Abstract
Recent evidence suggests that the malfunctioning disposal system of cell protein called ubiquitin–proteasome system (UPS) plays an important role in the development of disorders, including cancer and neurodegenerative diseases. Accumulating evidence suggests that the abnormal regulation of the E3 ubiquitin ligases, essential components of the UPS, contributes to uncontrolled proliferation, genomic instability and cancer, since these ligases and their substrates are involved in the regulation of cell cycle progression, gene transcription, signal transduction, DNA replication and others. Through selective degradation of specific substrates, E3 ligases regulate different biological processes. Cullins are a family of proteins that confer substrate specificity to multimeric complex of E3 ligases acting as scaffold proteins. So far, seven members of the cullin family of proteins have been identified. Interestingly, the data generated by several groups indicate that cullin 3 (Cul3) has begun to emerge as a protein involved in the etiopathology of multiple diseases. In this paper we examine the latest advances in basic research on the biology of Cul3 and how it could help to direct drug discovery efforts on this target.
Keywords: Cullin 3, Proteasome, Nedd8, Nrf2, Oxidative stress, Cell cycle
Graphical abstract
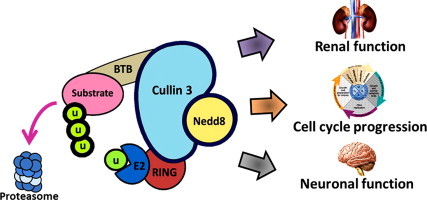
Highlights
-
•
The most important system for protein degradation is the ubiquitin–proteasome system.
-
•
The specific substrate for ubiquitination is highly specific and this activity can be provided by the E3 ubiquitin ligases.
-
•
The E3 ubiquitin ligases based on cullins are the type of ubiquitin ligases more studied.
-
•
The cullin 3 complex has emerged as a target due to its interaction with a wide range of BTB-proteins.
-
•
Cullin 3 could be a molecule with a high therapeutic potential.
Protein degradation: the proteasomal system
In order to maintain cell functionality and viability, it is important that damaged, altered, misfolded or unnecessary proteins can be recognized and degraded. One of the most important systems for protein degradation is the ubiquitin–proteasome system (UPS) [1,2]. This system is found in both the nucleus and the cytoplasm [3], where it regulates vital cellular processes. For example, the cell cycle progression is regulated by the degradation of cyclins and inhibitors of cyclin-dependent kinases [4]. Moreover, UPS is involved in the regulation of cell growth and gene expression by degrading transcriptional regulators as c-Jun, nuclear factor-kB (NFkB) [5], p53 [6] and β-catenin [7], as well as in the termination of signaling cascades through protein degradation of activated kinases. Thus, the interest in the study of the involvement of UPS in the degradation of proteins has become more relevant since the deregulation of this system has been implicated in the pathogenesis of human diseases, such as malignancies and neurodegenerative processes [8,9].
Degradation via the UPS system is mediated by a process called ubiquitination [2], where ubiquitin is attached to the target protein by a peptide linkage, allowing its transport to the 26S proteasome complex. Ubiquitination is a complex process that involves several steps, and is mediated by a series of enzymes, including an ubiquitin-activating enzyme (E1, UAE, also known as UBA1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3) [10]. The substrate recognition for ubiquitination is highly specific and this activity can be provided by the E3 ubiquitin ligases.
Ubiquitin ligases (E3)
The E3 ubiquitin ligases are the largest group of enzymes involved in the UPS. In contrast to the E1 and E2 enzymes, ubiquitin ligases (E3) are highly diverse; so far over 500 ubiquitin ligases have been identified, and this has allowed the suggestion that E2 may be associated with different E3 ligases, resulting in a large amount of E2/E3 functional complexes, and consequently, in a high number of specific target substrates [1,11]. The ubiquitin ligases are classified into two main classes: the first one is the E3 ligase with HECT domain (homologous to the E6-AP carboxyl terminus domain), whereas the second one is the E3 ligase with RING finger domain (a really interesting new gene) [12,13]. Ligases of this second class are based on cullin (CRL: cullin-RING ubiquitin ligases), which are the common type of ubiquitin ligases known so far [14].
Cullin family proteins: cullin-based ubiquitin ligases
CRLs are multimeric complexes having a catalytic center in a family member of proteins. Cullins are proteins that play a role in post-translational modification of proteins, including ubiquitination. The cullin family is highly conserved among species [15]; seven different cullins have been identified in mammals (Cul1, 2, 3, 4A, 4B, 5 and 7) [14,16].
Structure of CRLs complexes
Each cullin forms a distinct class of CRL complex consisting of different adapters and/or substrate recognition subunits [14,17]. Table 1 presents a list of the adapters associated with different cullins.
Table 1.
Components and adapters of CRLs complexes.
| Cullin | E3 complex components | Adapter |
|---|---|---|
| 1 | Skp1-Cul1-F-box-ROC1/Rbx1 | Skp1 |
| 2 | Elongin BC-Cul2-Protein SOCS-ROC1/Rbx1 | ElonginC/ElonginB |
| 3 | BTB-Cul3-ROC1/Rbx1 Protein | Protein BTB, BTB domain |
| 4A, 4B | DDB1-Cul4A/4B-DDB2 o CSA-ROC1/Rbx1 | DDB1 |
| 5 | Elongin BC-Cul5-Protein SOCS-ROC1/Rbx1 | ElonginC/ElonginB |
| 7 | Skp1-Cul7-Fbx29-ROC1/Rbx1 | Skp1 |
Skp1: S-phase kinase-associated protein 1; F-box: motif that functions as a site of protein–protein interaction; ROC1/Rbx1: RING-box protein; BTB: bric-a-brac/tramtrack/broad-complex; DDB1: damage-specific DNA binding protein 1; SOCS: suppressors of cytokine signaling protein; CSA: cockayne syndrome group A protein.
The properties and the biological role of cullins are very different, and while there are structural differences between these complexes, collected evidence suggests that CRLs exert a key role in the regulation and cell cycle control. In addition, these multimeric complexes are involved in various cellular processes, including multiple aspects of quality control of proteins, cell growth, signal transduction, transcription and DNA replication, among others [14]. This diversity of functions is given by each of the adapters present in the complex. For example, the adapter S-phase kinase-associated protein 1 (Skp1) mediates binding of F-box proteins to cullin 1 and cullin 7 [15], and its molecular targets are p27, cyclin E and β-catenin, that are involved in cell cycle regulation and signal transduction. In CRL4A, the substrate recognition unit is composed of protein 1 damaged DNA binding (DDB1), so its main function is to activate ribonucleotide reductase (RNR). Cullins that require ElonginC or B protein as the adapter are implicated in regulating the cell cycle by mediating degradation of p53, a protein involved in the regulation of intracellular oxygen concentration, and so, in the regulation of cell redox homeostasis [18] (Table 1).
Regulating the activity of CRLs
The activity of E3 ligases based on cullin is regulated through the reversible conjugation of a small ubiquitin-type protein known as Neural precursor cell expressed developmentally down-regulated protein 8 (Nedd8). Covalent modifications of these cullins are essential for its association with the E2 enzyme, so it can promote ubiquitination of target proteins [19].
Nedd8 is a ubiquitin-like protein and is classified as type 1, which means that it can combine with proteins to direct them to secondary processes, such as ubiquitination. It is known that Nedd8 is mainly bound to the family members of the cullins, and thus, it controls vital biological events such as proteolysis via UPS [20]. The Nedd8 conjugation (neddylation process) to different cullins is similar to ubiquitination, and is an ATP-dependent process that is catalyzed by an E1 (Uba3/APP-BP1), where activated Nedd8 is transferred to an E2 (Ubc12), which in turn is responsible for the polypeptide binding of cullin to Nedd8 [21,22].
Neddylation is given by the formation of an isopeptide bond, linking the carboxyl-end of Nedd8 Gly-76 to the amino group of a conserved cullin lysine residue, thereby increasing the conjugation of ubiquitin ligase activity for promoting the recruitment of E2, stimulating other conformational rearrangement in the B winged-helix domain of the C-terminal subdomain, allowing RING cullin to form a more “dynamic form”, and promoting the transfer of ubiquitin to the substrate (Fig. 1) [20].
Fig. 1.

Schematic illustration of the conformational change of cullin 3 by neddylation. Cul3: cullin 3 protein; Nedd8: Neural precursor cell expressed developmentally down-regulated protein 8; Nrf2: nuclear factor erythroid 2 related factor 2; Keap1: BTB-Kelch-like ECH-associated protein 1; Rbx1: RING-box protein; E2: ubiquitin conjugating enzyme; Ub: ubiquitin.
CRL complex based on cullin 3 (CRL3)
Compared to other cullin-based complexes, the cullin complex 3 is quirky: it does not require different adapters in order to recognize its target protein, but only requires a protein with a bric-a-brac/tramtrack/broad-complex (BTB) domain to recognize it; this property makes it even more interesting to study. Despite the fact that BTB domains were originally found in transcription factors of Drosophila melanogaster, it is now known that all eukaryotic species express a variety of BTB domain proteins [23]. The establishment of structural homology between BTB proteins with Skp1 or Elongin C (subunits of some CRLs) has served to hypothesize that this type of BTB protein could also interact with members of the cullin family, suggesting that various BTB proteins interact with a member of this family, with cullin 3 (Cul3), but not with other members [24,25]. This feature of BTB protein binding allows Cul3 to participate in biological processes; however, in recent years, the complex based on cullin 3 has been implicated especially in processes such as the cell cycle regulation of redox homeostasis, a process that must be of major interest since it is related to chronic degenerative diseases where oxidative stress plays a key role.
The function of Cul3 in cell cycle regulation
Since the identification of BTB domains initially found in Drosophila melanogaster, BTB domain proteins have also been found in humans, where there are approximately 500 proteins identified; its nomenclature is varied but the BTB protein family can be subdivided into BTB-zinc finger (BTB-ZF), BTB-BACK-kelch (BBK), MATH-BTB, BTB-NPH3, Kelch family (KLHL), and Kelch repeat and BTB domain-containing proteins (KBTBD) subfamilies of proteins, among others [23,26]. Many of these proteins have been specifically related with Cul3 and its biological function is related with cell cycle regulation. For example, one of the most studied proteins in this field is the maternal lethal effect-26 protein (MEL26), which is a MATH-BTB subfamily member (47 kDa) [27] that functions as a specific adapter substrate for the E3 ligase. Cul3 draws the complex meiosis inhibitor protein 1 (MEI-1) for degradation via interaction with its meprim and TRAF homology domain (MATH). MEI-1 is a member of the AAA ATPase family, and it is believed that short microtubules during the mitosis/meiosis transition facilitate polar body extrusion. Thus, MEL26 plays a dual role in early stages of development: it acts as an adapter for Cul3 ubiquitin ligase to degrade MEI-1, while, on the other hand, it functions as an activator of cytokinesis and is required for efficient actomyosin ring contraction, thereby controlling the initiation and lasting of the cleavage furrow in Caenorhabditis elegans [27,24]. The relevance of this process lies in the fact that the complex based on Cul3 is required to degrade MEI-1 after meiosis, which is essential for the assembly of a functional spindle [28]. It has been observed that poor Cul3 ligase activity leads to an accumulation of MEL26, and these elevated levels were correlated with MEL26 embryos containing short microtubules [27], so the Cul3-based E3 ligase functions as a regulator of MEL26 for cytokinesis, allowing the accumulation only in the place and time. Hence, increasing the activity of this protein could cause severe problems in the cytokinetic process. Proper control of cell cycle progression is critical for the maintenance of any cell; a special case are neurons because although they are in G0, they have the ability to reactivate their cell cycle in response to damage, despite this promoted re-entry of cell cycle of postmitotic cells induces cell death instead of proliferation, which is a common pathway shared in various neurodegenerative disorders [29]. The mechanism by which the cell cycle is reactivated in such cells is not well understood yet, but there is additional evidence about the regulation of the Cul3/MEL-26 complex, pointing out the importance of the cullin in cellular regulation of this process; for example, Moghe and colleagues [30] have shown that depletion of Cul3 or KLHL18 proteins causes the cells to enter into mitosis. In addition, Cul3 overexpression promotes the ubiquitination of Aurora-A both in vivo and in vitro. Thus, Cul3 is able to regulate the entrance to mitosis in an Aurora-A-dependent manner by interacting with KLHL18 protein, hence mediating the activation of Aurora-A in centrosome. This regulation is an important process because it has been shown that there is deregulation in tumors of different kinases, including Aurora-A, and pollo-like kinase 1 (PLK1) and the never in mitosis gene a-related kinase 2 (Nek2) [30–33]. In turn, PLK1 is a critical regulator of mitosis through its dynamic localization at kinetochores, centrosomes and the middle zone; PLK1 has been proven to be a target for protein E3 ubiquitin ligase complex based on Cul3, and it is also recognized by a BTB protein called KLHL22. In the absence of the adapter protein KLHL22, PLK1 is accumulated at kinetochores, promoting the activation of the spindle assembly checkpoint (SAC), which ensures that chromosome segregation takes place correctly [34,35].
Another important molecule in regulating the cell cycle is cyclin E. This is a highly conserved protein and is essential to promote the transition from G1 to S phase in the cell cycle [36]. Cyclin E binds to cyclin dependent kinase 2 (Cdk2) in the G1 phase of the cell cycle, because it is necessary for the G1/S. Cyclin E/Cdk2 complex phosphorylates p27, an inhibitor of cyclin D, inducing its degradation, promoting the expression of cyclin A, and allowing the entrance and progression of S phase after cyclin E is rapidly degraded by the UPS system [37]. The overexpression of cyclin E has often been seen in human diseases, particularly in breast and ovarian cancer [38]. The regulating cyclin E degradation is mediated by two cullin-dependent mechanisms, one based on Cul1 and the other based on the complex Cul3. Evidence shows the importance of Cul3 in cyclin E degradation, since the loss of Cul3 is likely to contribute to tumor progression, a process attributed to the deregulation of the cell cycle [39] (Table 2).
Table 2.
Cul3-associated proteins and their function in different cellular processes.
| Associated protein | Function | Reference |
|---|---|---|
| KEL-8 | Regulates off and on glutamate receptor at synapses. | [40] |
| Gigaxonin | Promotes degradation MAP1B and cofactor MAP8B of tubulin. | [41,42] |
| NAC-1 | Translocates UPS from the nucleus to the cytoplasm in dendritic spines. | [43] |
| Keap1 | Regulates proteasomal degradation of Nrf2. | [44,13] |
| Cyclin E | Controls the S phase entry and maintains cells in a quiescent state. | [36,37] |
| MEL-26 | Regulates the activity of MEI-1 in forming the cytoskeleton in meiosis and mitosis. | [45] |
| KLHL-7 | A mutation in the KHLH-7 gene affects protein binding to Cul3 and causes retinitis pigmentosa. | [46] |
MAP1B: Microtubule-associated protein 1B; MAP8B: Microtubule-associated protein 8; NAC1: nucleus accumbens-1.
Cullin 3 and its importance in the response to cellular stress
In recent years, it has been accepted that oxidative stress is implicated in the development of various disorders, such as cancer and neurodegenerative diseases. Therefore, the search for molecules possessing cytoprotective properties against oxidative damage to induce the expression of genes encoding enzymes of phase II by the nuclear factor erythroid 2 related factor 2 (Nrf2)-mediated pathway has proven to be a major research area (Fig. 2).
Fig. 2.
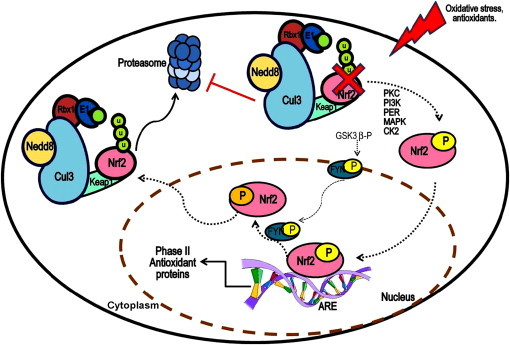
Mechanism of Nrf2 regulation. Cul3: cullin 3 protein; Nedd8: Neural precursor cell expressed developmentally down-regulated protein 8; Nrf2: nuclear factor erythroid 2 related factor 2; Keap1: BTB-Kelch-like ECH-associated protein 1; Rbx1: RING-box protein; E2: ubiquitin conjugating enzyme; Ub: ubiquitin; PKC: protein kinase C; PI3K: phosphoinositol 3-kinase; PERK: protein kinase RNA-like endoplasmic reticulum kinase; MAPK: mitogen-activated protein kinase; CK2: Casein kinase 2; GSK3β: Glycogen synthase kinase 3; FYN: non-receptor tyrosine kinase; ARE: antioxidant response element.
Nrf2 is constitutively expressed in most cells, so its activity remains strictly regulated by the cell to maintain a particular redox status, remaining in homeostasis. The most widely accepted model that describes the regulation of Nrf2 function proposes that, under homeostatic conditions, this factor is not free and active all the time, but only when oxidative stress conditions are generated. Also, this model suggests that Nrf2 is maintained primarily in the cytoplasm, where it remains bound to the BTB-Kelch-like ECH-associated protein 1 (Keap1 or KLHL19); therefore, low levels remain in the rest of the cell [47–50] (Fig. 2). Its localization in the cytoplasm is mainly due to an interaction between Neh2 domain of Nrf2 with a domain of Keap1 referred to as DGR domain, which is comprised of six repeats of double glycine [44,51].
Keap1 is a Kelch-like protein with the BTB domain, which is bound to Cul3 [52,53]. Numerous studies refer to the importance of Cul3 in the regulation of Nrf2 activity, since it has been found that this transcription factor is present in Cul3 complexes in vivo, constitutively leading to Nrf2 degradation via UPS [51,54] (Fig. 3). Also, it has been found that alterations in both Keap1 BTB domain and modified Cys151 of Keap1 Cul3 affect the binding of the latter, thereby decreasing the ubiquitination of Nrf2 [54,55]. In addition, it has been shown that Cul3 overexpression is associated with increased susceptibility to carcinogenes and oxidative stress in breast cancer because this overexpression contributes to the depletion of Nrf2 levels [56]. Altogether, this evidence has served to suggest that, far from being just a protein involved in a complex that directs the degradation of Nrf2 via UPS, Cul3 could be a determining factor in the regulation of the activity of Nrf2 during stress responses. Evidence also suggests that deregulation of Keap1 activity is strongly implicated in the development of resistance to chemotherapeutic drugs [57,29], or in the development of some cancers [58,59] through Nrf2 activation, thus allowing the expression of regulatory molecules with high capacity, including Cul3, since this is intimately related to the ability to regulate the levels of Keap1 and Nrf2 [60].
Fig. 3.
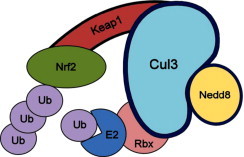
Schematic representation of the Keap1/Cul3/Nrf2 complex. Cul3: cullin 3 protein; Nedd8: Neural precursor cell expressed developmentally down-regulated protein 8; Nrf2: nuclear factor erythroid 2 related factor 2; Keap1: BTB- Kelch-like ECH-associated protein 1; Rbx: RING-box protein; E2: ubiquitin conjugating enzyme; Ub: ubiquitin.
Recently, it was found that nucleus accumbens-1 (NAC1), a transcription factor that belongs to the bric-a-brac, tramtrack, broad complex/Pox virus and zinc finger family (BTB/POZ), responds to cellular stress and modulates the process of autophagy. NAC1 inactivates autophagy and suppresses apoptosis in tumor cells treated with cisplatin, which places NAC1 as an important regulator of these cellular processes [61]. It has been observed that NAC1 is highly regulated in several tumor types, such as breast cancer, renal cell carcinoma and hepatocellular carcinoma, and it has been identified as a chemo-resistance gene [62,63]. Besides this, NAC1 is relevant because it regulates the UPS translocation from the nucleus to the cytoplasm in dendritic spines, a process mediated by Cul3 binding [43]. This evidence shows that Cul3 is a protein involved in diverse cellular processes, including the consolidation of the UPS functions.
Another protein that can be indirectly regulated by Cul3 through the action of Nrf2 is the multidrug resistance-associated protein-1 (MRP1), which has emerged as an important contributor to chemoresistance. MRP1 is induced by oxidative stress [64,65]; the molecular mechanism for the induction of this protein has not been clarified yet, but evidence indicates that the induction of phase II enzymes and the multidrug resistance proteins (MRPs) is similar, confirming that Nrf2 is required for the constitutive and inducible expression of MRP1; however, how Nrf2 mediates the transcriptional activation of MRP1 remains unclear [64,66].
Moreover, in Arabidopsis, the reduction of Cul3 function leads to increased protein ATHB6 (a HD-ZIP type transcription factor) accumulation, reducing plant growth and fertility, and affecting the stomatal behavior and responses to abscisic acid (ABA), a plant hormone which response is crucial for biotic and abiotic stress. This clearly shows that Cul3 is important in signaling pathways for rapid adaptation that allows growth and cell survival [67].
Currently, the study of the involvement of Cul3 in disease development is booming. Including the latest research, Cul3 has been linked to the development of pseudohypoaldosteronism type II (Gordon's hypertension syndrome/PHAII); in this disease, mutations have been identified in KLHL3 [68], which is a protein family BTB-BACK-Kelch, which serves as a substrate for Cul3 adapter KLHL3 and is highly expressed in the distal tubules of the nephron, where it is involved in the regulation of the electrolyte homeostasis and blood pressure [69]. Cul3 could be a new candidate protein presenting development-related PHAII type mutations. The existence of these mutations was confirmed and coupled with Cul3 ubiquitous expression in all segments of the nephron, suggesting that CRL E3 is critical in regulating hypertension [70,68]. Chronic kidney disease has been associated with hypertension, mitochondrial dysfunction and oxidative stress [71], so the study of the function of Cul3 in these conditions is critical. Distal myopathy is another example where Cul3 activity has been linked to the development of pathologies. In this case, a mutation has been identified in the gene coding for the BTB Kelch-like protein 9 (KLHL9), which is capable of interfering with the interaction with Cul3 [69,72], thereby affecting the ubiquitination of their substrates [73] (Table 3).
Table 3.
Cul3-associated proteins and their relevance to the renal system.
| Associated protein | Function | Reference |
|---|---|---|
| KLHL3/WNK4 | Loss in the ability of the complex to ubiquitinate WNK4 KLHL3/Cul3 and therefore, to regulate the levels of ROMK carried PHAII. WNK4 regulates the activity of the Na+/Cl- co-transporter (NCC), the epithelial Na+ channel (ENaC), the K+ channel (ROMK) and the Cl-/HCO3- exchanger. ROMK / H+ ATPase are required for secretion of renal K+ and H+. Patients with Cul3 mutations have severe hyperkalemia metabolic acidosis. | [73,68] |
| KLHL3/WNK1 | WNK1 exerts its effect on blood pressure through its ability to phosphorylate and activate kinases like SPAK/STK39 and OSR1. Increased expression of WNK4 stimulates an abnormal retention of salt in the kidney by the activation of NCC and NKCC2 (Na–K–Cl) co-transporters. | [70] |
| KLHL3/WNK4/OSR1 | Activation of NKCC1 co-transporters, and NCC/NKCC2 by OSR1, plays a critical role in the regulation of blood pressure and renal resorption of NaCl. | [74] |
WNK4: With-no-lysine (K) kinase-4; WNK1: With-no-lysine (K) kinase-1; OSR1: oxidative stress-responsive 1 protein; ROMK: renal outer medullary potassium channel; PHAII: pseudohypoaldosteronism type II; SPAK: STE20/SPS1-related, proline alanine-rich kinase; STK39: serine threonine kinase 39.
Cullin 3 as a therapeutic target
Through selective degradation of these substrates, E3 ligases regulate many biological processes and are considered as molecules with a high therapeutic potential for regulation of events that are mainly related to the development of cancer and chronic degenerative diseases.
In recent years, several groups have investigated the ability of modulating the neddylation pathway of CRLs, in order to reduce the toxicity caused by drugs that inhibit proteasome activity as a therapeutic strategy. This strategy has been used to treat a variety of different diseases related to the alteration in the UPS activity, such as the initiation or progression of tumors (astrocytomas, hemangioblastomas and medulloblastomas), as well as some neurodegenerative diseases [8,75]. Because of the emergence of resistance to proteasome inhibitors, selective novel inhibitors are being developed [75,76]. This is the case for new inhibitors directed against specific components of the UPS, like the Nedd8-activating enzyme, which is a protein required for the neddylation process, thus promoting the activity of CRLs [20,77], a strategy currently implemented for the treatment of several types of cancer [78–81].
Therefore, studying the Cul3 protein complexes is relevant to direct therapeutic strategies not only against cancer, but also against disorders that have been largely related with oxidative stress by regulating the Nrf2 pathway. Also, it is relevant in the treatment of neurodegenerative diseases by regulating the cell cycle progression, and in renal pathologies by regulating kinases activity involved in the normal renal system function, two areas that have not yet been widely explored.
It is noteworthy that one of the advantages for Cul3 targeted therapy is that it would be more specific for the regulation of certain substrates, as well as the reduction of toxicity because it does not inhibit bulk proteasomal degradation. However, the activity of Cul3 must be carefully analyzed to understand the mechanisms by which it regulates some cellular processes abovementioned. Of note is that a bad therapeutic manipulation of this target could trigger cell abnormalities that could lead to cell death.
Conclusion
The importance to highlight the evidence aforementioned is because it allows conducting research to identify other BTB-proteins involved in diseases, thereby suggesting a shared participation of Cul3 in the development of pathologic conditions. The deregulation of Cul3 activity could be a mechanism involved in pathologies mainly associated with oxidative stress and cell cycle deregulation. Therefore, strategies oriented against Cul3 activity comprise a high therapeutic potential for regulation of cellular processes related to the development of several pathologies.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Jung T., Catalgol B., Grune T. The proteasomal system. Molecular Aspects of Medicine. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Ubiquitin and intracellular protein degradation. Current Opinion in Cell Biology. 1992;4(6):1024–1031. doi: 10.1016/0955-0674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- 3.Peters J.M., Franke W.W., Kleinschmidt J.A. Distinct 19S and 20S subcomplexes of 26S proteasome and their distribution in the nucleus and the cytoplasm. The Journal of Biological Chemistry. 1994;269(10):7709–7718. [PubMed] [Google Scholar]
- 4.Thompson SJ, Loftus L.T., Ashley M.D., Meller R. Ubiquitin–proteasome system as a modulator of cell fate. Current Opinion in Pharmacology. 2008;8(1):90–95. doi: 10.1016/j.coph.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothwarf D.M., Karin M. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Science’s Signal Transduction Knowledge Environment. 1999;(5):RE1. doi: 10.1126/stke.1999.5.re1. [DOI] [PubMed] [Google Scholar]
- 6.Lim M.S., Elenitoba-Johnson K.S. Ubiquitin ligases in malignant lymphoma. Leukemia and Lymphoma. 2004;45(7):1329–1339. doi: 10.1080/10428190410001663635. [DOI] [PubMed] [Google Scholar]
- 7.Miller J.R., Moon R.T. Signal transduction through beta-catenin and specification of cell fate during embryogenesis. Genes and Development. 1996;10(20):2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 8.Lehman N.L. The ubiquitin proteasome system in neuropathology. Acta Neuropathologica. 2009;118:329–347. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segref A., Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Reports. 2009;10(1):44–50. doi: 10.1038/embor.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedford L., Lowe J., Dick L.R., Mayer R.J., Brownell J.E. Ubiquitin-like protein conjugation and the ubiquitin–proteasome system as drug targets. Nature Reviews. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich H.D. Natural substrates of the proteasome and their recognition by the ubiquitin system. Current Topics in Microbiology and Immunology. 2002;268:137–174. doi: 10.1007/978-3-642-59414-4_6. [DOI] [PubMed] [Google Scholar]
- 12.Pintard L., Willems A., Peter M. Cullin-based ubiquitin ligases: Cul3–BTB complexes join the family. The EMBO Journal. 2004;23(8):1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa M., Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3–Roc1 ligase. Journal of Molecular Cell Biology. 2005;25(1):162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petroski M.D., Deshaies R.J. Function and regulation of cullin-RING ubiquitin ligases. Nature Reviews. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 15.Sarikas A., Hartmann T., Pan Z.Q. The cullin protein family. Genome Biology. 2011;12(4):220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosu D.R., Kipreos E.T. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Division. 2008;3:7–11. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marín I. Diversification of the cullin family. BMC Evolutionary Biology. 2009;9:267–278. doi: 10.1186/1471-2148-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R.C., Conaway J.W., Nakayama K.I. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes and Development. 2004;18(24):3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J.T., Lin H.C., Hu Y.C., Chien C.T. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nature Cell Biology. 2005;7(10):1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 20.Boh B.K., Smith P.G., Hagen T. Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. Journal of Molecular Biology. 2010;409(2):136–145. doi: 10.1016/j.jmb.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Mori F., Nishie M., Piao Y.S., Kito K., Kamitani T., Takahashi H., Wakabayashi K. Acumulation of Nedd8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathology and Applied Neurobiology. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 22.Parry G., Estelle M. Regulation of cullin-based ubiquitin ligases by the Nedd8/RUB ubiquitin-like proteins. Seminars in Cell and Developmental Biology. 2004;15:221–229. doi: 10.1016/j.semcdb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Privé G.G. Sequence and structural analysis of BTB domain proteins. Genome Biology. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furukawa M., He Y.J., Borchers C., Xiong Y. Targeting of protein ubiquitination by BTB-cullin 3-Roc1 ubiquitin ligases. Nature cell biology. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 25.Xu L., Wei Y., Reboul J., Vaglio P., Shin T.H., Vidal M., Elledge S.J., Harper J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425(6955):316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 26.Canning P., Cooper C.D., Krojer T., Murray J.W., Pike A.C., Chaikuad A, Keates T., Thangaratnarajah C., Hojzan V., Marsden B.D., Gileadi O., Knapp S., von Delft F., Bullock A.N. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. The Journal of Biological Chemistry. 2013;288(11):7803–7814. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luke-Glaser S., Pintard L., Lu C., Mains P.E., Peter M. The BTB protein MEL-26 promotes cytokinesis in C. elegans by a CUL-3 independent mechanism. Current Biology. 2005;15(18):1605–1615. doi: 10.1016/j.cub.2005.07.068. [DOI] [PubMed] [Google Scholar]
- 28.Pintard L., Willis J.H., Willems A., Johnson J.L., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., Peter M. The BTB protein MEL-26 is a substrate-specific adapter of the CUL-3 ubiquitin-ligase. Nature. 2003;425(6955):311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 29.Wang XJ, Sun Z, Villeneuve N.F., Zhang S., Zhao F, Li Y., Chen W., Yi X., Zheng W., Wondrak G.T., Wong P.K., Zhang D.D. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29(6):1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moghe S., Jiang F., Miura Y., Cerny R.L., Tsai M.Y., Furukawa M. The CUL3–KLHL18 ligase regulates mitotic entry and ubiquitylates Aurora-A. Biology Open. 2012;1(2):82–91. doi: 10.1242/bio.2011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meraldi P., Honda R., Nigg E.A. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Current Opinion in Genetics and Development. 2004;14(1):29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Strebhardt K., Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nature Reviews Cancer. 2006;6(4):321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 33.Hayward D.G., Clarke R.B., Faragher A.J., Pillai M.R., Hagan I.M., Fry A.M. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Research. 2004;64(20):7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 34.Beck J., Maerki S., Posch M., Metzger T., Persaud A., Scheel H., Hofmann K., Rotin D., Pedrioli P., Swedlow J.R., Peter M., Sumara I. Ubiquitylation-dependent localization of PLK1 in mitosis. Nature Cell Biology. 2013;15(4):430–439. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maerki S., Olma M.H., Staubli T., Steigemann P., Gerlich D.W., Quadroni M., Sumara I, Peter M. The Cul3–KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. Journal of Cell Biology. 2009;187(6):791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer J.D., Gurian-West M., Clurman B., Roberts J.M. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Development. 1999;13(18):2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEvoy J.D., Kossatz U., Malek N., Singer J.D. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Molecular and Cellular Biology. 2007;27(10):3651–3666. doi: 10.1128/MCB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todd M.C., Spruill S.C., Meerbrey K.L. Small interference RNA-mediated suppression of overexpressed cyclin E protein restores G1/S regulation in NIH-OVCAR-3 ovarian cancer cells. International Journal of Oncology. 2009;35(2):375–380. [PubMed] [Google Scholar]
- 39.Kossatz U., Breuhahn K., Wolf B., Hardtke-Wolenski M., Wilkens L., Steinemann D., Singer S., Brass F., Kubicka S., Schlegelberger B., Schirmacher P., Manns M.P., Singer J.D., Malek N.P. The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. The Journal of Clinical Investigation. 2010;120(11):3820–3833. doi: 10.1172/JCI41959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer H., Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Molecular Biology of the Cell. 2006;17(3):1250–1260. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Ding J., Allen E., Zhu P., Zhang L., Vogel H., Yang Y. Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin–proteasome pathway. Current Biology. 2005;15(22):2050–2055. doi: 10.1016/j.cub.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 42.Dequen F., Bomont P., Gowing G., Cleveland D.W., Julien J.P. Modest loss of peripheral axons, muscle atrophy and formation of brain inclusions in mice with targeted deletion of gigaxonin exon 1. Journal of Neurochemistry. 2008;107(1):253–264. doi: 10.1111/j.1471-4159.2008.05601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen H., Korutla L., Champtiaux N., Toda S., LaLumiere R., Vallone J., Klugmann M., Blendy J.A., Mackler S.A., Kalivas P.W. NAC1 regulates the recruitment of the proteasome complex into dendritic spines. Journal of Neuroscience. 2007;27(33):8903–8913. doi: 10.1523/JNEUROSCI.1571-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cullinan SB, Gordan JD, Jin J., Harper J.W., Diehl J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Molecular and Cellular Biology. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson J.L., Lu C., Raharjo E., McNally K., McNally F.J., Mains P.E. Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis. Developmental Biology. 2009;330(2):349–357. doi: 10.1016/j.ydbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman J.S., Ray J.W., Waseem N., Johnson K., Brooks M.J., Hugosson T., Breuer D., Branham K.E., Krauth D.S., Bowne S.J., Sullivan L.S., Ponjavic V., Gränse L., Khanna R., Trager E.H., Gieser L.M., Hughbanks-Wheaton D, Cojocaru R.I., Ghiasvand N.M., Chakarova CF, Abrahamson M., Göring H.H., Webster A.R., Birch D.G., Abecasis G.R., Fann Y., Bhattacharya S.S., Daiger S.P., Heckenlively J.R., Andréasson S., Swaroop A. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. American Journal of Human Genetics. 2009;84(6):792–800. doi: 10.1016/j.ajhg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Königsberg-Fainstein M. Nrf2: La historia de un nuevo factor de transcripción que responde a estrés oxidativo. Revista de Educación Bioquímica. 2007;26(1):18–25. [Google Scholar]
- 48.Stewart D., Killeen E., Naquin R., Alam S., Alam J. Degradation of transcription factor Nrf2 via the UBIQuitin–proteasome pathway and stabilization by cadmium. The Journal of Biological Chemistry. 2003;278(4):2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 49.Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radical Biology and Medicine. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itoh K., Wakabayashi N, Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes and Development. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo S.C., Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Molecular and Cellular Biology. 2006;26(4):1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motohashi H., Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends in Molecular Medicine. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Copple I.M., Goldring C.E., Kitteringham N.R., Park B.K. The Nrf2-Keap1 defence pathway: role in protection against drug-induced toxicity. Toxicology. 2008;246:24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 54.Sekhar K.R., Rachakonda G., Freeman M.L. Cysteine-based regulation of the CUL3 adaptor protein Keap1. Toxicology and Applied Pharmacology. 2010;244:21–26. doi: 10.1016/j.taap.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eggler A.L., Small E., Hannink M., Mesecar A.D. Cul3-mediated Nrf2 ubiquitination and antioxidant response element (ARE) activation are dependent on the partial molar volume at position 151 of Keap1. The Biochemical Journal. 2009;422:171–180. doi: 10.1042/BJ20090471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loignon M., Miao W., Hu L, Bier A., Bismar T.A., Scrivens P.J., Mann K., Basik M., Bouchard A., Fiset P.O., Batist Z., Batist G. Cul3 overexpression depletes Nrf2 in breast cancer and is associated with sensitivity to carcinogens, to oxidative stress, and to chemotherapy. Molecular Cancer Therapeutics. 2009;8(8):2432–2440. doi: 10.1158/1535-7163.MCT-08-1186. [DOI] [PubMed] [Google Scholar]
- 57.Okawa H., Motohashi H., Kobayashi A., Aburatani H., Kensler T.W., Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochemical and Biophysical Research Communications. 2006;339(1):79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 58.Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S, Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., Brock M.V., Biswal S. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Medicine. 2006;3(10):e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M.I., Kobayashi A., Yokoyama S., Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Molecular Cell. 2006;21(5):689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Kaspar J.W., Jaiswal A.K. An autoregulatory loop between Nrf2 and Cul3-Rbx1 controls their cellular abundance. The Journal of Biological Chemistry. 2010;285(28):21349–21358. doi: 10.1074/jbc.M110.121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y., Yang J.W., Ren X., Yang J.M. NAC1 and HMGB1 enter a partnership for manipulating autophagy. Autophagy. 2011;7(12):1557–1568. doi: 10.4161/auto.7.12.17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakayama K., Nakayama N., Davidson B., Sheu J.J., Jinawath N., Santillan A, Salani R., Bristow R.E., Morin P.J., Kurman R.J, Wang T.L., Shih IeM. A. BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(49):18739–18744. doi: 10.1073/pnas.0604083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeasmin S., Nakayama K., Ishibashi M., Katagiri A., Iida K., Purwana I.N., Nakayama N., Miyazaki K. Expression of the bric-a-brac tramtrack broad complex protein NAC-1 in cervical carcinomas seems to correlate with poorer prognosis. Clinical Cancer Research. 2008;14(6):1686–1691. doi: 10.1158/1078-0432.CCR-07-4085. [DOI] [PubMed] [Google Scholar]
- 64.Ji L., Li H., Gao P., Shang G., Zhang D.D., Zhang N., Jiang T. Nrf2 pathway regulates multidrug-resistance-associated protein 1 in small cell lung cancer. PLoS One. 2013;8(5):e63404. doi: 10.1371/journal.pone.0063404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamane Y., Furuichi M., Song R., Van N.T., Mulcahy R.T., Ishikawa T., Kuo M.T. Expression of multidrug resistance protein/GS-X pump and gamma-glutamylcysteine synthetase genes is regulated by oxidative stress. The Journal of Biological Chemistry. 1998;273:31075–31085. doi: 10.1074/jbc.273.47.31075. [DOI] [PubMed] [Google Scholar]
- 66.Hayashi A., Suzuki H., Itoh K., Yamamoto M., Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochemical and Biophysical Research Communications. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 67.Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. The EMBO Journal. 2002;21(12):3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyden L.M., Choi M, Choate K.A., Nelson-Williams C.J., Farhi A., Toka H.R., Tikhonova I.R., Bjornson R., Mane S.M., Colussi G., Lebel M., Gordon R.D., Semmekrot B.A., Poujol A., Välimäki M.J., De Ferrari M.E., Sanjad S.A., Gutkin M., Karet F.E., Tucci J.R., Stockigt J.R., Keppler-Noreuil K.M., Porter C.C., Anand S.K., Whiteford M.L., Davis I.D., Dewar S.B., Bettinelli A., Fadrowski J.J., Belsha C.W., Hunley T.E., Nelson R.D., Trachtman H., Cole T.R., Pinsk M., Bockenhauer D., Shenoy M., Vaidyanathan P, Foreman J.W., Rasoulpour M., Thameem F, Al-Shahrouri H.Z., Radhakrishnan J., Gharavi A.G., Goilav B., Lifton R.P. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji A.X., Privé G.G. Crystal structure of KLHL3 in complex with Cullin3. PLoS One. 2013;8(4):e60445. doi: 10.1371/journal.pone.0060445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohta A., Schumacher F.R., Mehellou Y., Johnson C., Knebel A., Macartney T.J., Wood N.T., Alessi D.R., Kurz T. The CUL3–KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. The Biochemical Journal. 2013;451(1):111–122. doi: 10.1042/BJ20121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruilope L.M., Bakris G.L. Renal function and target organ damage in hypertension. European Heart Journal. 2011;32(13):1599–1604. doi: 10.1093/eurheartj/ehr003. [DOI] [PubMed] [Google Scholar]
- 72.Cirak S., von Deimling F, Sachdev S., Errington W.J., Herrmann R., Bönnemann C., Brockmann K., Hinderlich S., Lindner T.H., Steinbrecher A., Hoffmann K., Privé G.G., Hannink M., Nürnberg P., Voit T. Kelch-like homologue 9 mutation is associated with an early onset autosomal dominant distal myopathy. Brain. 2010;133(Pt 7):2123–2135. doi: 10.1093/brain/awq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shibata S., Zhang J., Puthumana J., Stone K.L., Lifton R.P. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7838–7843. doi: 10.1073/pnas.1304592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin S.H., Yu I.S., Jiang S.T., Lin S.W., Chu P., Chen A., Sytwu H.K., Sohara E., Uchida S., Sasaki S., Yang S.S. Impaired phosphorylation of Na(+)–K(+)–2Cl(−) cotransporter by oxidative stress-responsive kinase-1 deficiency manifests hypotension and Bartter-like syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(42):17538–17543. doi: 10.1073/pnas.1107452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edelmann M.J., Nicholson B., Kessler B.M. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Reviews in Molecular Medicine. 2011;13:e35. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruschak A.M., Slassi M., Kay L.E., Schimmer A.D. Novel proteasome inhibitors to overcome bortezomib resistance. Journal of the National Cancer Institute. 2011;103(13):1007–1017. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- 77.Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov I.V., Oberoi P., Li C.C., Kenten J.H., Beutler J.A., Vousden K.H., Weissman A.M. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Research. 2007;67(19):9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 78.Soucy T.A., Smith P.G., Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clinical Cancer Research. 2009;15(12):3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 79.Nawrocki S.T., Kelly KR, Smith P.G., Espitia C.M., Possemato A., Beausoleil SA, Milhollen M., Blakemore S., Thomas M., Berger A., Carew J.S. Disrupting protein NEDDylation with MLN4924 is a novel strategy to target cisplatin resistance in ovarian cancer. Clinical Cancer Research. 2013 doi: 10.1158/1078-0432.CCR-12-3212. ([Epub ahead of print]) [DOI] [PubMed] [Google Scholar]
- 80.Luo Z., Yu G., Lee H.W., Li L., Wang L, Yang D, Pan Y., Ding C., Qian J., Wu L., Chu Y., Yi J, Wang X., Sun Y., Jeong L.S., Liu J., Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Research. 2012;72(13):3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 81.Zhao Y., Sun Y. Cullin-RING Ligases as attractive anti-cancer targets. Current Pharmaceutical Design. 2013;19(18):3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]


