Abstract
4-Hydroxy-2-nonenal (HNE) is one of the most studied products of phospholipid peroxidation, owing to its reactivity and cytotoxicity. It can be formed by several radical-dependent oxidative routes involving the formation of hydroperoxides, alkoxyl radicals, epoxides, and fatty acyl cross-linking reactions. Cleavage of the oxidized fatty acyl chain results in formation of HNE from the methyl end, and 9-oxo-nonanoic acid from the carboxylate or esterified end of the chain, although many other products are also possible. HNE can be metabolized in tissues by a variety of pathways, leading to detoxification and excretion. HNE-adducts to proteins have been detected in inflammatory situations such as atherosclerotic lesions using polyclonal and monoclonal antibodies, which have also been applied in ELISAs and western blotting. However, in order to identify the proteins modified and the exact sites and nature of the modifications, mass spectrometry approaches are required. Combinations of enrichment strategies with targetted mass spectrometry routines such as neutral loss scanning are now facilitating detection of HNE-modified proteins in complex biological samples. This is important for characterizing the interactions of HNE with redox sensitive cell signalling proteins and understanding how it may modulate their activities either physiologically or in disease.
Abbreviations: DHN-MA, 1,4-Dihydroxynonane-mercapturic acid; DNPH, 2,4-Dinitrophenylhydrazine; ESI, Electrospray ionization; FT-ICR, Fourier transform ion cyclotron resonance; HNE, 4-Hydroxy-2-nonenal; HODA, 9-Hydroxy-12-oxo-10(E)-dodecenoic acid; HPETE, Hydroperoxyeicosatetraenoic acid; HPODE, Hydroperoxyoctadecadienoic acid; KODA, 9-Keto-12-oxo-10(E)-dodecenoic acid; Mab, Monoclonal antibody; MALDI, Matrix assisted laser desorption ionization; MDA, Malondialdehyde; MS, Mass spectrometry; ONA, 9-Oxo-nonanoic acid; ONE, 9-Oxo-2-nonenal; PETE, Peroxyeicosatetraenoate; PODE, Peroxyoctadecadienoate
Keywords: Anti-HNE antibodies, Hydroxyalkenal, HNE-protein adducts, Mass spectrometry, Neutral loss scanning, Redox signalling
Graphical Abstract
4-Hydroxy-2-nonenal can form Schiffs base or Michael adducts with residues in proteins, which can be analyzed by tandem mass spectrometry (MSMS) to identify the protein and characterize the location and nature of the modification.
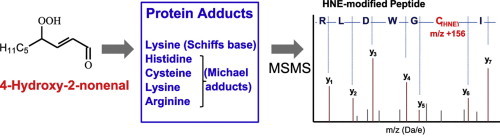
Highlights
► Summary of proposed mechanisms of 4-hydroxy-2-nonenal formation. ► Overview of the development of antibodies for immunodetection of 4-hydroxy-2-nonenal. ► Advances in detection of 4-hydroxy-2-nonenal adducts of proteins by mass spectrometry.
Discovery and importance of 4-hydroxy-2-nonenal
Phospholipid peroxidation generates a large number of diverse products, including breakdown products resulting from cleavage of the oxidized fatty acyl chain. One of the most studied products, which results from oxidation of phospholipids containing ω-6 polyunsaturated fatty acyl chains, is 4-hydroxy-trans-2-nonenal (HNE). This compound falls into the class of α, β-unsaturated aldehydes, which are bi-reactive compounds able to undergo electrophilic reactions such as Michael addition, as well as Schiff base formation. The compound now known to be HNE was first detected and characterized as a 4-hydroxyalkenal by Schauenstein et al. in the early 1960 s [1], although unfortunately it was initially mis-identified as 4-hydroxyoctenal, based on inconclusive microanalysis data that suggested the presence of between eight and nine carbons. At the time there was little interest in these hydroxyalkenals, which were thought to be uninteresting by-products of lipid autoxidation during rancidification, despite the fact that they showed potential tumoristatic activity and selective toxicity in tumour cell lines compared to non-cancerous cells [1,2]. Subsequently the structure of HNE compound was correctly characterized, and it was found to occur in rat liver subjected to oxidative damage by carbon tetrachloride [3]. The hydroxyalkenals were found to be the major low molecular weight products of biological lipid peroxidation and of these, HNE was found to be the most cytotoxic, with biological effects in the low micromolar range [4], as well as the most abundant following oxidation of arachidonic acid [5]. The ensuing collaboration between the Austrian and Italian research groups marked a turning point in this research [6]; subsequently scientific interest in this molecule expanded exponentially to address all aspects of its biochemistry and roles in pathophysiology.
Mechanisms of 4-hydroxy-2-nonenal generation
For many years, the chemical reaction mechanisms by which HNE or analogous 4-hydroxyalkenals could be formed were not well understood. Early experiments demonstrated that ω-6 polyunsaturated fatty acyl chains were required for HNE formation, whereas oxidation of ω-3 polyunsaturated chains gave rise to its analogue 4-hydroxyhexenal, indicating that these compounds are derived from the methyl end of the fatty acyl chain following C3C bond cleavage [7–9]. The majority of studies carried out to elucidate the mechanisms used purified lipids, especially linoleic acid and linoleate methyl ester, although γ-linolenic and arachidonic acids were also found to be good sources of HNE. Over the years, many different mechanisms have been proposed, with either two or three O2 additions required, and with several possible sites of oxidation owing to the radical rearrangements following hydrogen abstraction at bis-allylic sites. The chemical mechanisms that lead to cleavage of a carbon–carbon bond in the fatty acyl chain can be broadly classified into the following types: reduction of the hydroperoxide to an alkoxy radical by Fe2+ followed by β-scission; a Hock rearrangement of a hydroperoxide, entailing an acidified hydroperoxide (3OH2+) acting as a leaving group, and migration of a C3C to a C3O bond and cleavage; or dioxetane formation and cleavage, especially via peroxycyclization reactions (Fig. 1).
Fig. 1.
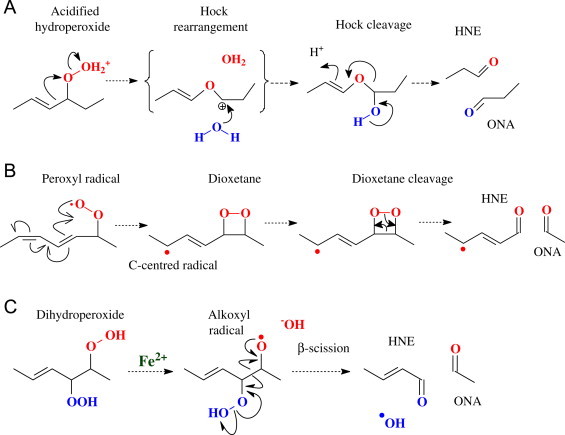
Generic mechanisms for cleavage of oxidized fatty acyl chains to yield HNE and ONA. (A) Protonation of the hydroperoxide yields a good leaving group and rearrangement of a C3C to C3O bond. The resulting carbonium ion is unstable, and hydrolysis occurs. (B) A peroxy radical cyclizes to form a dioxetane and carbon-centred radical. The dioxetane can rearrange by a 2-electron process resulting in cleavage of the dioxetane ring. The C-centred radical is susceptible to attack by oxygen to form a 2nd hydroperoxide, probably before fragmentation occurs. (C) In the presence of transition metals, hydroperoxides are converted to alkoxy radicals, which initiate a radical rearrangement resulting in β-scission.
The earliest suggestions for mechanisms for the formation of HNE were based on the observations of hydroperoxyperoxides as intermediates (9/13-hydroperoxyoctadecadienoate [HPODE] from methyl-linoleate (Fig. 2) or 11/15-hydroperoxyeicosatetraenoate [HPETE] from methyl-arachidonate) and enhanced HNE and oxo-nonanoic acid (ONA) formation in the presence of transition metal ions, such as Fe2+, which are known to convert hydroperoxides to alkoxy radicals [7,9,10]. A mechanism involving addition of three O2 via 13-HPODE to form an epoxyhydroperoxide intermediate that underwent a Hock cleavage was proposed by Pryor and Porter [10], while Loidl-Stahlhofen et al. reported an Fe2+-dependent mechanism with a hydroxyhydroperoxy intermediate that underwent β-scission [11]. This mechanism generated HNE plus a vinylperoxy radical that readily converted to a known product, ONA, and was considered feasible because an analogous enzymatic route was discovered in plants, dependent on lipoxygenase and a hydroperoxide lyase [12]. An alternative mechanism reported a few years later involves cyclization of a peroxy radical (9-PODE or 11-PETE) to form a 9,10-dioxetane that could rearrange to cleave the chain [13]; interestingly, this route requires the addition of only two O2. Further support for a mechanism similar to hydrohydroperoxy fragmentation was presented by Schneider et al. [14], who found that 9S-HPODE is cleaved following a Hock rearrangement to yield 3Z-nonenal that is then oxidized further to 4-HPNE, whereas 13S-HPODE forms an unstable dihydroperoxide that undergoes Hock cleavage to give 4-HPNE directly. However, more recently the same group have proposed that cross-chain peroxyl radical addition and decomposition can occur, with formation of dimeric or even trimeric oxidized fatty acyl chains [15] and have obtained evidence for its occurrence in tetralinoleoyl cardiolipin [16]. It is argued that this route represents a competition with bis-allylic hydrogen abstraction, and therefore it seems most likely for linoleic acid, where there is no bis-allylic site at the critical stage of the mechanism. Finally, earlier this year evidence was presented for the role of a diepoxycarbinyl radical that can interconvert between 9-allyloxy and 13-allyloxy forms, and undergo β-scission to either HNE and ONE/ ONA, or hexanal plus KODA/HODA [17]. The structures of the several of these reactive intermediates are shown in Fig. 2. In conclusion, sound evidence has been obtained for several different routes from linoleate and arachidonate to HNE plus ONE, and hexanal plus HODA, which incidentally shows that hexanal is not formed only from ω-3 fatty acids.
Fig. 2.
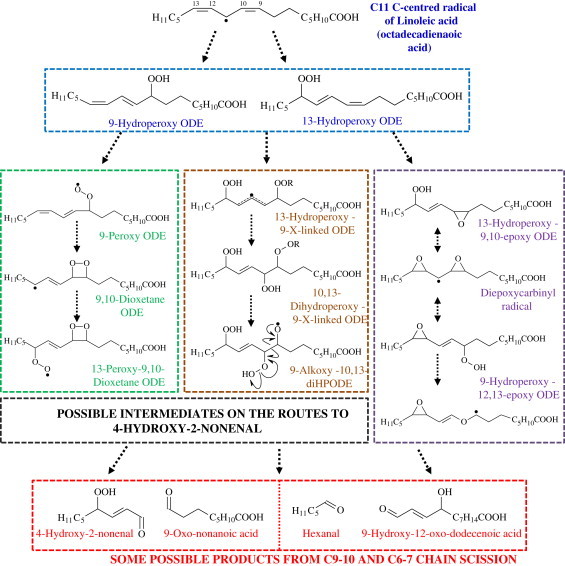
Formation of some intermediates on the pathways from linoleic acid (C18:2) to HNE. Analogous pathways exist for arachidonic acid (C20:4) with starting materials 11-hydroperoxy-eicosatetraenoic acid and 15-hydroperoxy-eicosatetraenoic acid.
Metabolism of 4-hydroxy-2-nonenal
In many tissues, HNE can be metabolized to less cytotoxic compounds, which are also more water-soluble, facilitating their excretion [18]. The use of tritiated HNE enabled early studies of the metabolic pathways in several laboratories, and hepatocytes, thymocytes and enterocytes from small intestine are known to be particularly efficient at metabolizing HNE [19]. An important pathway for detoxification of HNE involves conjugation to the thiol group of the antioxidant glutathione by glutathione-S-transferases, especially GSTA4–4 [20], which results in loss of the α,β-unsaturation and formation of HNE-GSH adduct or its lactone form. This can be reduced by aldose reductases to 1,4-dihydroxynonene-GSH, although the enzymes are also capable of reducing free HNE to 1,4-dihydroxynonene. Alternatively, aldehyde dehydrogenases (ALDH) are able to oxidize HNE to HNA, either in free or conjugated form, and HNA may be hydroxylated by cytochrome P450 4A to form 9-hydroxy-HNA. Studies of rats in vivo have found that HNE metabolites in urine are most commonly in the mercapturic acid form, with the major component reported to be 1,4-dihydroxynonane-mercapturic acid (DHN-MA) [21], and consequently an immunoassay for this urinary metabolite has been developed [22]. The importance of aldehyde dehydrogenase and aldose reductase for metabolism of HNE has been demonstrated in rat cardiac myocytes, using [3H]HNE [23]. More recently studies of myocardial ischaemia have shown that mitochondria contribute significantly to HNE metabolism by direct oxidation to HNA, rather than conjugation to GSH; in ischaemia this reaction is inhibited and increased formation of HNE-protein adducts occurs in the mitochondria. The mitochondrial oxidation of HNE was found to be regulated primarily by the NAD+/NADH ratio [24].
Analysis of free 4-hydroxy-2-nonenal
In understanding the formation and biological roles of HNE, an important aspect is its detection and measurement in tissues. There are a variety of methods available for analysis of HNE (Fig. 3), but in view of its reactivity a fundamental issue is whether to measure free aldehyde, or adducts of HNE. Methods for detection and quantification of free HNE have been reviewed by Esterbauer et al. [2,25]. HNE absorbs in the UV range (220–223 nm), and in protein-free samples with levels of HNE in the micromolar range it can be detected directly following separation from other aldehydes by HPLC. However, in biological samples, more specific and sensitive detection can be achieved by the use of aldehyde-reactive probes, of which 2,4-dinitrophenylhydrazine (DNPH) and 1,3-cyclohexandione (CHD) have been most used, but many others exist [26]. HPLC and use of standards is still required to identify HNE within the complex mixtures of alkanals, alkenals and modified alkenals that can occur, as the probes per se do not distinguish between the aldehydes. Alternatively, mass spectrometric analyses can be carried out, which have the advantage that the mass to charge ratio and fragmentation pattern can be used to identify the compound. GC–MS requires derivatization of the sample, for example with pentafluorobenzyl (PFB) oxime followed by silylation, and the derivatized HNE can be detected with negative chemical ionization and quantified by comparison with deuterated HNE as internal standard [27,28]. LCMS approaches to detecting HNE have the advantage that derivatization to obtain a volatile compound is not required, so the sample can be analyzed with minimal handling [29,30]. However, analysis of free HNE may have limited value: owing to its reactivity a large part of the HNE formed may exist as adducts with other biomolecules, such as proteins, DNA or aminophospholipids. The structures of HNE adducts with amino acid residues, including Schiffs base formation with lysine and Michael adducts with cysteine, histidine, lysine and arginine, have been nicely described in several reviews [31–33]. Consequently, many studies have focused on the detection of HNE adducts with proteins, and the two main approaches available are immunochemical techniques and mass spectrometric analysis. Although Schiffs base adducts are generally regarded as more readily reversible than Michael adducts, the latter can also be reversible, especially the lysine-HNE Michael adduct [34,35]. It is important to bear this in mind, as it can influence the adducts that are observed: for example, it has been reported that HNE Michael adducts represent more than 99% of the protein modifications detected [36], and that His-Michael adducts are most stable, although the most favourable reaction is Michael addition to lysine [37]. Consequently, many studies include a reduction step with borohydride, which stabilizes Schiffs bases and some Michael adducts, prior to analysis.
Fig. 3.
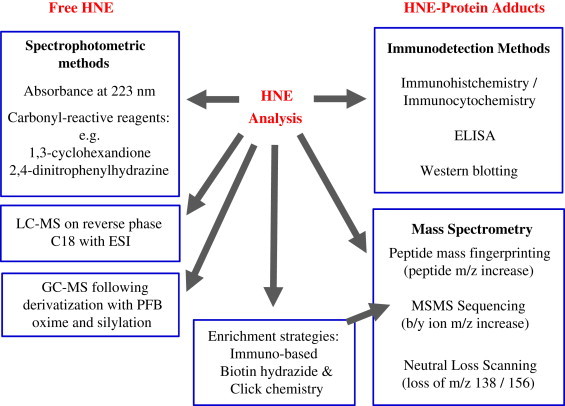
Overview of most common methods for detecting and analyzing HNE and its protein adducts.
Analysis of 4-hydroxy-2-nonenal adducts by immunoassay
The development of antibodies specific for HNE bound to proteins or other biomolecules has undoubtedly been extremely useful in establishing the range of pathologies where HNE is produced and showing its tissue distribution by immunohistochemistry. The earliest report of anti-HNE and anti-malondialdehyde (MDA) antibodies is by Palinski et al. [38], although antisera against other forms of modified LDL had been reported previously [39]. Polyvalent and monoclonal antibodies were raised against cyanoborohydride-reduced HNE-treated LDL, and it was reported that the antibodies recognized a 4-HNE-lysine adduct on LDL and other proteins, but were unreactive with native LDL [38,40]. In subsequent work in Austria, antibodies were raised against a non-reduced HNE-LDL adducts, and found that there was cross-reactivity with HNE-albumin, and to a lesser extent, HNE-HDL [41,42]. A solid phase fluorescent immunoassay was used to characterize the antiserum, and competition assays with HNE-modified poly-(l-amino acids) indicated that HNE-poly-(l-lysine) and (in decreasing order of effectiveness) HNE-poly-(l-tyrosine), HNE-poly-(l-arginine), and HNE-poly-(l-histidine) could be recognized. There was also cross-reactivity with LDL modified with 2,4-heptadienal and hexanal, but not with MDA-LDL.
Polyclonal serum against HNE-treated keyhole limpet haemocyanin (KLH) was produced by Uchida et al. [43], and this group went on to use this immunogen to produce five monoclonal antibodies with high specificity for HNE-histidine adducts on GAPDH, of which HNEJ-2 was the most effective [44]. Further monoclonal antibodies were produced by immunizing rabbits with HNE-conjugated heptapeptide (Gly3–His–Gly3 amide) coupled to KLH [45]. These antibodies showed no significant cross-reactivity with proteins treated with MDA, hexanal, 2-hexenal or 2-trans-nonenal, but there was some reactivity towards proteins treated with other hydroxyalkenals, namely 4-hydroxy-2-octenal and 4-hydroxy-2-decenal, as well as with HNE-cysteine and HNE-lysine. It was therefore concluded that the epitope recognized was the 2-CH3(CH2)n-5-hydroxytetrahydrofuran moiety, which is formed by cyclization of the Michael adduct [45]. Essentially in parallel, a series of monoclonal antibodies from mice were also generated by Waeg et al. [46], using BSA and KLH treated with 4-hydroxyalkenals. One of the clones, Ig4, was found to be highly specific for HNE-histidine adducts, with little cross-reaction with HNE-lys or HNE-cys, or with shorter hydroxyalkenals (e.g. 4-hydroxyhexanal). Later, a new monoclonal antibody with reactivity versus a fluorescent derivative of HNE with the ε-amino group of lysine was developed by conjugation of the hydroxyiminodihydropyrrole (HIDP) to KLH [47]; HIDP is known to form in LDL following HNE modification. The monoclonal antibody bound both HIDP and 4-oxo-nonenal readily, showed much lower reactivity against HNE-adducts, and very little activity versus all other aldehydes. Another group produced rabbit polyclonals with specificity for HNE adducts of cysteine and DNPH-labelled HNE-SH adducts [48], which are useful as DNPH is often used as a carbonyl-specific labelling reagent. The first antibody to a hydroxyalkenal cross-linked product was prepared by Cohn et al. [49], who raised polyclonal antibodies to HNE-cross-linked lysine residues (reduced Schiffs base and a Michael addition) on glucose-6-phosphate dehydrogenase. Interestingly, a mAb raised against the R-HNE-histidine adduct was found to have sequence homology with anti-DNA autoantibodies detected in systemic lupus erythematosus, and cross-reactivity with 4-oxo-2-nonenal-treated DNA, thus showing dual specificity of some anti-HNE antibodies [50].
There has been interest from the early stages in the development of ELISAs as quantitative techniques for analyzing HNE-protein adducts. In 1995, an “HNE-trapping ELISA” designed to quantify free HNE by its reaction with protein coating the plate, and HNE-His adducts were detected using mAbHNEJ-2 [51]. A rapid competition ELISA for detecting pre-formed HNE-His adducts on proteins was subsequently reported by the same group, and used to detect immunoreactive components in rat serum and urine [52]. More recently, a kit for detecting HNE-his adducts in lysates of cultured cell subjected to slight or severe oxidative stress has been developed [53], and shows good sensitivity for cell samples although it requires refinement for use with plasma. An enzyme immunoassay for quantitative analysis of the urinary HNE metabolite 1,4-dihydroxynonane-mercapturic acid (DHN-MA) has also been reported, which uses polyclonal serum raised against DHN-MA linked to KLH [22].
Many of the anti-HNE adduct antibodies developed were used for immunocytochemistry and immunohistochemistry, and demonstrated the occurrence of HNE-modified LDL and proteins in inflammatory loci such as human atheromatous lesions [38,45,47] or in oxidatively stressed mitochondria [49], cultured cells [33], apoptotic hippocampal neurones [54], or ischaemic rat heart [55]. There are also a number of more recent studies, for example in neurodegenerative disease [56,57]. Anti-HNE antibodies were used for western blotting of extracts of Alzheimers disease brain, and HNE-adducts were found on a number of energy metabolism proteins (e.g. malate dehydrogenase and enolase) as well as Mn superoxide dismutase. The neuronal communication protein dihydropyrimidinase-related protein 2 (DRP-2) also showed increased HNE adducts in early Alzheimer's disease [58,59]. However, the exact sites of adduction were not identified in these studies, limiting confidence in the hypothesis that these specific proteins were modified.
Labelling approaches to detect 4-hydroxy-2-nonenal-protein adducts
3H-labelled HNE has been used to determine the metabolic fate of this lipid peroxidation product in tissues and animal studies, as described above, and has allowed some basic assessments of the cellular locations of protein-bound HNE [24]. An alternative approach to identifying cellular targets of attack by electrophilic lipid products is to use electrophilic compounds pre-labelled with either biotin or a fluorescent tag such as bodipy. Cells or tissues are treated with the tagged compounds, which allow the proteins covalently bound to the lipids to be identified [26]. While initially used for phospholipids [60,61], a similar approach for HNE involving click chemistry has been reported for HNE [62]. Azido-tagged HNE or alkynyl-tagged HNE was incubated with cultured colorectal cells to allow adduction to target proteins, and after extraction biotin-triphenylphosphine or the appropriate biotin label for click chemistry were added to label the lipid covalently. While this is a smart and informative protocol, unfortunately it cannot be translated to identify lipoxidized proteins in vivo in humans, owing to the requirement for artificially labelled lipid in the tissue of interest. Ideally, it also requires the use of mass spectrometry techniques to identify the tagged protein.
Analysis of 4-hydroxy-2-nonenal adducts by mass spectrometry
While the use of HNE-specific antibodies in ELISAs and immunostaining allows detection and quantitation of adducts, there can be questions over the exact nature of adducts and the protein modified. The most informative, though at the same time the most challenging, method for characterizing HNE adducts is undoubtedly mass spectrometry (MS). MALDI-TOF-MS is often used for peptide mass fingerprinting and adducts can be detected by the increase in mass to charge ratio of the peptide compared to the native form. MSMS allows sequencing of proteins and identification of the site of post-translational modifications, including the formation of adducts by lipid peroxidation breakdown products (Fig. 4). Samples are usually reduced with dithiothreitol and treated with iodoacetamide to block cysteines and prevent cross-linking, before subjecting to digestion with MS grade trypsin. The tryptic peptides are separated by (nano)LC and sequenced by fragmentation in the mass spectrometer. The data acquired is usually processed through a software programme called Mascot, which matches it to in silico tryptic peptide data calculated using proteomic databases, by a statistical process. The masses for HNE modifications to the various amino acids, for example by Schiffs base (138 Da) or Michael addition (156 Da) [32,63], can be added to the search database as variable modifications. However, owing to the statistical nature of the processing algorithm it is important to check the MS data manually to confirm the modifications. The analysis is complicated further by the possibility of reduced products (corresponding to addition of 2 Da) following treatment of the samples with sodium borohydride or cyanoborohydride to stabilize the adducts, or structural rearrangements, such as the cyclization to a pyrrole form. MS methodologies have been used both to investigate the reaction of HNE (or other aldehydes) in vitro with individual proteins, and to identify HNE-modified proteins in biological samples.
Fig. 4.
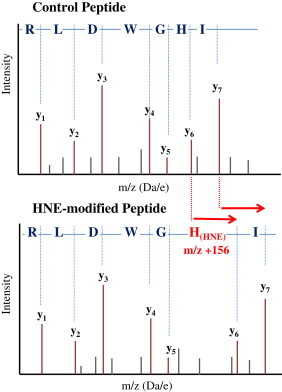
Schematic mass spectra showing how the formation of a Michael adduct at a histidine residue affects the fragmentation pattern of a peptide during MSMS sequencing by adding m/z 156 to the mass of the corresponding y6 and y7 fragment ions. The y ions are shown in brown; the grey peaks represent b ions or other minor fragment ions.
Liu et al. [64] conducted a thorough investigation of myoglobin modification by HNE and ONE adducts using LC-MSMS and identified covalent adducts on several peptides, and this work was expanded later to the reactions of other electrophilic oxidation products of linoleic acid with β-lactoglobulin as a model proteins; mass shifts for many the adducts were reported [35]. Formation of adducts between HNE and six histidine residues of myoglobin, resulting in instability has also been reported [65]. Other examples of characterization of HNE modifications in vitro include the detection of adducts at histidine, lysine and arginine residues of cytochrome c [66], and histidine and cysteine residues of creatine kinase analyzed by FT-ICR MS, which correlated with decreased activity of the protein [67]. This is similar to an earlier report on the loss of GAPDH activity following treatment in vitro with HNE, which was linked to the modification of lysine, histidine and cysteine residues observed by LC-MSMS, although the active site cys149 was found not to be affected [68].
There have also been studies of HNE adduct formation in vivo. MALDI-TOF-MS analysis has been used to show modification of erythrocyte catalase by HNE in systemic lupus erythematosus, but the actual site of modification was not reported [69]. In a rat model of chronic alcoholic liver diseased Hsp90 was identified by immunoblot with anti-HNE-antibody as a target of HNE, although Cys572 was only identified as the site of adduction by treatment of Hsp90 in vitro and LC/MS/MS [70]. Similar work in mice investigated modification of proteins from obese adipose tissue, except that carbonyl-containing proteins were enriched using biotin hydrazide, and A-FABP (a cytoplasmic fatty acid carrier protein) was studied further in vitro and found to be modified HNE at Cys117 [71]. Enrichment approaches have also been used by other groups to improve the yield of HNE-modified proteins in analyses of biological sample. For example, biotin hydrazide and avidin capture followed by LC/MS/MS found 24 modifications in 14 proteins from human plasma. However, many of the modifications corresponded to direct oxidations of residues and only one adduct of 4-HNE was observed on Apolipoprotein B-100 [72,73]. Better results were obtained using a combined protocol involving peptide enrichment with an anti-HNE-antibody affinity column, mass spectrometry and de novo sequencing, which identified HNE-adducts of several membrane proteins in healthy human red blood cells [74]. A similar approach was applied with success to healthy cardiac myocytes [75].
Targetted approaches to finding HNE-adducts are also beginning to emerge as useful methodologies. Rauniyar et al. [36,76] also reported the use of neutral loss scanning for m/z 138 or 156 with HNE-treated cytochrome c oxidase as a model protein, using a neutral loss driven MS3 scan to sequence any peptides that demonstrated these losses, and were able to identify a substantial number of Michael adducts mainly of histidine but also lysine and cysteine. The same group also obtained good results with solid phase hydrazide enrichment and a combination of collision-induced dissociation and electron capture dissociation to detect HNE elimination (loss of m/z 156) as a signature tag, although this was also carried out with a model protein, rather than in vivo [77]. The value of neutral loss scanning for m/z 156 and 138 was also demonstrated in blood samples to select specifically peptides containing Michael adducts or Schiffs bases of HNE respectively; HNE Michael adducts formed at lys497 and his494 of β-spectrin were detected by this method [74]. An isotope-code tagging protocol for quantitative analysis of HNE-modified peptides has also been reported and applied to a complex mixture of digested plasma proteins [78].
Conclusion and future perspectives on 4-hydroxy-2-nonenal
Nearly 50 years on from the initial discovery of bioactives 4-hydroxyalkenal products, there is a large body of knowledge on the chemistry and biochemistry of HNE. While questions do still remain about the chemical mechanisms by which HNE is formed during lipid peroxidation, several critical intermediates have been isolated and identified, and it is clear that more than one pathway exists. The local membrane and cellular environment during the lipid peroxidation process, such as fatty acyl composition and pH, most likely determine the flux through the possible pathways. Mass spectrometric analysis has been instrumental in understanding the intermediates and pathways during studies of lipid peroxidation in vitro, and is now the main approach to identifying sites of HNE modification on proteins and characterizing the structure of adducts both in vitro and in vivo. The MS technologies available for this purpose are continually improving, and will underpin advancements in understanding of the functional consequences of protein modification by HNE (and other reactive lipid peroxidation products). Increasing evidence is emerging for the action of HNE in modulating protein activity and function, and in redox regulation of cell signalling pathways, as has been discussed recently in many publications and reviews [8,79–84]. The next objectives of research in this field are likely to be a molecular understanding of the role of HNE in physiological signal regulation, and therapeutic strategies targetting formation and effects of reactive aldehydes [85,86].
Acknowledgements
CMS would like to acknowledge the COST Action CM1001 for scientific discussion and support, as well as Jörg Schaur for the historical perspective and Andy Pitt for assistance with manuscript preparation.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-Non Commercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Schauenstein E., Esterbauer H., Jaag G., Taufer M. The effect of aldehydes on normal and malignant cells. 1st report: hydroxy-octenal, a new fat-aldehyde. Monatshefte Fur Chemie. 1964;95:180–183. [Google Scholar]
- 2.Esterbauer H., Schaur R.J., Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biology & Medicine. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 3.Benedetti A., Comporti M., Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochimica et Biophysica Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 4.Benedetti A., Comporti M., Fulceri R., Esterbauer H. Cytotoxic aldehydes originating from the peroxidation of liver microsomal lipids. Identification of 4,5-dihydroxydecenal. Biochimica et Biophysica Acta. 1984;792:172–181. doi: 10.1016/0005-2760(84)90219-4. [DOI] [PubMed] [Google Scholar]
- 5.Mlakar A., Spiteller G. Previously unknown aldehydic lipid peroxidation compounds of arachidonic acid. Chemistry and Physics of Lipids. 1996;79:47–53. doi: 10.1016/0009-3084(95)02506-5. [DOI] [PubMed] [Google Scholar]
- 6.Comporti M. Lipid peroxidation and biogenic aldehydes: from the identification of 4-hydroxynonenal to further achievements in biopathology. Free Radical Research. 1998;28:623–635. doi: 10.3109/10715769809065818. [DOI] [PubMed] [Google Scholar]
- 7.Esterbauer H., Benedetti A., Lang J., Fulceri R., Fauler G., Comporti M. Studies on the mechanism of formation of 4-hydroxynonenal during microsomal lipid peroxidation. Biochimica et Biophysica Acta. 1986;876:154–166. doi: 10.1016/0005-2760(86)90329-2. [DOI] [PubMed] [Google Scholar]
- 8.Poli G., Schaur R.J. 4-Hydroxynonenal in the pathomechanisms of oxidative stress. IUBMB Life. 2000;50:315–321. doi: 10.1080/713803726. [DOI] [PubMed] [Google Scholar]
- 9.Esterbauer H., Zollner H., Schaur R.J., editors. Aldehydes Formed by Lipid Peroxidation: Mechanisms of Formation, Occurrence and Determination. CRC Press; Boca Raton FL: 1990. [Google Scholar]
- 10.Pryor W.A., Porter N.A. Suggested mechanisms for the production of 4-hydroxy-2-nonenal from the autoxidation of polyunsaturated fatty acids. Free Radical Biology & Medicine. 1990;8:541–543. doi: 10.1016/0891-5849(90)90153-a. [DOI] [PubMed] [Google Scholar]
- 11.Loidl-Stahlhofen A., Hannemann K., Spiteller G. Generation of alpha-hydroxyaldehydic compounds in the course of lipid peroxidation. Biochimica et Biophysica Acta. 1994;1213:140–148. doi: 10.1016/0005-2760(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 12.Takamura H., Gardner H.W. Oxygenation of (3Z)-alkenal to (2E)-4-hydroxy-2-alkenal in soybean seed (Glycine max L.) Biochimica et Biophysica Acta. 1996;1303:83–91. doi: 10.1016/0005-2760(96)00076-8. [DOI] [PubMed] [Google Scholar]
- 13.Kaur K., Salomon R.G., O'Neil J., Hoff H.F. Carboxyalkyl)pyrroles in human plasma and oxidized low-density lipoproteins. Chemical Research in Toxicology. 1997;10:1387–1396. doi: 10.1021/tx970112c. [DOI] [PubMed] [Google Scholar]
- 14.Schneider C., Tallman K.A., Porter N.A., Brash A.R. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. The Journal of Biological Chemistry. 2001;276:20831–20838. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- 15.Schneider C., Porter N.A., Brash A.R. Routes to 4-hydroxynonenal: fundamental issues in the mechanisms of lipid peroxidation. The Journal of Biological Chemistry. 2008;283:15539–15543. doi: 10.1074/jbc.R800001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W., Porter N.A., Schneider C., Brash A.R., Yin H. Formation of 4-hydroxynonenal from cardiolipin oxidation: intramolecular peroxyl radical addition and decomposition. Free Radical Biology & Medicine. 2011;50:166–178. doi: 10.1016/j.freeradbiomed.2010.10.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu X., Salomon R.G. Fragmentation of a linoleate-derived gamma-hydroperoxy-alpha,beta-unsaturated epoxide to gamma-hydroxy- and gamma-oxo-alkenals involves a unique pseudo-symmetrical diepoxycarbinyl radical. Free Radical Biology & Medicine. 2012;52:601–606. doi: 10.1016/j.freeradbiomed.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alary J., Gueraud F., Cravedi J.P. Fate of 4-hydroxynonenal in vivo: disposition and metabolic pathways. Molecular Aspects of Medicine. 2003;24:177–187. doi: 10.1016/s0098-2997(03)00012-8. [DOI] [PubMed] [Google Scholar]
- 19.Siems W.G., Pimenov A.M., Esterbauer H., Grune T. Metabolism of 4-hydroxynonenal, a cytotoxic lipid peroxidation product, in thymocytes as an effective secondary antioxidative defense mechanism. Journal of Biochemistry (Tokyo) 1998;123:534–539. doi: 10.1093/oxfordjournals.jbchem.a021969. [DOI] [PubMed] [Google Scholar]
- 20.Balogh L.M., Atkins W.M. Interactions of glutathione transferases with 4-hydroxynonenal. Drug Metabolism Reviews. 2011;43:165–178. doi: 10.3109/03602532.2011.558092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alary J., Debrauwer L., Fernandez Y., Paris A., Cravedi J.P., Dolo L., Rao D., Bories G. Identification of novel urinary metabolites of the lipid peroxidation product 4-hydroxy-2-nonenal in rats. Chemical Research in Toxicology. 1998;11:1368–1376. doi: 10.1021/tx980068g. [DOI] [PubMed] [Google Scholar]
- 22.Gueraud F., Peiro G., Bernard H., Alary J., Creminon C., Debrauwer L., Rathahao E., Drumare M.F., Canlet C., Wal J.M., Bories G. Enzyme immunoassay for a urinary metabolite of 4-hydroxynonenal as a marker of lipid peroxidation. Free Radical Biology & Medicine. 2006;40:54–62. doi: 10.1016/j.freeradbiomed.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava S., Chandra A., Wang L.F., Seifert W.E., Jr, DaGue B.B., Ansari N.H., Srivastava S.K., Bhatnagar A. Metabolism of the lipid peroxidation product, 4-hydroxy-trans-2-nonenal, in isolated perfused rat heart. The Journal of Biological Chemistry. 1998;273:10893–10900. doi: 10.1074/jbc.273.18.10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill B.G., Awe S.O., Vladykovskaya E., Ahmed Y., Liu S.Q., Bhatnagar A., Srivastava S. Myocardial ischaemia inhibits mitochondrial metabolism of 4-hydroxy-trans-2-nonenal. The Biochemical Journal. 2009;417:513–524. doi: 10.1042/BJ20081615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esterbauer H., Zollner H. Methods for determination of aldehydic lipid peroxidation products. Free Radical Biology & Medicine. 1989;7:197–203. doi: 10.1016/0891-5849(89)90015-4. [DOI] [PubMed] [Google Scholar]
- 26.Yan L.J., Forster M.J. Chemical probes for analysis of carbonylated proteins: a review. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2011;879:1308–1315. doi: 10.1016/j.jchromb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selley M.L. Determination of the lipid peroxidation product (E)-4-hydroxy-2-nonenal in clinical samples by gas chromatography negative-ion chemical ionisation mass spectrometry of the O-pentafluorobenzyl oxime. Journal of Chromatography B. 1997;691:263–268. doi: 10.1016/s0378-4347(96)00446-x. [DOI] [PubMed] [Google Scholar]
- 28.Van kuijk F.J.G.M., Siakotos A.N., Fong L.G., Stephens R.J., Thomas D.W. Quantitative measurement of 4-hydroxyalkenals in oxidized low-density-lipoprotein by gas-chromatography mass-spectrometry. Analytical Biochemistry. 1995;224:420–424. doi: 10.1006/abio.1995.1060. [DOI] [PubMed] [Google Scholar]
- 29.Gioacchini A.M., Calonghi N., Boga C., Cappadone C., Masotti L., Roda A., Traldi P. Determination of 4-hydroxy-2-nonenal at cellular levels by means of electrospray mass spectrometry. Rapid Communications in Mass Spectrometry. 1999;13:1573–1579. doi: 10.1002/(SICI)1097-0231(19990815)13:15<1573::AID-RCM675>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 30.Zanardi E., Jagersma C.G., Ghidini S., Chizzolini R. Solid phase extraction and liquid chromatography-tandem mass spectrometry for the evaluation of 4-hydroxy-2-nonenal in pork products. Journal of Agricultural and Food Chemistry. 2002;50:5268–5272. doi: 10.1021/jf020201h. [DOI] [PubMed] [Google Scholar]
- 31.Aldini G., Dalle-Donne I., Colombo R., Maffei Facino R., Milzani A., Carini M. Lipoxidation-derived reactive carbonyl species as potential drug targets in preventing protein carbonylation and related cellular dysfunction. ChemMedChem. 2006;1:1045–1058. doi: 10.1002/cmdc.200600075. [DOI] [PubMed] [Google Scholar]
- 32.Carini M., Aldini G., Facino R.M. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrometry Reviews. 2004;23:281–305. doi: 10.1002/mas.10076. [DOI] [PubMed] [Google Scholar]
- 33.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Progress in Lipid Research. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 34.Fritz K.S., Petersen D.R. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chemical Research in Toxicology. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X., Tang X., Anderson V.E., Sayre L.M. Mass spectrometric characterization of protein modification by the products of nonenzymatic oxidation of linoleic acid. Chemical Research in Toxicology. 2009;22:1386–1397. doi: 10.1021/tx9000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauniyar N., Prokai L. Detection and identification of 4-hydroxy-2-nonenal Schiff-base adducts along with products of Michael addition using data-dependent neutral loss-driven MS3 acquisition: method evaluation through an in vitro study on cytochrome c oxidase modifications. Proteomics. 2009;9:5188–5193. doi: 10.1002/pmic.200900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X., Sayre L.M., Tochtrop G.P. A mass spectrometric analysis of 4-hydroxy-2-(E)-nonenal modification of cytochrome c. Journal of Mass Spectrometry: JMS. 2011;46:290–297. doi: 10.1002/jms.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palinski W., Rosenfeld M.E., Yla-Herttuala S., Gurtner G.C., Socher S.S., Butler S.W., Parthasarathy S., Carew T.E., Steinberg D., Witztum J.L. Low density lipoprotein undergoes oxidative modification in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:1372–1376. doi: 10.1073/pnas.86.4.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinbrecher U.P., Fisher M., Witztum J.L., Curtiss L.K. Immunogenicity of homologous low density lipoprotein after methylation, ethylation, acetylation, or carbamylation: generation of antibodies specific for derivatized lysine. Journal of Lipid Research. 1984;25:1109–1116. [PubMed] [Google Scholar]
- 40.Palinski W., Yla-Herttuala S., Rosenfeld M.E., Butler S.W., Socher S.A., Parthasarathy S., Curtiss L.K., Witztum J.L. Antisera and monoclonal antibodies specific for epitopes generated during oxidative modification of low density lipoprotein. Arteriosclerosis (Dallas, Tex) 1990;10:325–335. doi: 10.1161/01.atv.10.3.325. [DOI] [PubMed] [Google Scholar]
- 41.Jurgens G., Ashy A., Esterbauer H. Detection of new epitopes formed upon oxidation of low-density lipoprotein, lipoprotein(a) and very-low-density lipoprotein—use of an antiserum against 4-hydroxynonenal-modified low-density lipoprotein. Biochemical Journal. 1990;265:605–608. doi: 10.1042/bj2650605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q., Esterbauer H., Jurgens G. Studies on epitopes on low-density lipoprotein modified by 4-hydroxynonenal. Biochemical characterization and determination. The Biochemical Journal. 1992;288:249–254. doi: 10.1042/bj2880249. Pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida K., Szweda L.I., Chae H.Z., Stadtman E.R. Immunochemical detection of 4-hydroxynonenal protein adducts in oxidized hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8742–8746. doi: 10.1073/pnas.90.18.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyokuni S., Miyake N., Hiai H., Hagiwara M., Kawakishi S., Osawa T., Uchida K. The monoclonal antibody specific for the 4-hydroxy-2-nonenal histidine adduct. FEBS Letters. 1995;359:189–191. doi: 10.1016/0014-5793(95)00033-6. [DOI] [PubMed] [Google Scholar]
- 45.Uchida K., Itakura K., Kawakishi S., Hiai H., Toyokuni S., Stadtman E.R. Characterization of epitopes recognized by 4-hydroxy-2-nonenal specific antibodies. Archives of Biochemistry and Biophysics. 1995;324:241–248. doi: 10.1006/abbi.1995.0036. [DOI] [PubMed] [Google Scholar]
- 46.Waeg G., Dimsity G., Esterbauer H. Monoclonal antibodies for detection of 4-hydroxynonenal modified proteins. Free Radical Research. 1996;25:149–159. doi: 10.3109/10715769609149920. [DOI] [PubMed] [Google Scholar]
- 47.Itakura K., Oya-Ito T., Osawa T., Yamada S., Toyokuni S., Shibata N., Kobayashi M., Uchida K. Detection of lipofuscin-like fluorophore in oxidized human low-density lipoprotein. 4-hydroxy-2-nonenal as a potential source of fluorescent chromophore. FEBS Letters. 2000;473:249–253. doi: 10.1016/s0014-5793(00)01539-8. [DOI] [PubMed] [Google Scholar]
- 48.Hartley D.P., Kroll D.J., Petersen D.R. Prooxidant-initiated lipid peroxidation in isolated rat hepatocytes: detection of 4-hydroxynonenal- and malondialdehyde-protein adducts. Chemical Research in Toxicology. 1997;10:895–905. doi: 10.1021/tx960181b. [DOI] [PubMed] [Google Scholar]
- 49.Cohn J.A., Tsai L., Friguet B., Szweda L.I. Chemical characterization of a protein-4-hydroxy-2-nonenal cross-link: immunochemical detection in mitochondria exposed to oxidative stress. Archives of Biochemistry and Biophysics. 1996;328:158–164. doi: 10.1006/abbi.1996.0156. [DOI] [PubMed] [Google Scholar]
- 50.Akagawa M., Ito S., Toyoda K., Ishii Y., Tatsuda E., Shibata T., Yamaguchi S., Kawai Y., Ishino K., Kishi Y., Adachi T., Tsubata T., Takasaki Y., Hattori N., Matsuda T., Uchida K. Bispecific abs against modified protein and DNA with oxidized lipids. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6160–6165. doi: 10.1073/pnas.0600865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida K., Osawa T., Hiai H., Toyokuni S. 4-Hydroxy-2-nonenal-trapping ELISA: direct evidence for the release of a cytotoxic aldehyde from oxidized low density lipoproteins. Biochemical and Biophysical Research Communications. 1995;212:1068–1073. doi: 10.1006/bbrc.1995.2078. [DOI] [PubMed] [Google Scholar]
- 52.Satoh K., Yamada S., Koike Y., Igarashi Y., Toyokuni S., Kumano T., Takahata T., Hayakari M., Tsuchida S., Uchida K. A 1-hour enzyme-linked immunosorbent assay for quantitation of acrolein- and hydroxynonenal-modified proteins by epitope-bound casein matrix method. Analytical Biochemistry. 1999;270:323–328. doi: 10.1006/abio.1999.4073. [DOI] [PubMed] [Google Scholar]
- 53.Borovic S., Rabuzin F., Waeg G., Zarkovic N. Enzyme-linked immunosorbent assay for 4-hydroxynonenal-histidine conjugates. Free Radical Research. 2006;40:809–820. doi: 10.1080/10715760600693422. [DOI] [PubMed] [Google Scholar]
- 54.Kruman I., Bruce-Keller A.J., Bredesen D., Waeg G., Mattson M.P. Evidence that 4-hydroxynonenal mediates oxidative stress-induced neuronal apoptosis. The Journal of Meuroscience: The Official Journal of the Society for Neuroscience. 1997;17:5089–5100. doi: 10.1523/JNEUROSCI.17-13-05089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eaton P., Li J.M., Hearse D.J., Shattock M.J. Formation of 4-hydroxy-2-nonenal-modified proteins in ischaemic rat heart. The American Journal of Physiology. 1999;276:H935–943. doi: 10.1152/ajpheart.1999.276.3.H935. [DOI] [PubMed] [Google Scholar]
- 56.Butterfield D.A., Reed T., Sultana R. Roles of 3-nitrotyrosine- and 4-hydroxynonenal-modified brain proteins in the progression and pathogenesis of Alzheimer's disease. Free Radical Research. 2011;45:59–72. doi: 10.3109/10715762.2010.520014. [DOI] [PubMed] [Google Scholar]
- 57.Smith M.A., Sayre L.M., Anderson V.E., Harris P.L.R., Beal M.F., Kowall N., Perry G. Cytochemical demonstration of oxidative damage in Alzheimer disease by immunochemical enhancement of the carbonyl reaction with 2,4-dinitrophenylhydrazine. Journal of Histochemistry & Cytochemistry. 1998;46:731–735. doi: 10.1177/002215549804600605. [DOI] [PubMed] [Google Scholar]
- 58.Reed T., Perluigi M., Sultana R., Pierce W.M., Klein J.B., Turner D.M., Coccia R., Markesbery W.R., Butterfield D.A. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer's disease. Neurobiology of Disease. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 59.Reed T.T., Pierce W.M., Markesbery W.R., Butterfield D.A. Proteomic identification of HNE-bound proteins in early Alzheimer disease: insights into the role of lipid peroxidation in the progression of AD. Brain Research. 2009;1274:66–76. doi: 10.1016/j.brainres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Higdon A.N., Dranka B.P., Hill B.G., Oh J.Y., Johnson M.S., Landar A., Darley-Usmar V.M. Methods for imaging and detecting modification of proteins by reactive lipid species. Free Radical Biology & Medicine. 2009;47:201–212. doi: 10.1016/j.freeradbiomed.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landar A., Zmijewski J.W., Dickinson D.A., Le Goffe C., Johnson M.S., Milne G.L., Zanoni G., Vidari G., Morrow J.D., Darley-Usmar V.M. Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. American Journal of Physiology Heart and Circulatory Physiology. 2006;290:H1777–1787. doi: 10.1152/ajpheart.01087.2005. [DOI] [PubMed] [Google Scholar]
- 62.Vila A., Tallman K.A., Jacobs A.T., Liebler D.C., Porter N.A., Marnett L.J. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chemical Research in Toxicology. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doorn J.A., Petersen D.R. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chemical Research in Toxicology. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 64.Liu Z., Minkler P.E., Sayre L.M. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chemical Research in Toxicology. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 65.Alderton A.L., Faustman C., Liebler D.C., Hill D.W. Induction of redox instability of bovine myoglobin by adduction with 4-hydroxy-2-nonenal. Biochemistry. 2003;42:4398–4405. doi: 10.1021/bi0271695. [DOI] [PubMed] [Google Scholar]
- 66.Isom A.L., Barnes S., Wilson L., Kirk M., Coward L., Darley-Usmar V. Modification of Cytochrome c by 4-hydroxy- 2-nonenal: evidence for histidine, lysine, and arginine-aldehyde adducts. Journal of the American Society for Mass Spectrometry. 2004;15:1136–1147. doi: 10.1016/j.jasms.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 67.Eliuk S.M., Renfrow M.B., Shonsey E.M., Barnes S., Kim H. Active site modifications of the brain isoform of creatine kinase by 4-hydroxy-2-nonenal correlate with reduced enzyme activity: mapping of modified sites by Fourier transform-ion cyclotron resonance mass spectrometry. Chemical Research in Toxicology. 2007;20:1260–1268. doi: 10.1021/tx7000948. [DOI] [PubMed] [Google Scholar]
- 68.Ishii T., Tatsuda E., Kumazawa S., Nakayama T., Uchida K. Molecular basis of enzyme inactivation by an endogenous electrophile 4-hydroxy-2-nonenal: identification of modification sites in glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 2003;42:3474–3480. doi: 10.1021/bi027172o. [DOI] [PubMed] [Google Scholar]
- 69.D'Souza A., Kurien B.T., Rodgers R., Shenoi J., Kurono S., Matsumoto H., Hensley K., Nath S.K., Scofield R.H. Detection of catalase as a major protein target of the lipid peroxidation product 4-HNE and the lack of its genetic association as a risk factor in SLE. BMC Medical Genetics. 2008;9:62. doi: 10.1186/1471-2350-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carbone D.L., Doorn J.A., Kiebler Z., Petersen D.R. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chemical Research in Toxicology. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 71.Grimsrud P.A., Picklo M.J., Sr, Griffin T.J., Bernlohr D.A. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Molecular & Cellular Proteomics: MCP. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Madian A.G., Regnier F.E. Proteomic identification of carbonylated proteins and their oxidation sites. Journal of Proteome Research. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madian A.G., Regnier F.E. Profiling carbonylated proteins in human plasma. Journal of Proteome Research. 2010;9:1330–1343. doi: 10.1021/pr900890k. [DOI] [PubMed] [Google Scholar]
- 74.Mendez D., Hernaez M.L., Kamali A.N., Diez A., Puyet A., Bautista J.M. Differential carbonylation of cytoskeletal proteins in blood group O erythrocytes: potential role in protection against severe malaria. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases. 2012;12:1780–1787. doi: 10.1016/j.meegid.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 75.Chavez J.D., Wu J.Y., Bisson W., Maier C.S. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. Journal of Proteomics. 2011;74:2417–2429. doi: 10.1016/j.jprot.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rauniyar N., Stevens S.M., Prokai-Tatrai K., Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Analytical Chemistry. 2009;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rauniyar N., Prokai-Tatrai K., Prokai L. Identification of carbonylation sites in apomyoglobin after exposure to 4-hydroxy-2-nonenal by solid-phase enrichment and liquid chromatography-electrospray ionization tandem mass spectrometry. Journal of Mass Spectrometry: JMS. 2010;45:398–410. doi: 10.1002/jms.1725. [DOI] [PubMed] [Google Scholar]
- 78.Rauniyar N., Prokai L. Isotope-coded dimethyl tagging for differential quantification of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. Journal of Mass Spectrometry. 2011;46:976–985. doi: 10.1002/jms.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poli G., Biasi F., Leonarduzzi G. 4-hydroxynonenal-protein adducts: a reliable biomarker of lipid oxidation in liver diseases. Molecular Aspects of Medicine. 2008;29:67–71. doi: 10.1016/j.mam.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 80.Poli G., Schaur R.J., Siems W.G., Leonarduzzi G. 4-hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Medicinal Research Reviews. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 81.Forman H.J., Fukuto J.M., Miller T., Zhang H., Rinna A., Levy S. The chemistry of cell signalling by reactive oxygen and nitrogen species and 4-hydroxynonenal. Archives of Biochemistry and Biophysics. 2008;477:183–195. doi: 10.1016/j.abb.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castello L., Marengo B., Nitti M., Froio T., Domenicotti C., Biasi F., Leonarduzzi G., Pronzato M.A., Marinari U.M., Poli G., Chiarpotto E. 4-hydroxynonenal signalling to apoptosis in isolated rat hepatocytes: the role of PKC-delta. Biochimica et Biophysica Acta-Molecular and Cell Biology of Lipids. 1737;83–93:2005. doi: 10.1016/j.bbalip.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 83.Noguchi N. Role of oxidative stress in adaptive responses in special reference to atherogenesis. Journal of Clinical Biochemistry and Nutrition. 2008;43:131–138. doi: 10.3164/jcbn.2008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaudhary P., Sharma R., Sharma A., Vatsyayan R., Yadav S., Singhal S.S., Rauniyar N., Prokai L., Awasthi S., Awasthi Y.C. Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signalling. Biochemistry. 2010;49:6263–6275. doi: 10.1021/bi100517x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Menini S., Iacobini C., Ricci C., Scipioni A., Blasetti Fantauzzi C., Giaccari A., Salomone E., Canevotti R., Lapolla A., Orioli M., Aldini G., Pugliese G. D-Carnosine octylester attenuates atherosclerosis and renal disease in ApoE null mice fed a Western diet through reduction of carbonyl stress and inflammation. British Journal of Pharmacology. 2012;166:1344–1356. doi: 10.1111/j.1476-5381.2012.01834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aldini G., Dalle-Donne I., Facino R.M., Milzani A., Carini M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Medicinal Research Reviews. 2007;27:817–868. doi: 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]


