI] [PubMed] [Google Scholar]
- 8.Demiryilmaz I, Cetin N, Altuner D, Akcay F, Suleyman H. A comparative investigation of biochemical and histopathological effects of thiamine and thiamine pyrophosphate on ischemia-reperfusion induced oxidative damage in rat ovarian tissue. Arch Pharm Res. doi: 10.1007/s12272-013-0173-8. In press. [DOI] [PubMed] [Google Scholar]
- 9.Gangolf M, Czerniecki J, Radermecker M, Detry O, Nisolle M, Jouan C, et al. Thiamine status in humans and content of phosphorylated thiamine derivatives in biopsies and cultured cells. PLoS One. 2010;5:e13616. doi: 10.1371/journal.pone.0013616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem. 1979;95:351–8. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
- 12.Sedlak J, Lindsay RH. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman's reagent. Anal Biochem. 1968;25:192–205. doi: 10.1016/0003-2697(68)90092-4. [DOI] [PubMed] [Google Scholar]
- 13.Shigenaga MK, Aboujaoude EN, Chen Q, Ames BN. Assays of oxidative DNA damage biomarkers 8-oxo-22 -deoxyguanosine and 8-oxoguanine in nuclear DNA and biological fluids by high-performance liquid chromatography with electrochemical detection. Methods Enzymol. 1994;234:16–33. doi: 10.1016/0076-6879(94)34073-0. [DOI] [PubMed] [Google Scholar]
- 14.Kaur H, Halliwell B. Measurement of oxidized and methylated DNA bases by HPLC with electrochemical detection. Biochem J. 1996;318(Pt 1):21–3. doi: 10.1042/bj3180021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asami S, Hirano T, Yamaguchi R, Tomioka Y, Itoh H, Kasai H. Increase of a type of oxidative DNA damage, 8-hydroxyguanine, and its repair activity in human leukocytes by cigarette smoking. Cancer Res. 1996;56:2546–9. [PubMed] [Google Scholar]
- 16.Nath KA, Norby SM. Reactive oxygen species and acute renal failure. Am J Med. 2000;109:665–78. doi: 10.1016/s0002-9343(00)00612-4. [DOI] [PubMed] [Google Scholar]
- 17.Paller MS, Hoidal JR, Ferris TF. Oxygen free radicals in ischemic acute renal failure in the rat. J Clin Invest. 1984;74:1156–64. doi: 10.1172/JCI111524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serafini M, Del Rio D. Understanding the association between dietary antioxidants, redox status and disease: Is the total antioxidant capacity the right tool? Redox Report. 2004;9:145–52. doi: 10.1179/135100004225004814. [DOI] [PubMed] [Google Scholar]
- 19.Sehirli AO, Sener G, Satiroglu H, Ayanoðlu-Dülger G. Protective effect of N-acetylcysteine on renal ischemia/reperfusion injury in the rat. J Nephrol. 2003;16:75–80. [PubMed] [Google Scholar]
- 20.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–60. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 21.Mandel LJ, Schnellmann RG, Jacobs WR. Intracellular glutathione in the protection from anoxic injury in renal proximal tubules. J Clin Invest. 1990;85:316–24. doi: 10.1172/JCI114440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003;17:1195–214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 23.Halliwell B, Gutteridge JM. 3rd ed. Oxford, UK: Oxford University press; 1999. Free radicals in biology and medicine. [Google Scholar]
- 24.Zastawny TH, Altman SA, Randers-Eichhorn L, Madurawe R, Lumpkin JA, Dizdaroglu M, et al. DNA base modifications and membrane damage in cultured mammalian cells treated with iron ions. Free Radic Biol Med. 1995;18:1013–22. doi: 10.1016/0891-5849(94)00241-b. [DOI] [PubMed] [Google Scholar]
- 25.Lukienko PI, Mel’nichenko NG, Zverinskii IV, Zabrodskaya SV. Antioxidant properties of thiamine. Bull Exp Biol Med. 2000;130:874–6. [PubMed] [Google Scholar]


