Abstract
The main machinery responsible for cellular protein maintenance is the ubiquitin-proteasomal system, with its core particle the 20S proteasome. The main task of the system is a fast and efficient degradation of proteins not needed anymore in cellular metabolism. For this aim a complex system of regulators evolved, modifying the function of the 20S core proteasome. Here we summarize shortly the structure of the 20S proteasome as well as its associated regulator proteins.
Keywords: 20S proteasome, 26S proteasome, Proteasomal regulators
Graphical abstract
Highlights
► Form and function of the 20S proteasome are described. ► Comparison between Archaeal and eukaryotic 20S proteasome are pointed out. ► Structure and function of proteasomal regulators are highlighted.
Introduction
The 20S proteasome is a multisubunit protease that can be found even in the oldest bacteria (archaea) as well as in “modern” plants and animals. Evolutionary the proteasome gained new functions supported by the development of several inducible subunits and regulatory proteins. Those regulatory protein complexes are able to bind to the 20S proteasome, changing its substrate specificity and activity. In mammalian cells this complex system contains the 20S proteasome, several regulators and inhibitors and is involved in almost every cellular function as well as in maintenance of cellular functionality.
The 20S proteasome is a cylindrical structure, built of four rings, containing seven subunits each. The rings are arranged in the sequence α-β-β-α, while the α-rings contain only α-, the β-rings only β-subunits, accumulating a mass of about 700 kDa (Fig. 1). In the archaeal form, the α-rings contain only one type of α-subunits, as well as the β-rings contain only one type of β-subunits. Substrate access to the inner proteolytic chamber is regulated by the α-rings, while the proteolytic activity is localized on the β-subunits, pointing to the inside of the proteolytic chamber that is formed between the β-rings (Fig. 2). In order to change substrate specificity, the archaeal 20S proteasome can bind to PAN (proteasome-activating nucleotidase) [1,2], a regulator protein, that enables the 20S proteasome to degrade natively folded proteins in an ATP-dependent way (Fig. 3).
Fig. 1.
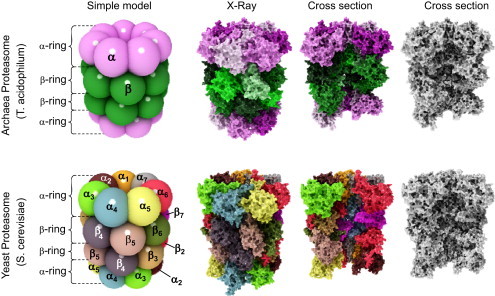
Structure of the archaeal and “modern” proteasome.
The proteasome (both the archaeal and the “evolutionary advanced” form) is a cylindrical structure formed of four rings, containing seven subunits, each. The rings are arranged in the sequence α-β-β-α.
The upper row of this figure shows the structure of the archaeal 20S proteasome, the row below the structure of the eukaryotic form. In both cases, the most left images show a simplified ball-model, different subunits are color coded. The archaeal proteasome contains only one type of alpha-subunits (pink) and only one type of beta-subunits (green). The “modern” proteasome contains 7 different alpha- and beta-subunits (left image of the bottom row). Thus, the whole complex adds up an overall mass of about 700 kDa and 28 subunits (both in the ancient and the “modern” form). In the case of the archaeal proteasome, there are only two (α and β), in the case of the “modern” proteasome, there are 14 (α1–α7 and β1–β7) different subunits.
In both rows the second image shows a high-detailed structure computed from data of X-ray-diffraction. For a better visualization, the different subunits of the archaeal proteasome are color-coded, too (pink-shades for alpha- and green-shades for beta-subunits). The different subunits of the “modern” proteasome have the same color as in the simple ball-model. The third image in both rows shows the inner structure after removing of several subunits from every single ring, exposing the inner structure. The last image in both rows shows the same as the third one, but in grayscale for a better visibility of the inner structure.
The proteolytic activity is located on the beta-subunits; thus, the archaeal proteasome has an overall of 14 proteolytic centers, the eukaryotic proteasome has only two sets of three (β1, β2, and β5). The alpha rings regulate substrate access to the proteolytic centers facing the inside of the proteasome (see Fig. 2 for more detailed information) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Fig. 2.
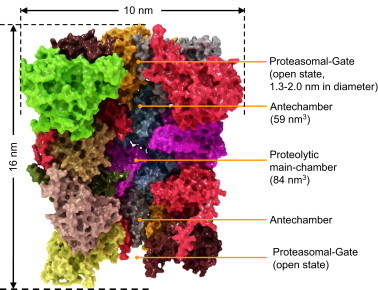
Structure of the eukaryotic proteasome.
In this image the single subunits are color coded (same colors as in Fig. 1). The whole proteasome is about 16 nm in height and has a diameter of about 10 nm [13].
The inside of the proteasome is subdivided into three chambers: two antechambers (with a volume of about 59 nm3, each) formed between an alpha- and a beta-ring, as well as the main proteolytic chamber (volume of about 84 nm3) between the two beta-rings [14,15]. The centers of the proteolytic active subunits face the inside of the main chamber, while substrate access is regulated by the alpha-rings, mediated by the N-terminal ends (last nine amino acids) of its single subunits. The exact function of the antechambers is still unclear. In the eukaryotic proteasome, proteolytic activity is localized at the subunits β1, β2, and β5, thus 6 different proteolytic sides are available.
β1 has a peptidyl-glutamyl-peptide-hydrolyzing-activity (caspase-like, cleaving after acidic amino acids, also termed as “post-glutamyl-peptide hydrolytic”-activity), β2 provides a trypsin-like activity (cleaves after basic amino acids) and β5 a chymotrypsin-like activity (cleavage after neutral amino acids) [16]. The individual proteolytic activity is β5⪢β2>β1. Mouse mutants with both β5 and β2 knocked out are lethal, while β5/β1- and β1/β2-knockout-combinations are viable.
In the normal (non-activated) state, the proteasomal activity is very low. The N-terminal ends of the alpha-subunits block (gate) the channel (annulus) leading to the proteolytic main chamber. In this state, substrate access is not possible. The main subunits involved in gating of the proteasome are α2, α3, and α4, while the gate is formed by the last 10 amino acids of their N-terminal ends. Those N-terminal ends show a common motif, the so-called YDR-motif (Tyr8-Asp9-Arg10) that may act as a “joint”, bending away the blocking N-termini from the annulus [17]. α3ΔN-mutants in yeast, missing the last 9 amino acids reveal a constantly high proteolytic activity, while the activity of α7ΔN-mutants is not significantly increased [18]. The key element is the α3-subunit, virtually blocking the gate with its N-terminus like a bar.
As mentioned, the alpha-rings recognize proteasomal substrates or hydrophobic patches in substrates of unfolded proteins. The exact mechanism of substrate-recognition is still unclear. It may be possible that the exposed hydrophobic sequences of soluble proteins bind to the alpha-rings and induce a conformational change, opening the gate to the main chamber. A similar mechanism of recognition (exposed hydrophobic sequences) is found in the heat shock proteins Hsp70 and Hsp90 [19]. The gate of the 20S proteasome in the “non-activated” state has a diameter of about 0.9, after activation the annulus is extended to about 1.3–2.0 nm, sufficient for the entrance of a protein chain. The products of proteolytic degradation of unfolded proteins show different lengths in a range from 2–35 amino acids with three maxima: 2–3, 8–10, and 20–30 amino acids [20].
Fig. 3.
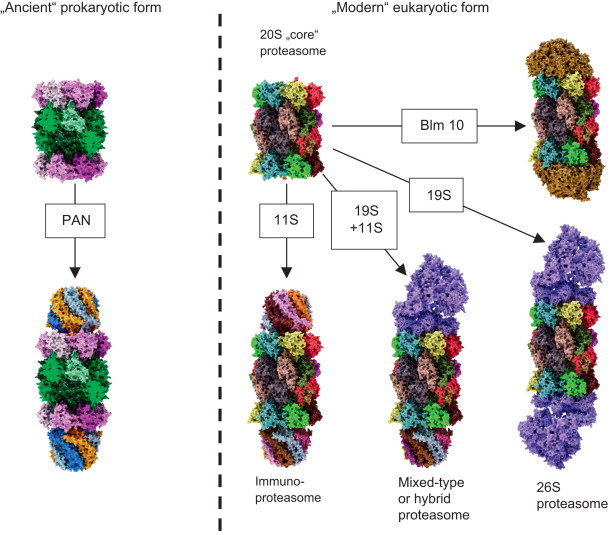
Regulator proteins of the archaeal and “modern” proteasome.
This figure shows the diversity of complexes of the archaeal (left part of the figure) and eukaryotic (right part of the figure) 20S proteasome with different regulator proteins. PAN: the archaeal 20S proteasome (left side of the image) can only bind to a single type of regulator protein, the so-called PAN, an AAA-ATPase [21]. PAN is built of six identical nucleotidases, forming a dome-shaped structure of about 650 kDa that binds to one or both alpha-rings. PAN enables the ATP-consuming degradation of natively folded proteins, while the “uncapped” 20S can only degrade already unfolded substrates in an ATP-independent way. A special labeling of the substrate proteins with ubiquitin is not necessary. The energy, provided by ATP, is needed only for unfolding the substrate that takes place at the surface of PAN, for gate-opening of 20S and for substrate translocation. For this, a consumption of about 300–400 ATP-molecules was estimated by Benaroudj et al. [22] per processing of a whole native protein (in this case Benaroudj et al. used a GFP with a C-terminal extension, summarizing about 250 amino acids). The degradation of the substrate itself by 20S is actually ATP-independent. Just the binding of ATP to PAN turned out to be sufficient for formation of the PAN–20S-complex, opening of the annulus, translocation and proteolytic degradation of unfolded substrate proteins; in contrast, unfolding and degradation of native proteins by the PAN–20S-complex were dependent from ATP-hydrolysis.
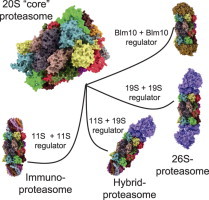
The eukaryotic form of 20S can bind to different regulator caps (right part of this figure). Sometimes 20S binds to different regulators at the same time. 11S regulator: the so-called immunoproteasome can be induced by IFN-γ, TNF-α, or lipopolysaccharides [23]. In this case instead of the constitutive proteolytic subunits (β1, β2, and β5) of the constitutive 20S proteasome (c20S) the inducible forms (iβ1, iβ2, and iβ5) are used for during proteasomal synthesis. Due to a higher affinity of iβ5 compared to the constitutive β5 to Ump1 – this complex is the initiator of 20S proteasome-assembly – in the presence of iβ5 the formation of immunoproteasomes has a higher probability. Though, inducible and constitutive subunits are coexpressed, resulting in formation of proteasomes that contain between a single one and the maximum of six of the inducible subunits, while the other subunits are the constitutive ones. The inducible form of the proteasome shows a significant shorter half-life of about 27 h compared to the constitutive form (8–12d) [24]. Thus, the cell is able to respond quickly to pathogens or inflammatory processes and the inducible proteasome is also removed quickly, too. During aging, the amount of immunoproteasomes increases, especially in postmitotic aging cells as in skeletal muscles, neurons, and astrocytes. Together with the inducible proteasomal subunits, the 11S regulator protein (also termed as “PA28” or “REG”) is induced. Until now, three different 11S subunits are known: PA28α, PA28β, and PA28γ. Both PA28α and PA28α are mainly found in the cytosol, mainly in immune tissues. PA28γ is found in the nuclei of cells. These subunits are able to form different hexa- and heptamers. In cells PA28α3β3, PA28α4β3 and PA28α3β4 (the subunits are arranged in an alternating way) occur. PA28γ (not inducible by IFN-γ) forms PA28γ7-homohepamers. The 11S-complex is 9 nm high, showing a diameter of about 6 nm; its central channel has a diameter of 2–3 nm and induces proteasomal activity significantly: β2-activity is increased 10-fold, both β1 and β5 50-fold, each [25,26]. PA28γ increases only the proteolytic activity of β2, while the other two activities are inhibited (to about 50%). The main task of the immunoproteasome is the production of short oligopeptides with hydrophobic C-termini, that can be presented by MHC-I-molecules (immunohistocompatibility complex-I) on the surface of the cell. After recognition of unknown antigens by CD8+ T-cells, the presenting cell is destroyed. As the constitutive proteasome, the immunoproteasome is only able to degrade (already unfolded) substrates in an ATP-independent way. 19S regulator: the 26S proteasome is technically a 20S “core” proteasome that binds a single 19S subunit (19S–20S), in contrast to the 30S proteasome (19S-20S-19S). Though, both conformations are referred to as “26S” in the literature. 19S is the homolog of the archaeal PAN regulator protein (see above). The 19S regulator contains at least 19 different (known) subunits: 6 subunits show an ATPase activity (Rpt1-Rpt6), the others do not (Rpn1–12 and Rpn15). The Rpt-subunits form a hexameric ring, that binds the alpha-ring of the proteasome, the Rpn-subunits form a lid-like structure, that enables recognition and binding (Rpn10 and -13) of substrates. Binding of 19S activates the 20S “core” proteasome (Rpt2, -3, and -5 are involved) and is dependent of the presence of both ATP and Mg2+. The whole 19S-complex has a mass of about 700 kDa. The 26S proteasome is the only conformation of the eukaryotic proteasomal form that is able to degrade natively folded and fully functional substrate proteins in an ATP-consuming way. Labeling of those substrates is mediated by polyubiquitination: a very complex system, involving four different types of E-enzymes (E1-E4), recognizes substrates and attaches a chain of ubiquitin (Ub) molecules that provide the degradation signal to be recognized by the 19S regulator. Hybrid proteasomes (19S-20S-11S): binding of both an 11S and 19S regulator cap forms the “mixed”-type (or “hybrid”) proteasome, that can both degrade ATP-dependent and -independent. Its exact function is still not clear, it may be possible that it processes oligopeptides both ATP-dependent and -independent, contributing to a widespread pool of oligopeptides for MHC-I presentation. Blm10/PA200: the third regulator of the proteasome is Blm10 (in yeast); the mammalian homolog is termed as “PA200” (for protein activator 200 kDa). PA200 is only found in the nucleus of mammalian cells and its exact function is still unclear, though it turned out to be involved in spermatogenesis [27] and DNA-repair [28]. Three different isoforms are known: PA200i, ii, and iii, while only PA200i really seems to bind the proteasome. In contrast to other regulators it is a monomer, a dome-like shaped protein 10 nm in diameter and about 6 nm high with a weight of about 200 kDa. Blm10 binds to the alpha-ring of the proteasome interacting with all alpha-subunits (while the mammalian PA200 does not interact with α7). After binding, gate-opening is induced and the degradation of small protein fragments is increased. Besides its role as a proteasomal regulator, Blm10 is induced after DNA-damage by ionizing radiation and accumulates as Blm10–20S complex on chromatin [29]. A further complex inducible by ionizing radiation, too, is 19S-20S-Blm10, that accumulates 24 h after exposure and shows a 19-fold increased β1-activity and a 6-fold of β2. The 20S concentration at the chromatin increases 8-fold.
In contrast to the archaeal form, the “modern” eukaryotic form of the 20S proteasome contains both seven different α-(α1–α7) and β-subunits (β1–β7) (Fig. 1). Proteolytic activity is localized only on three (instead of all) β-subunits (β1, β2, and β5). Furthermore, in mammals, a different set of active β-subunits can be used in de novo synthesized proteasomes to form the so-called inducible proteasome, playing a major role in MHCI-mediated presentation of antigens during immune response. The expression of the inducible fo5rms can be triggered by TNF-α, IFN-γ or lipopolysaccharides [3,4]. Together with the inducible form of the 20S proteasome, the 11S regulator protein is expressed, forming the “immunoproteasome” via binding to the inducible form of 20S (i20S). The immunoproteasome shows increased activity and produces short oligopeptides (8–10 amino acids) with hydrophobic C-termini, ideal for MHC-I-presentation on the cells surface to CD8+-cells (cytotoxic T-lymphocytes).
While the 20S “core” proteasome (Fig. 2) is only able to recognize and to degrade damaged/misfolded proteins that are already unfolded, the 26S proteasome, main protease of the ubiquitin–proteasome-system (UPS) is able to degrade natively folded and functional proteins in an ATP- and ubiquitin-dependent way. The 26S proteasome is formed by a 20S “core” proteasome, that binds one or two so-called 19S regulators (also termed as PA700) [5,6] (Figs. 3 and 4). Binding to this regulator increases proteasomal activity and enables the proteasome to degrade substrate proteins that have been labeled by a complex enzymatic machinery via the attachment of a chain of ubiquitin (Ub) molecules [7].
Fig. 4.
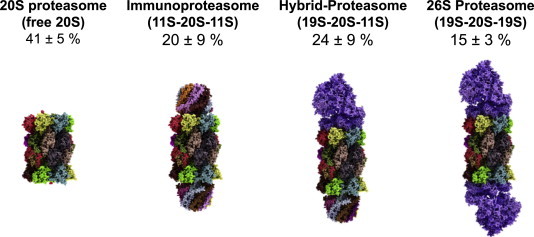
Relative amounts of free and regulator-bound 20S (in HeLa-cells).
In this figure one can see the relative amounts of free and regulator-bound proteasomes according to Tanahahsi et al. [30]. It turned out that proteasomes with only a single regulator cap attached are very short-lived intermediates, undetectable via immunochemical methods. Thus, in the shown image actually two regulator caps are attached and the 26S proteasome (right image) represents the 19S-20S-19S-configuration (the 30S proteasome, to be exactly), not the 19S-20S-form. The immunoproteasome (second image from the left) is also fit out with two 11S-regulators.
Labeling of natively folded proteins by ubiquitin is realized by four types of enzymes that work in a consecutive way [8]: the first step is the ATP-dependent ubiquitin-activation by E1 (“ubiquitin-activating enzyme”), followed by transfer of Ub to E2 (“ubiquitin-conjugating enzyme”). Some eight different E1- and a few dozen of E2-enzymes are known today. Substrate specificity is delivered by the E3-enzymes (about 650 known today): E3-Enzymes are specific for a single or just a very few target proteins that are bound to enable Ub-transfer from E2 to the substrate. Two different types of E3-mediated Ub-transfer are known: RING (direct Ub-transfer from E2 to the substrate) and HECT (Ub-transfer from E2 to E3 and from E3 to the substrate). About 600 E3-enzymes count among the RING-form, the rest to the HECT forms. After transfer of the first Ub to the substrate, the Ub-chain is quickly prolonged (a Ub4-chain provides the maximal signal for 26S-mediated proteolysis). The existence of E4-enzymes, responsible for the prolongation of ubiquitin chains, is still under discussion. It was claimed that E4 enzymes are a new class of enzymes, whereas others state that E4 enzymes are just a subclass of E3. Degradation of polyubiquitinated substrates follows the binding of the tagged substrate to the 19S-regulator, that unfolds the target protein (needing ATP), and threads the protein chain into the 20S “core” protein. The polyubiquitin chain is degraded into its monomers by deubiqinating proteins (DUBs) to be re-used by the E1-enzymes again. In addition to that the DUBs provide a delicate balance between polyubiquitination (a common posttranslational protein modification) and deubiquitination, permanently adjusting the steady state of the cellular protein pool in a very dynamic manner. Besides the degradation of not required proteins the UPS is also responsible for the “quality control” of protein folding and the endoplasmic-reticulum-associated degradation (ERAD) [9], since 30–80% [10] of all newly synthesized proteins are misfolded.
The proteasome has become a target in therapeutic treatment: proteasomal inhibitors are used in certain cancer forms, including multiple myeloma. Interestingly, similar strategies developed during evolution: the HIV-I virus expresses a protein (HIV-I Vpu) that targets C4D for ERAD-like degradation [11] and the human cytomegalovirus codes two proteins that transport MHC-I from the ER into the cytosol followed by 26S-mediated degradation. Prions (PrPSc) are actually able to inhibit the proteasome [12].
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Rabl J., Smith D.M., Yu Y., Chang S.C., Goldberg A.L., Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Molecular Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith D.M., Chang S.C., Park S., Finley D., Cheng Y., Goldberg A.L. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Molecular Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piccinini M., Mostert M., Croce S., Baldovino S., Papotti M., Rinaudo M.T. Interferon-gamma-inducible subunits are incorporated in human brain 20S proteasome. Journal of Neuroimmunology. 2003;135:135–140. doi: 10.1016/s0165-5728(02)00439-3. [DOI] [PubMed] [Google Scholar]
- 4.Stratford F.L., Chondrogianni N., Trougakos I.P., Gonos E.S., Rivett A.J. Proteasome response to interferon-gamma is altered in senescent human fibroblasts. FEBS Letters. 2006;580:3989–3994. doi: 10.1016/j.febslet.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 5.Demartino G.N., Moomaw C.R., Zagnitko O.P., Proske R.J., Chu-Ping M., Afendis S.J. PA700, an ATP-dependent activator of the 20S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. Journal of Biological Chemistry. 1994;269:20878–20884. [PubMed] [Google Scholar]
- 6.Liu C.W., Millen L., Roman T.B., Xiong H., Gilbert H.F., Noiva R. Conformational remodeling of proteasomal substrates by PA700, the 19S regulatory complex of the 26S proteasome. Journal of Biological Chemistry. 2002;277:26815–26820. doi: 10.1074/jbc.M201782200. [DOI] [PubMed] [Google Scholar]
- 7.Thrower J.S., Hoffman L., Rechsteiner M., Pickart C.M. Recognition of the polyubiquitin proteolytic signal. EMBO Journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung T., Catalgol B., Grune T. The proteasomal system. Molecular Aspects of Medicine. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Raasi S., Wolf D.H. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Seminars in Cell & Developmental Biology. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Rivett A.J., Hearn A.R. Proteasome function in antigen presentation: immunoproteasome complexes, peptide production, and interactions with viral proteins. Current Protein & Peptide Science. 2004;5:153–161. doi: 10.2174/1389203043379774. [DOI] [PubMed] [Google Scholar]
- 11.Gallegos J.R., Litersky J., Lee H., Sun Y., Nakayama K., Nakayama K. SCF TrCP1 activates and ubiquitylates TAp63gamma. Journal of Biological Chemistry. 2008;283:66–75. doi: 10.1074/jbc.M704686200. [DOI] [PubMed] [Google Scholar]
- 12.Deriziotis P., Andre R., Smith D.M., Goold R., Kinghorn K.J., Kristiansen M. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO Journal. 2011;30:3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomisugi Y., Unno M., Mizushima T., Morimoto Y., Tanahashi N., Tanaka K. New crystal forms and low resolution structure analysis of 20S proteasomes from bovine liver. Journal of Biochemistry. 2000;127:941–943. doi: 10.1093/oxfordjournals.jbchem.a022709. [DOI] [PubMed] [Google Scholar]
- 14.Maupin-Furlow J.A., Gil M.A., Humbard M.A., Kirkland P.A., Li W., Reuter C.J. Archaeal proteasomes and other regulatory proteases. Current Opinion in Microbiology. 2005;8:720–728. doi: 10.1016/j.mib.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Maupin-Furlow J.A., Kaczowka S.J., Reuter C.J., Zuobi-Hasona K., Gil M.A. Archaeal proteasomes: potential in metabolic engineering. Metabolic Engineering. 2003;5:151–163. doi: 10.1016/s1096-7176(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 16.Groll M., Huber R. Inhibitors of the eukaryotic 20S proteasome core particle: a structural approach. Biochimica et Biophysica Acta. 1695;33-44:2004. doi: 10.1016/j.bbamcr.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Groll M., Bajorek M., Kohler A., Moroder L., Rubin D.M., Huber R. A gated channel into the proteasome core particle. Nature Structural & Molecular Biology. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 18.Bajorek M., Finley D., Glickman M.H. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Current Biology. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 19.Pratt W.B., Morishima Y., Peng H.M., Osawa Y. Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Experimental Biology and Medicine. 2010;235:278–289. doi: 10.1258/ebm.2009.009250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler A., Cascio P., Leggett D.S., Woo K.M., Goldberg A.L., Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Molecular Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 21.Bar-Nun S., Glickman M.H. Proteasomal AAA-ATPases: structure and function. Biochimica et Biophysica Acta. 1823;67-82:2012. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Benaroudj N., Zwickl P., Seemuller E., Baumeister W., Goldberg A.L. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Molecular Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 23.Nelson J.E., Loukissa A., Altschuller-Felberg C., Monaco J.J., Fallon J.T., Cardozo C. Up-regulation of the proteasome subunit LMP7 in tissues of endotoxemic rats. Journal of Laboratory and Clinical Medicine. 2000;135:324–331. doi: 10.1067/mlc.2000.105615. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka K., Ichihara A. Half-life of proteasomes (multiprotease complexes) in rat liver. Biochemical and Biophysical Research Communications. 1989;159:1309–1315. doi: 10.1016/0006-291x(89)92253-5. [DOI] [PubMed] [Google Scholar]
- 25.Di C.D. Human erythrocyte contains a factor that stimulates the peptidase activities of multicatalytic proteinase complex. Italian journal of Biochemistry. 1992;41:213–224. [PubMed] [Google Scholar]
- 26.Kuehn L., Dahlmann B. Proteasome activator PA28 and its interaction with 20 S proteasomes. Archives of Biochemistry and Biophysics. 1996;329:87–96. doi: 10.1006/abbi.1996.0195. [DOI] [PubMed] [Google Scholar]
- 27.Khor B., Bredemeyer A.L., Huang C.Y., Turnbull I.R., Evans R., Maggi L.B., Jr. Proteasome activator PA200 is required for normal spermatogenesis. Molecular and Cellular Biology. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ustrell V., Hoffman L., Pratt G., Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO Journal. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blickwedehl J., McEvoy S., Wong I., Kousis P., Clements J., Elliott R. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiation Research. 2007;167:663–674. doi: 10.1667/RR0690.1. [DOI] [PubMed] [Google Scholar]
- 30.Tanahashi N., Murakami Y., Minami Y., Shimbara N., Hendil K.B., Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. Journal of Biological Chemistry. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]


