Abstract
The salamander is the only tetrapod that regenerates complex body structures throughout life. Deciphering the underlying molecular processes of regeneration is fundamental for regenerative medicine and developmental biology, but the model organism had limited tools for molecular analysis. We describe a comprehensive set of germline transgenic strains in the laboratory-bred salamander Ambystoma mexicanum (axolotl) that open up the cellular and molecular genetic dissection of regeneration. We demonstrate tissue-dependent control of gene expression in nerve, Schwann cells, oligodendrocytes, muscle, epidermis, and cartilage. Furthermore, we demonstrate the use of tamoxifen-induced Cre/loxP-mediated recombination to indelibly mark different cell types. Finally, we inducibly overexpress the cell-cycle inhibitor p16INK4a, which negatively regulates spinal cord regeneration. These tissue-specific germline axolotl lines and tightly inducible Cre drivers and LoxP reporter lines render this classical regeneration model molecularly accessible.
Highlights
-
•
A set of germline transgenic axolotls for spatiotemporal control of gene expression
-
•
Controlled gene expression in cartilage and neural stem cells via Cre/loxP system
-
•
p16INK4a represses spinal cord regeneration and induces regeneration phenotype
Introduction
Generations of biologists have been fascinated by salamander limb and tail regeneration. Important grafting experiments delineated the fundamental tissue interactions involved that provide a rich basis for a molecular understanding of regeneration (Bryant and Iten, 1977; Dunis and Namenwirth, 1977; Steen, 1968; Stocum, 1975; Wallace and Wallace, 1973) (for reviews, see Nacu and Tanaka, 2011; Stocum and Cameron, 2011). For example, after limb amputation, cells from the muscle, connective tissue, skeletal, peripheral nerve, and epidermis all contribute to the progenitor cell zone, called the blastema, which will reconstitute all limb tissues. The progenitors from these diverse tissues remain largely separate and show divergent behaviors, for example, during limb patterning (Kragl et al., 2009; Nacu et al., 2013). Therefore, a mechanistic understanding of regeneration requires the control of gene expression in specific cell types. Furthermore, because genes involved in regeneration may also be implemented during development, inducible gene expression is important.
Although several vertebrate models of regeneration are available, there are compelling reasons to study regeneration in the salamander. First, the graftability of salamander tissue provides a uniquely powerful approach to lineage trace cells and to test cell-autonomous versus -nonautonomous events (Muneoka et al., 1986; Pescitelli and Stocum, 1980). Second, the distinct anatomy of the zebrafish fin from the tetrapod limb limits what we can infer from fins to limbs (Sordino et al., 1995). Xenopus, a valuable molecular model of regeneration, regenerates the developing hindlimb only prior to metamorphosis (for review, see Yakushiji et al., 2009). This regeneration occurs before full differentiation and could involve progenitor cell sources distinct from those utilized to regenerate a fully differentiated limb (Dent, 1962). Therefore, it is important to study regeneration in multiple vertebrates.
Detailed molecular analysis of regeneration has been difficult because it is an adult phenotype with a complex starting point involving the response of many tissues to injury. It is likely that only a subset of cells in any given adult tissue contributes to the blastema. Electroporation of plasmid DNAs and morpholinos has been used to analyze important phenotypes during salamander limb regeneration, but this technique provides transient, variable transfection (Echeverri and Tanaka, 2005; Kumar et al., 2007; Mercader et al., 2005). Adenovirus, vaccinia virus, and pseudotyped retrovirus have also been applied, but the limited cargo size and toxicity have limited their use and have not yet been widely adopted (Kawakami et al., 2006; Morrison et al., 2010; Roy et al., 2000; Whited et al., 2013). Sustained expression from genomic integration is extremely important for the study of salamander limb regeneration. Successful germline transgenesis has been reported for axolotl (Sobkow et al., 2006) and has been applied to the Japanese newt and Iberian ribbed newt (Casco-Robles et al., 2010; Hayashi et al., 2013). Moreover, germline EGFP and nuclear Cherry-expressing transgenic axolotls were used in combination with tissue grafting to provide important proof that the regenerating limb blastema is a mixed population of tissue-restricted progenitors (Kragl et al., 2009). For further molecular dissection of regeneration, however, conditional induction of gene expression in a time and/or cell-type-dependent fashion is essential. Recently, Whited and colleagues described a system to induce gene expression in axolotls using an IPTG system, whereas this system is promising for acute control of gene expression, the continuous presence of inducer is required to sustain expression that could limit its use for cell tracking during regeneration (Whited et al., 2012). It is not yet known how well the system can be combined with cell-type-specific control of gene expression. Furthermore, the effectiveness of the system was not shown in germline-transmitted F1 animals where full-genomic integration of the transgene may alter its expression properties. In larval axolotls, full-limb regeneration occurs over 2–3 weeks, whereas in fully adult animals, the process can take over 10 weeks. Therefore, a sustainable gene induction system such as the Cre/loxP system would be valuable for regeneration studies, but tamoxifen-inducible systems were characterized as too leaky for axolotl transgenics (Whited et al., 2012).
Here, we describe tissue- and time-dependent control of gene expression in germline transgenic axolotls via Cre/loxP methodology. We provide a set of germline transgenic animals driving the GFP reporter behind different cell-type-specific promoters relevant for regeneration research. We further employ Cre/loxP to conditionally initiate gene expression in a time and cell-type-specific manner. As a proof of principle for phenotype analysis, we demonstrate the inducible overexpression of the tumor suppressor p16INK4a. It has been proposed that the p16INK4a/ARF locus first arose in mammals and is not present in the salamander, a potential basis for the restricted regeneration capacities of mammals (Pajcini et al., 2010). By CRE-mediated induction of p16INK4a, we show that it suppresses axolotl spinal cord regeneration. These tools make the axolotl amenable for molecular analysis of regeneration.
Results
Tissue-Specific Germline Transgenic Reporter Strains
Because grafting experiments indicated that each tissue contributes uniquely to regeneration (Kragl et al., 2009), a molecular analysis of regeneration requires controlling gene expression specifically in different tissues. We implemented tissue-specific promoters from different animal species to successfully establish germline transgenic axolotl strains showing faithful, cell-type-specific expression of the EGFP gene (Table 1; Figure 1). Transgenic strains were produced by injection of plasmid or BAC DNA into one- to two-cell embryos as previously described (Khattak et al., 2009; Sobkow et al., 2006). Injected animals were grown to larval stages and examined via fluorescence protein expression for the extent of expression. Only animals with apparently greater than 80% expression along the body were raised to sexual maturity and then mated to nontransgenic mates (see Table 1 for numerical details for each transgenic strain). The F1 progenies were examined for EGFP expression in the expected cell/tissue types as live whole mounts. To confirm specificity and penetrance of expression, limbs and tails were cross-sectioned and immunostained for respective proteins. We only propagated strains that showed strict colocalization of the EGFP with the respective marker proteins and no extraneous expression. We found this to be an important aspect of the screening process because random integration of the transgenic constructs leads to position effects that can yield mosaicism in the progeny.
Table 1.
Summary of Germline Transgenic Axolotls
| Genotypes | Germline Transmission Rate (No. of Founders Raised and Mated/No. that Went Germline) | Details of Constructs | References |
|---|---|---|---|
| Ubiquitous Transgenic Lines | |||
| CAGGs: EGFP | 15/6 | CAGGs promoter driving EGFP | Sobkow et al. (2006) |
| CAGGs: CherryNuc | 19 Raised and 4 mated/4 | CAGGs promoter driving nuclear Cherry | Kragl et al. (2009) |
| CAGGs: LP-EGFP-LP-Cherry | 11 Raised and 7 mated/7 | CAGGs promoter driving floxed EGFP cassette followed by Cherry | |
| CAGGs: LP-EGFP-LP-Tomato | 20 Raised and 2 mated/2 | CAGGs promoter driving floxed EGFP cassette followed by Tomato | |
| CAGGs: LP-EGFP-LP-p16-T2A-Cherry | 9 Raised and 4 mated/2 | CAGGs promoter driving floxed EGFP cassette followed by p16INK4A and Cherry | |
| CAGGs: ER-Cre-ER-T2A-EGFP-nuc | 3/3 | CAGGs promoter driving ert2-Cre-ert2 and nuclear EGFP | |
| Tissue-Specific Transgenic Lines | |||
| B3Tubulin: EGFP | 6/1 | Mouse βIII-tubulin BAC with membrane localized EGFP | Attardo et al. (2008) |
| CNP: EGFP | 7 Raised and 3 mated/3 | Mouse 2′3′-cyclic nucleotide 3′-phosphodiesterase (CNP) promoter driving EGFP | Glaser et al. (2005) |
| Col2a1: EGFP | 6/2 | Xenopus collagen 2 α 1 promoter driving EGFP | Kerney et al. (2010) |
| KRT12: EGFP | 4/1 | Xenopus keratin 12 promoter fragment driving EGFP | Suzuki et al. (2010) |
| CarAct: EGFP | 8 Raised and 3 mated/3 | Xenopus cardiac actin promoter driving EGFP | Ogino et al. (2006) |
| AxSox2:cre-ert2-T2A-GFP | 5/1 | Axolotl Sox2 genomic clone with cre-ert2-GFP-nuc integrated instead of the ORF | |
| Col2A1:ER-Cre-ER-T2A-EGFP-nuc | 5/1 | Xenopus Col2A1 promoter driving ert2-Cre-ert2 and nuclear EGFP | Kerney et al. (2010) |
Figure 1.
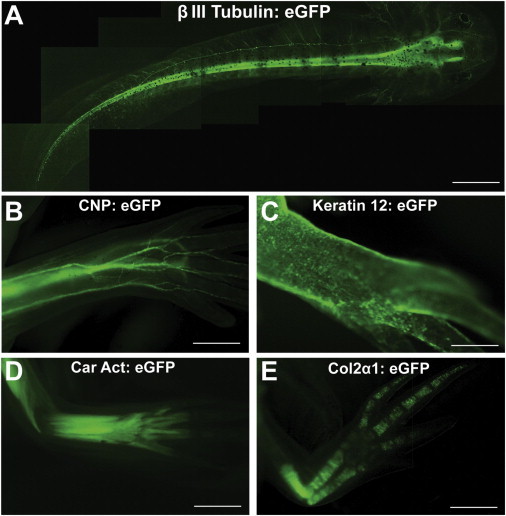
Transgenic Axolotls with Cell-type-Specific Transgene Expression of EGFP
(A) Live axolotl larva expressing membrane-tethered EGFP in neurons under the control of mouse βIII-tubulin promoter.
(B) Limb of live transgenic axolotl expressing EGFP in Schwann cells. The transgenic animal is expressing EGFP under the control of mouse CNP promoter and marking all myelin-positive cells in the body.
(C) Xenopus Keratin 12 promoter driving EGFP in epidermis of live axolotl. Here, a limb image is shown.
(D) Fluorescence image of limb on a live animal showing EGFP expression in muscle driven by the Xenopus Cardiac Actin (Car Act) promoter.
(E) Limb of live transgenic axolotl where Xenopus Col2α1 promoter is driving EGFP in cartilage.
Scale bars, 1 mm (B, D, and E) and 2 mm (A and C). See also Figure S1.
To generate germline transgenic lines expressing EGFP in neurons, we implemented the mouse βIII-tubulin regulatory sequences by injecting a previously characterized mouse BAC construct in which the gene for a GAP43-EGFP fusion protein sequence had been integrated with its own poly(A) signal downstream of the coding sequence (Attardo et al., 2008). Injection was performed without SceI meganuclease, and 12 animals out of 343 injected eggs showed neural-specific expression over large portions of the body (approximately >80% of cells) (Table 1). Six animals were grown up for germline transmission and mated to a nontransgenic mate. One of the animals displayed a germline transmission rate of 24%. Figure 1A shows whole-mount images of a live F1 progeny showing GFP signal in nerve fibers. To confirm the specificity of expression to neurons, we immunostained tail and limb sections for βIII-tubulin (Figures S1A and S1B available online). Excellent (100%) colocalization of EGFP signal with immunofluorescence signal was observed, confirming the specific expression of the transgene. Abundant expression was observed in the spinal cord (arrow), as well as the dorsal root ganglion (arrowhead) and peripheral nerve tracts (asterisks) (Figure S1A), indicating that central and peripheral neurons expressed the transgene.
For marking of glial cells including oligodendrocytes and Schwann cells, we implemented the mouse CNP promoter (Glaser et al., 2005). After coinjection of the construct with SceI meganuclease, we obtained 7 strongly expressing F0 animals out of 758 originally injected. Three were mated, and all showed germline transmission. Figure 1B shows the nerve tracts expressing EGFP in the limb of a typical live F1 animal. Out of the three mated founders, the F1 progeny of one had uniform EGFP expression in putative Schwann cells and oligodendrocytes, whereas the progeny of the other two founders showed mosaic EGFP expression. The limbs and tails of F1 progeny from the uniform founder were cross-sectioned and immunostained with an anti-MBP antibody confirming high (100%) colocalization of EGFP signal with MBP+ structures (Figures 1B, S1C, and S1D). EGFP expression was observed in both the central spinal cord (arrow) (Figure S1C), as well as in DRG (arrowhead) and peripheral nerve tracts (asterisks) of the tail and limb (Figures S1C and S1D), indicating that oligodendrocytes and Schwann cells expressed the transgene. Higher-magnification confocal imaging of a nerve tract in the limb confirmed expression of EGFP in the nucleus and cytoplasm of MBP+ cells wrapping around the axon (Figure S1E).
To label other tissues important in regeneration, such as epidermis, muscle, and cartilage, we employed previously characterized Xenopus promoters driving EGFP, namely Keratin12:EGFP, CarAct:EGFP, and Col2α1:EGFP (Kerney et al., 2010; Ogino et al., 2006; Suzuki et al., 2010). The Keratin:EGFP animals express EGFP exclusively in the skin (Figures 1C, S1F, and S1G), whereas the CarAct:EGFP transgenics show robust expression in the myosin heavy-chain-expressing muscle and not in the PAX7-positive satellite cells, indicating faithful muscle-specific expression (Figures 1D, S1H, and S1I). The Col2a1:EGFP line shows expression in cartilage cells, for example, of the larval limb (Figure 1E).
To further propagate these confirmed lines, F1 progenies were raised to sexual maturity and crossed with white nontransgenic hosts to check for germline transmission and specific uniform EGFP expression in F2 progeny. The F1 male and females were mated to establish “homozygous” lines that result in 100% transgenic progeny when mated to a nontransgenic animal (Table 1).
Application: Tracking Neurons and Muscle during Limb Regeneration
The ingrowth of nerves is an essential aspect of limb regeneration (Kumar and Brockes, 2012), but the process has so far been followed by quantitative electron microscopy and immunofluorescence. The optical clarity of the “white” axolotl strain allows visualization of fluorescent cells in live animals (Echeverri et al., 2001). Here, to demonstrate the feasibility of following axonal outgrowth during limb regeneration, we amputated limbs of larval βIIItub:GAP43-EGFP animals and followed them over time in the same live animal (Figures 2A–2C). During regeneration, we observed ingrowth of thin axonal fibers already in the early 6 day blastema (Figure 2A). Further infiltration of axons was observed by the Notch stage of regeneration (Figure 2B) and consolidation of larger axonal bundles infiltrating the forming digits at the Palette stage (Figure 2C). These data indicate that robust innervation occurs early during blastema formation, and fiber termini extend well toward the distal epithelium. In the future, more detailed imaging studies can elucidate the dynamics of nerve-epithelial interactions that are crucial for proper limb outgrowth (Kumar et al., 2007; Mullen et al., 1996).
Figure 2.
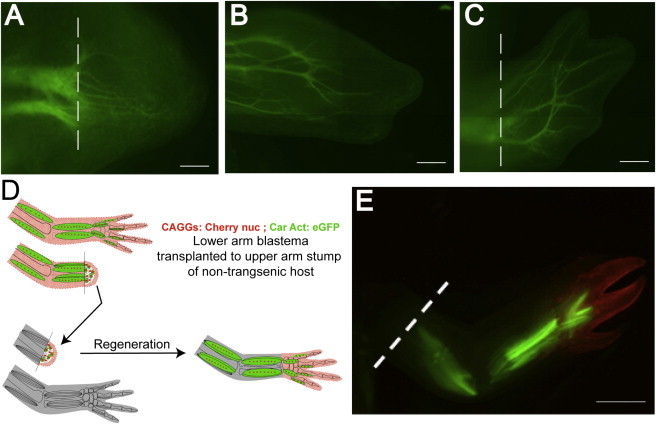
In Vivo Tracking of Nerves and Muscle during Normal and Intercalary Limb Regeneration
(A–C) Live tracking of growing nerves in a βIIItub:GAP43-EGFP transgenic animal. Nerves as seen in a 6 day limb blastema (A), early digit-forming (Notch) stage (B), and late digit (Palette)-regeneration stage (C) of the same animal. The dashed white lines in (A) and (C) represent the amputation plane.
(D) Scheme of hand blastema transplantation to follow EGFP muscle versus other cell types during intercalary regeneration. A double-transgenic (CAGGs:Cherrynuc;CarAct:EGFP) hand blastema was transplanted onto an upper-arm stump of a nontransgenic white mutant host and allowed to regenerate.
(E) Regenerated limb from the double-transgenic hand blastema graft. The white dashed line shows the amputation plane of the host stump (to where the hand blastema was transplanted). Muscle fibers (green) are present in upper-arm regions during intercalary regeneration, whereas the cherry-positive cells largely populate the hand region of the regenerate.
Scale bars, 100 μm (A), 200 μm (B), 300 μm (C), and 1 mm (E).
Another important aspect of limb regeneration is determination of proximal versus distal identity. It was recently shown that skeletal/connective tissue-derived blastema cells regenerate limb elements only more distal to the amputation plane, whereas muscle-derived blastema cells have the potential to regenerate muscle tissue of all limb segments (Nacu et al., 2013). In the previous studies, muscle labeling was performed by grafting of presomitic mesoderm from constitutively expressing CAGGs:EGFP donor embryos into white hosts. Here, we demonstrate the utility of the CarAct:EGFP transgenic animal for revealing the potency of muscle to regenerate cells in all limb segments. To track muscle versus other limb tissues, a CarAct:EGFP animal was first crossed to the CAGGs:Cherry nuc animal (expresses nuclear Cherry in all cells) to generate double-transgenic CarAct:EGFP; CAGGs:Cherry nuc that express both transgenes. Midbud hand blastemas were grafted onto the upper-arm stump of a white host animal, in a classic intercalation assay (Figure 2D) (Iten and Bryant, 1975; Stocum, 1975). Limbs of normal morphology regenerated (Figure 2E). Examination of fluorescence signal showed that the muscle-specific EGFP signal (that comes only from the transplanted wrist blastema) was observed in muscle of upper limb, lower limb, and hand (Figures 2D and 2E). In contrast, the nuclear Cherry signal, which in whole mount is most noticeable in the skin-derived cells, was largely confined to the hand. These results show that muscle progenitors from a hand blastema can form upper- and lower-arm muscle.
Use of Cre/loxP System for Tissue and Time-Dependent Control of Gene Expression
To study regeneration, an inducible expression system is important for two reasons. First, when using genetic fate mapping, irreversible activation of marker expression in a given cell type is important because the cell expressing a given marker gene in the mature tissue may, during regeneration, turn off the marker. Second, when studying gene function, inducible expression is important because genes functioning in regeneration may also have earlier roles in development. Considering the long timescales of regeneration, a system that induces sustained expression would be desirable. We therefore focused on testing the Cre/loxP system. We first generated a germline transgenic loxP reporter strain where the CAGGs promoter drives a floxed EGFP-STOP cassette followed by the Cherry gene- CAGGs:LP-EGFP-STOP-LP-Cherry (Figure S2). A total of 11 putative founders were raised to sexual maturity. Seven were mated to white nontransgenic hosts, and all yielded germline transmission with a frequency in the range of 5.4%–54%. Only one founder had mosaic EGFP expression as determined by cross-sectioning the limbs and tails of F1 progeny and staining the sections with DAPI to ascertain that EGFP is expressed in every cell.
To evaluate whether CRE-mediated recombination can work on the genomically inserted transgene, we electroporated a Cre expression plasmid into the mature limb and tail of the F1 progeny of loxP reporter. After 3 days, we observed robust Cherry expression only in animals that had received Cre plasmid (Figures S2D and S2H), and not in control unelectroporated animals (Figures S2B and S2F). No Cherry expression was observed when PBS and/or empty plasmid (pUC19) was electroporated into the tail and limb of the loxP reporter animal. Similarly, no spontaneous Cherry expression was observed after limb/tail amputation and during regeneration of the PBS/pUC19 electroporated animals (Figure S3).
Interestingly, coelectroporation of the split Cre constructs (Casanova et al., 2003) together also induced robust recombination and Cherry expression, whereas electroporation of the single constructs did not, bringing up the possibility of making CRE activity dependent on expression of two promoters (Figure S4).
Cell-type-Dependent Control of Cre Activity in Sox2+ Neural Stem Cells
We next sought to control Cre expression in a cell-type-specific manner. We isolated a 15 kb genomic clone for the axolotl Sox2 gene that is expressed in the neural stem cells of the CNS (Li et al., 1998; McHedlishvili et al., 2012; Zappone et al., 2000). The clone included 11.68 kb upstream of the coding sequence, the Sox2 coding sequence and 2.66 kb downstream sequence of the coding sequence. Recombineering was used to incorporate a Cre ERT2-T2A-nucGFP cassette by replacing the Sox2 coding sequence. The resulting construct was injected into eggs together with SceI meganuclease. Cre-ERT2 is a highly used method for induction of CRE activity via the tamoxifen metabolite, 4-hydroxytamoxifen (4-OHT) (Metzger and Chambon, 2001). F1 progeny from a germline-transmitting animal showed robust expression of nuclear GFP in the ventricular, SOX2+ cells of the brain (Figures 3C–3G′). When crossed to the CAGGs:LP-EGFP-STOP-LP-Cherry reporter, the F1 progeny displayed strong Cherry expression in the brain even in the absence of tamoxifen, indicating leaky but cell-type-specific activity of the CRE-ERT2 (Figures 3I and 3J). Because recombination was occurring continuously even in the absence of tamoxifen, the distribution of Cherry+ cells was broader than the nucGFP because the SOX2+ cells and their progeny expressed the Cherry gene (Figures 3J–3M′). From these experiments, we can conclude that cell-type-specific expression of CRE activity in neural stem cells is achievable but that the CRE-ERT2 is not sufficiently stringent for tamoxifen-induced activity, showing spontaneous recombination (Figure 3).
Figure 3.
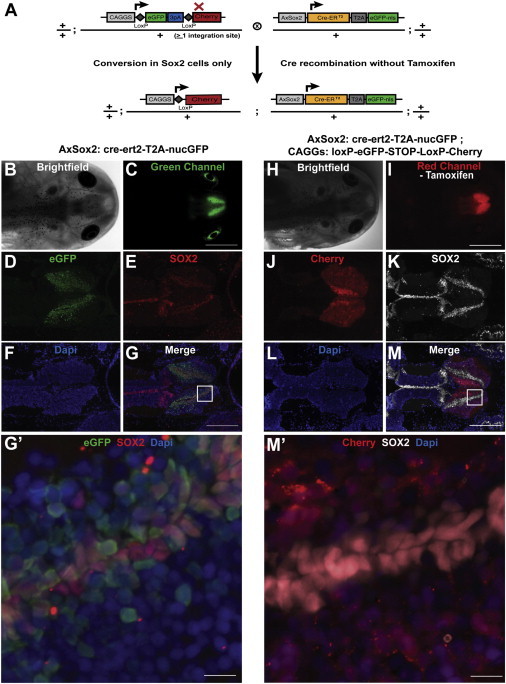
Neural Stem Cell Expression via AxSox2:Cre-ERT2-nucGFP and Marking of Neural Stem Cell Descendents via CRE-Mediated Recombination
(A) Scheme of double transgenic used to label SOX2+ cells in the brain. LoxP reporter (CAGGs:loxP-EGFP-loxP-Cherry) was crossed with a transgenic axolotl where the Cre-ERT2 gene is under the control of the axolotl Sox2 promoter.
(B) Bright-field image of head of Sox2: Cre-ERT2 transgenic animal line (F1s).
(C) EGFP fluorescence image of the head shown in (B) showing brain-specific expression.
(D–G) Immunostaining of head cross-section of Sox2: Cre-ERT2 driver animal. nucGFP expression is seen in (D), SOX2 immunofluorescence is in (E), nuclei are stained for DAPI in (F), and (G) shows the merged image. (G′) Higher-resolution image of inset in (G) showing colocalization of EGFP signal with immunostaining for SOX2 in the ventricular zone of the brain section.
(H) Bright-field image of head of double-transgenic animal, AxSox2:Cre-ERT2-nucGFP; CAGGs:loxP-EGFP-loxP-Cherry.
(I) Cherry fluorescence image of the head shown in (H). Clear Cherry fluorescence is seen in brain without tamoxifen administration.
(J–M) Cross-section of head of AxSox2:Cre-ERT2-nucGFP; CAGGs:loxP-EGFP-loxP-Cherry showing expression in Cherry (J), SOX2 immunofluorescence (K)-positive cells in the brain. (L) shows DAPI+ nuclei, whereas (M) is the merged image. A broader fluorescence is seen compared to the nucGFP signal in the AxSox2:Cre-ERT2-nucGFP driver, indicating that Sox2+ cells have contributed to new neurogenesis in the brain. (M′) Higher-resolution inset of (M) brain section showing Cherry-positive cells in SOX2+ cells as well as their putative descendants.
Scale bars, 2 mm (C and I), 200 μm (G and M), and 30 μm (G′ and M′).
ERT2-Cre-ERT2 Fusions Provide Nonleaky Tamoxifen-Inducible Cre-Mediated Gene Expression in Axolotl
To achieve tighter temporal induction of CRE activity, we assayed a number of induction systems for driving the Cre gene, including Tet-on/off, GBD, Gal80ts, Gal4VP16/UAS, and a double fusion to the ERT2 sequences (García-Otín and Guillou, 2006; Mallo, 2006; McGuire et al., 2003; Verrou et al., 1999), by electroporation of plasmids for these systems into limb tissue. Most systems displayed problems such as leakiness or toxicity. For example, dexamethasone was toxic to the animals. Two versions of the Tet-on system were tested: the Tet-on advanced transactivator (rtTA2S-M2) (Urlinger et al., 2000) yielded significant Cre recombination in the absence of doxycycline (Figures S5A and S5B). The improved reverse tetracycline transactivator (irtTA) fused to the ligand binding domain of mutated glucocorticoid receptor (irtTA-GBD*) (Anastassiadis et al., 2010) showed less leakiness than rtTA2S-M2, but dexamethasone was toxic (Figures S5C and S5D). The temperature-sensitive repressor Gal80Ts that binds and blocks the Gal4-mediated transcription at 18°C and allows transcription at 30°C has been used in Drosophila (Pavlopoulos and Akam, 2011). Electroporation of Gal80Ts into the axolotl limb of loxP reporter along with Gal4-UAS-Cre still yielded Cre recombination at 18°C (Figures S5E and S5F).
We therefore reexplored the control of CRE by the ERT2 sequences and tamoxifen. As expected, in limbs electroporated with a CAGGs:Cre-ERT2 expression plasmid, we observed strong Cherry expression in the absence of tamoxifen (Figures S5G and S5H). Therefore, we tried a number of means to attenuate the CRE-ERT activity including fusion with a nuclear export signal and fusion with a ubiquitinylation signal, but neither decreased the background activity sufficiently (Figures S5I–S5L). However, we found that the ERT2-Cre-ERT2 fusion (Zhang et al., 1996) did elicit tight, tamoxifen-inducible CRE activity, as seen by no Cherry expression after the electroporation of the construct in the absence of tamoxifen (Figures S5M and S5N) but robust expression of Cherry after electroporation and subsequent tamoxifen induction (Figures S6O and S6P). We therefore produced a germline transgenic CAGGs:ERT2-Cre-ERT2-T2A-nuc-EGFP (Cre-2xERT2) animal (Figures 4A and 4B). When double-CAGGs:ERT2-Cre-ERT2-T2A-nuc-EGFP/+;CAGGs:loxP-EGFP-STOP-loxP-Cherry/+ animals were bred (Figure 4C), we observed no recombination prior to 4-OHT induction (Figures 4D and 4F). After a single intraperitoneal injection of 4-OHT, we observed robust Cherry expression as assayed in limb and tail tissue (Figures 4E and 4G). Some EGFP expression persisted after induction even in Cherry-positive cells. This is presumably due to the loxP reporter harboring multiple integrated copies of the transgene or due to perdurance of the GFP. We conclude that the ERT2-Cre-ERT2 fusion provides tight, temporal control of gene expression in the axolotl.
Figure 4.
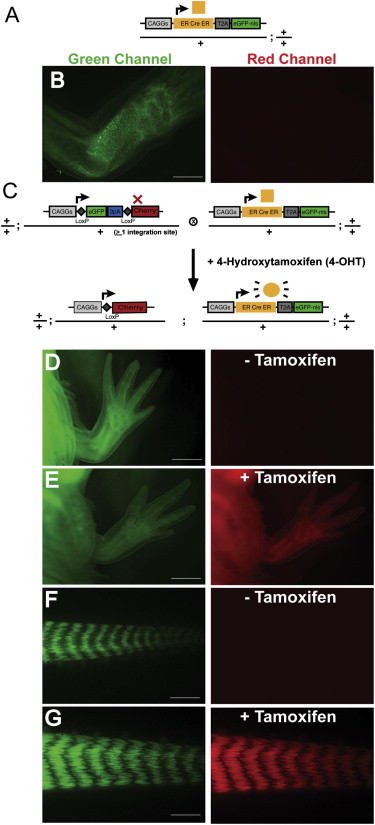
Tight Temporal Control of Cre/loxP-Mediated Gene Expression Using the ERT2-cre-ERT2 System
(A) Schematic diagram of the Cre driver. The CAGGs promoter is driving the ERT2-cre-ERT2-T2A-nuc-EGFP cassette.
(B) Limb of CAGGs:ERT2-Cre-ERT2-T2A-EGFP-nuc transgenic animal in green and red channel.
(C) Schema of mating between CAGGs:loxP-EGFP-loxP-Cherry and CAGGs:ERT2-Cre-ERT2-T2A-EGFP-nuc animals.
(D and F) Limb and tail of a double-transgenic (CAGGs:loxP-EGFP-loxP-Cherry; CAGGs:ERT2-Cre-ERT2-T2A-EGFP-nuc) animal showing EGFP and Cherry expression levels before tamoxifen induction.
(E and G) Robust Cherry expression is observed in limb and tail of double-transgenic (CAGGs:loxP-EGFP-loxP-Cherry; CAGGs:ERT2-Cre-ERT2-T2A-EGFP-nuc) animals after administration of 4-OHT.
Scale bars, 1 mm (B and D–F) and 500 μm (G). See also Figures S2, S3, S4, and S5.
Tamoxifen-Inducible Expression in CollagenII-Expressing Vertebral Cells
To combine cell-type-specific and temporal induction of gene expression, we placed the ERT2-Cre-ERT2-T2A-nuc-EGFP cassette behind the Xenopus Col2a1 promoter that drives expression in skeletal cells (see Figure 1E), and germline-transmitting animals were produced. Five putative founders were raised to sexual maturity, and one transmitted through the germline showing nuclear EGFP expression in cartilage of F1 progeny when crossed with white nontransgenic host. In this animal, the head and tail cartilage showed particularly strong expression of the transgene. The germline-transmitting animal was crossed to the loxP reporter animals (Figure 5A). In the F1, double-transgenic progeny, no Cherry expression was observed in the head or tail tissue prior to tamoxifen administration (Figures 5B and 5D). After tamoxifen induction, bright areas of Cherry-positive cells were observed in the head cartilage and in the notochord/cartilage ventral to the spinal cord (Figures 5C and 5E).
Figure 5.
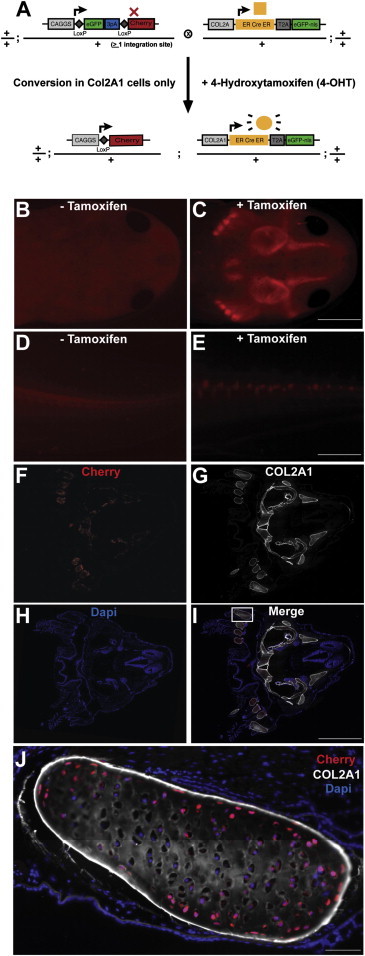
Temporal Induction of Gene Expression in Co2A1+ Cells Using Col2A1:ERT2-Cre-ERT2-T2A-EGFP-nuc Transgenic Axolotls
(A) Scheme of mating between loxP reporter animal (CAGGs:loxP-EGFP-STOP-loxP-Cherry) and the driver animal (Col2A1:ERT2-Cre-ERT2-T2A-EGFP-nuc).
(B and D) Double-transgenic animal shows no Cherry fluorescence in head (B) and tail (D) before tamoxifen induction. High-exposure times were used for imaging so that overall tissue architecture could be seen.
(C and E) Double-transgenic head (C) and tail (E) images after tamoxifen induction. Clear Cherry expression is observed only in the skeletal elements of the head and vertebral column in the tail. Half-exposure time from control (C and E) was used.
(F–I) Cross-section of head showing colocalization of Cherry (F) with Collagen type II antibody staining (G). DAPI delineates nuclei in blue. (I) represents merged image.
(J) High-resolution image of inset marked in (I) showing Cherry-positive cells associated with Col2A1 staining in head cartilage.
Scale bars, 2 mm (C, E, and I) and 100 μm (J).
In conclusion, we have demonstrated that cell-type and temporal control of gene expression is efficient and possible in germline transgenic axolotl lines. This will be useful to track cells during axolotl regeneration and development.
Use of Tamoxifen-Inducible Cre/loxP System to Overexpress p16INK4a
The Cre/loxP system is a potentially powerful tool to test the role of molecular pathways during regeneration by overexpression of genes. It has been proposed that one rationale for the robust regeneration in the salamander is its lack of the growth inhibitor gene products p16INK4a/ARF. We therefore aimed to apply Cre/loxP-based induction to determining the effect of p16INK4a overexpression during regeneration. Temporal control of gene expression was important because the p16INK4a may be expected to strongly repress cell division during normal development. To overexpress p16INK4a during regeneration, we fused the human p16INK4a sequences with T2A-Cherry and cloned the fusion construct behind the floxed GFP cassette (floxed p16-Cherry). In order to elicit induction of p16INK4A expression just prior to regeneration, this animal was crossed to a 4-OHT-inducible CAGGs:ERT2-Cre-ERT2-T2A-nuc-EGFP transgenic animal (Figure 6A). p16INK4A gene expression was initiated with a single intraperitoneal injection of 4-OHT. Cherry expression was observed 5 days after 4-OHT induction in double-transgenic animals (Figure 6C), and p16INK4A ectopic expression was independently confirmed using an antibody against human p16 (data not shown). Tails from Cherry-expressing and -nonexpressing control animals were amputated, followed, and the length of regenerated spinal cord was measured as a discrete indicator of regeneration (Figures 6B and 6C). At 4 days after tail amputation, a significant inhibition of spinal cord regeneration was observed in animals overexpressing human p16INK4A (Figure 6D).
Figure 6.
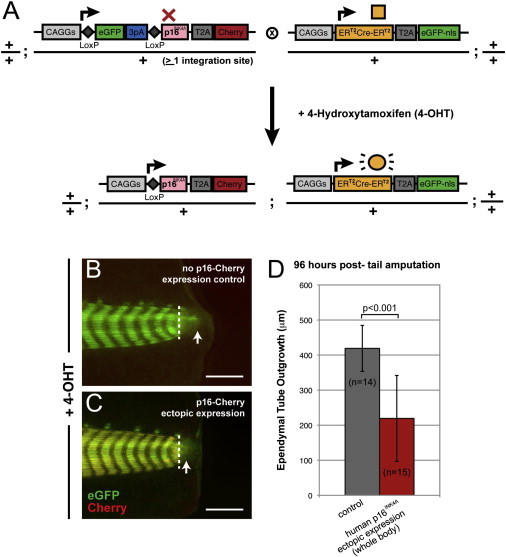
Inducible Overexpression of the Cell-Cycle Inhibitor p16INK4 Represses Spinal Cord Regeneration
(A) To overexpress human p16INK4A, a transgenic animal was made where the p16INK4A-T2A-Cherry gene was cloned behind a floxed GFP cassette (CAGGs:loxP-EGFP-STOP-loxP-p16INK4AT2A-Cherry). This animal was crossed with a transgenic animal where the 4-OHT-inducible Cre was driven by the ubiquitous CAGGs promoter (CAGGs:ERT2-Cre-ERT2-T2A-EGFP-nuc). The progenies of this mating were screened and injected intraperitoneally with 4-OHT and examined live for induction of Cherry expression 5 days later.
(B and C) Phenotype of single-transgenic control and double-transgenic experimental animals ectopically expressing p16INK4A, following 4-OHT intraperitoneal injection and tail amputation. No Cherry induction is seen in single-transgenic animals (B), whereas the double-transgenic animals induced Cherry expression (C). Images were taken 4 days after tail amputation. White dotted lines indicate the amputation plane; white arrows demarcate the extent of ependymal tube outgrowth.
(D) Quantitation of the regenerate spinal cord outgrowth in p16INK4A-expressing transgenic animals and nonexpressing controls. The length of the ependymal tube is significantly reduced in the p16INK4A-expressing animals (paired t-test and the Mann-Whitney U test, p <0.001).
Scale bars, 1 mm (B and C).
Discussion
Here, we have contributed a number of tools and insights into the use of germline transgenic animals for salamander regeneration research by generating germline transgenic animals for cell-type-specific control of gene expression. By employing BACs, heterologous promoters, and axolotl genomic sequences, we have generated a set of animals that drives EGFP in different cell types of the nervous system—neurons (βIII-tubulin), glia (Cnp), and neural stem cells (Sox2). These will provide an invaluable resource for studying brain and spinal cord, as well as peripheral nerve regeneration in these animals. We have also generated animals driving EGFP in muscle, cartilage, and epidermis.
Importantly, we have combined cell-type-specific expression with tight temporal control of gene expression using the Cre/loxP system. These are critical tools for the molecular analysis of regeneration because cells from several different tissues contribute to the blastema and remain as distinct progenitor cell pools during regeneration (Kragl et al., 2009). Furthermore, the marker used to initiate gene expression in the mature tissue may not be maintained during regeneration. Therefore, the Cre/loxP system is particularly valuable for regeneration studies. A critical aspect of the Cre/loxP or any induction system is leakiness of the inducer. We therefore scanned a number of means to tightly induce the CRE activity. In contrast to Whited et al. (2012), we observe strict tamoxifen-inducible gene expression when employing the doubly regulated ERT2-Cre-ERT2 sequences in F0 and after germline transmission to F1 in combination with ubiquitous promoters (CAGGs) or a tissue-specific promoter (COL2A1). We can speculate on several sources for the difference in leaky versus nonleaky expression in the two settings. First, we have observed that injection liquids present in the glass microinjection needle (such as plasmid DNAs for transgenesis or tamoxifen for injection into the animals) are often left behind in the micropipette holder after use, and these reagents can be carried over into subsequent injections with new glass microcapillaries via aerosol. Therefore, if several plasmids are screened in sequence, the presence of contaminating Cre-ERT2 DNA from previous injections remaining behind in the micropipette holder and then being transferred to new microcapillaries could have confounded the previous results. Similarly, use of the microinjection device for tamoxifen injection yielded “background” recombination when the DMSO control sample was injected after the tamoxifen samples without cleaning the micropipette holder in between, but not vice versa. We therefore clean the micropipette holder between each round of injections. A second potential source of apparent leakiness for ERT2-Cre-ERT2 systems could be the quality of the local water because the animals are aquatically raised and constantly exposed to water. The presence of estrogen mimics in the animal water supply could confound the results, and therefore, the use of a carefully controlled water supply is recommended.
Another important technical aspect for transgenesis in this system is careful, cellular level monitoring of the germline transmission. Because the transgenes integrate randomly in the genome, position effects result in differences in expression of the same construct among different integrants. We therefore screen the F1 progeny of germline-transmitting strains by histological/immunofluorescence analysis to confirm the specificity and completeness of the expression. For example, when generating constructs driven by the CAGGs promoter, which should be expressed in every cell type, we observe some F0 founders whose progeny show no expression in specific cell types such as satellite cells or neuronal cells. We therefore always drive a fluorescent reporter gene behind the transgene and analyze the tissues of interest for appropriate expression.
We have demonstrated the ability to interrogate gene function during tail regeneration via inducible overexpression of the human p16INK4a gene. It has been proposed that the p16INK4a/ARF locus first arose in mammals and is not present in animals like the salamander, a potential basis for the restricted regeneration capacities of mammals (Pajcini et al., 2010). Here, we have tested if expression of the p16INK4a gene represses regeneration when induced ubiquitously prior to regeneration onset. Indeed, we observed a significant retardation of spinal cord regeneration in overexpressing the p16INK4a gene, consistent with an antiregenerative function for this gene. These animals could in the future be used to understand the differing downstream responses in p16INK4a-expressing or -nonexpressing cells. These transgenic tools in combination with the growing amount of axolotl sequence data that incorporate next-generation sequencing results from our own and other labs for contig assemblies of the axolotl transcriptome and some genomic sequences (Habermann et al., 2004; Monaghan et al., 2009; Putta et al., 2004; Smith et al., 2009) open up new possibilities to parlay this classical regeneration system into a molecular genetic system to investigate the mechanistic basis of regeneration in a vertebrate and its restriction in other animals.
Experimental Procedures
Axolotl Care and Transgenesis
Animal experiments were performed after approval by the Landesdirektion Saxony, Board of Animal Welfare. Animals were bred in Dresden, Germany, and kept in local tap water at 18°C. The water quality was controlled every day for temperature, pH, ammonia levels, and water consumption. Juveniles and adult animals were kept in continuous flow towers outfitted with particle filters, charcoal filters, and UV filters. Larvae were kept in small plastic tubs with change of fresh water every second day. Larvae were fed Artemia daily, whereas juvenile and adult animals were fed fish pellets. Transgenic axolotls were generated as described (Khattak et al., 2009; Sobkow et al., 2006). Briefly, one-cell-stage embryos were collected and manually dejellied and kept at 4°C until injections were performed. Plasmids that harbored SceI meganuclease target sites flanking the expression cassette were coinjected with the SceI meganuclease enzyme (New England Biolabs). Although the SceI meganuclease increased efficiency of transgenesis as previously described, it was not absolutely necessary for generation of transgenics. Swimming larvae were anesthetized in 0.01% ethyl-p-aminobenzoate (benzocaine; Sigma-Aldrich) for screening based on fluorescence and were screened on an Olympus SZX16 stereomicroscope. Selected embryos were raised for sexual maturity (males 7–9 months and females 12–15 months) and mated with nontransgenic white animals to check for germline transmission. The vectors used to generate transgenic axolotls are described in Table 1.
Generation of AxSox2:Cre-ERT2 Transgenic Animals
An Ambystoma mexicanum (axolotl) lambda genomic library (Stratagene) was expanded in the lab using the manufacturer’s protocol. Axolotl Sox2 gene-specific primers were designed to screen the primary, secondary, and tertiary pools of the library as described by Israel (1993) with minor modifications. Briefly, the 440 pools (50,000 clones each) were screened by PCR, but no colony hybridization was done, rather the tertiary pools were further diluted down to isolate a single positive plaque. The isolated axolotl genomic DNA was recombined into a plasmid backbone, and the resulting plasmid was subjected to a second step of recombineering where the Sox2 open reading frame was replaced by the Cre-ERT2-T2A-EGFP-nls gene using a liquid-recombineering protocol (Sarov et al., 2006). The resulting plasmid AxSox2:Cre-ERT2-T2A-EGFP-nls was injected into one-cell-stage embryo with SceI meganuclease and allowed to develop normally. Larvae were screened for EGFP expression in brain and raised to sexual maturity.
Generation of CAGGs:loxP-EGFP-STOP-loxP-p16INK4A-T2A-Cherry and COL2A1:ERT2-Cre-ERT2-T2A-EGFP-nls Transgenic Animals
To generate CAGGs:loxP-eGFP-STOP-loxP-p16INK4A-T2A-Cherry and COL2A1:ERT2-Cre-ERT2-T2A-EGFP-nls transgenic axolotls, Tol2-CAGGs:loxP-EGFP-STOP-loxP-p16INK4A-T2A-Cherry and Tol2-COL2A1:ERT2-Cre-ERT2-T2A-EGFP-nls constructs were coinjected with Tol2 transposase mRNA into the fertilized one-cell-stage white axolotl eggs and selected using the methods described above. Immunostaining was performed to confirm expression of human p16Ink4A using a mouse monoclonal antibody against p16 (Santa Cruz Biotechnology; #sc-56330). To detect collagenII α1-specific expression, a mouse anti-chicken Collagen type II antibody was used (Millipore; # MAB 8887).
Distal Limb Blastema Transplantation
Blastema donors (5–6-cm axolotls) were double transgenics of the genotype: Car Act:EGFP; CAGGs:nucCherry. Recipients were their nontransgenic white siblings. Donors were amputated through the forearm wrist. Nine days later, recipients were amputated through the proximal half of the stylopod, and protruding bones were trimmed. Immediately, the donor’s wrist blastema was cut off and transferred in the same proximal-distal orientation onto the recipient’s stump. Animals were left asleep on a wet tissue with 0.01% benzocaine for the following 1–3 hr before placing them back into tap water to recover and regenerate lost limb structures.
DNA Electroporation and 4-OHT Injection
Circular Cre expression plasmid DNA (0.5 mg/ml) was electroporated in the limb and/or spinal cord as previously described (Echeverri and Tanaka, 2003; McHedlishvili et al., 2007). For 4-OHT injections, larvae were anesthetized in 0.01% benzocaine and weighed. 4-OHT (Sigma-Aldrich; catalog # H7904 or #H6278) was diluted in DMSO to obtain a stock of 10 mg/ml. Stock aliquots were stored at −80°C until use. Fast Green (Sigma-Aldrich; F7258) was added to the thawed solution just prior to injection. A total of 50 μg/g body weight of 4-OHT was injected intraperitoneally in the ventral trunk. After injection, larvae were covered with a blanket of wet tissue for hydration for 20–30 min at room temperature until returning them to tap water.
Immunohistochemical Analysis of Transgenic Animals
Tails and limbs of transgenic axolotl larvae were cut and fixed, cryosectioned, and kept at −20°C until immunostained. The cryosections were stained with respective antibodies as previously described by Kragl et al. (2009).
Acknowledgments
We gratefully acknowledge the dedicated animal care of Heino Andreas, Jitka Michling, Sabine Mögel, Christa Junghans, and Beate Gruhl. We thank Jessica Whitehead for ert-cre-ert construct and Andrea Meinhardt for help in confocal microscopy. This work was supported by grants from the DFG (TA274/2 [Collaborative Research Center 655] and TA274/3 and TA274/4 [SPP1365]), HFSP, Volkswagen Foundation, Central funds from the Max Planck Institute of Molecular Cell Biology and Genetics, the DFG Research Center for Regenerative Therapies, and the Technical University Dresden to E.M.T. S.L.H. was supported by a Postdoctoral Research Fellowship from the Alexander von Humboldt Foundation. S.K. and E.M.T. designed and analyzed most of the experiments. M.S. and T.R. generated transgenic animals. S.K., T.R., K.H., and R.K. generated constructs. S.K., D.K., S.L.H., A.D., and T.S.-G. contributed regeneration experiments with transgenic animals. S.K. and E.M.T. wrote the manuscript.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Anastassiadis K., Rostovskaya M., Lubitz S., Weidlich S., Stewart A.F. Precise conditional immortalization of mouse cells using tetracycline-regulated SV40 large T-antigen. Genesis. 2010;48:220–232. doi: 10.1002/dvg.20605. [DOI] [PubMed] [Google Scholar]
- Attardo A., Calegari F., Haubensak W., Wilsch-Brauninger M., Huttner W.B. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS One. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant S.V., Iten L.E. Intercalary and supernumerary regeneration in regenerating the mature limbs of Notophthalmus viridescens. J. Exp. Zool. 1977;202:1–16. doi: 10.1002/jez.1402020102. [DOI] [PubMed] [Google Scholar]
- Casanova E., Lemberger T., Fehsenfeld S., Mantamadiotis T., Schutz G. Alpha complementation in the Cre recombinase enzyme. Genesis. 2003;37:25–29. doi: 10.1002/gene.10227. [DOI] [PubMed] [Google Scholar]
- Casco-Robles M.M., Yamada S., Miura T., Chiba C. Simple and efficient transgenesis with I-SceI meganuclease in the newt, Cynops pyrrhogaster. Dev. Dyn. 2010;239:3275–3284. doi: 10.1002/dvdy.22463. [DOI] [PubMed] [Google Scholar]
- Dent J.N. Limb regeneration in larvae and metamorphosing individuals of the South African clawed toad. J. Morphol. 1962;110:61–77. doi: 10.1002/jmor.1051100105. [DOI] [PubMed] [Google Scholar]
- Dunis D.A., Namenwirth M. The role of grafted skin in the regeneration of x-irradiated axolotl limbs. Dev. Biol. 1977;56:97–109. doi: 10.1016/0012-1606(77)90157-9. [DOI] [PubMed] [Google Scholar]
- Echeverri K., Tanaka E.M. Electroporation as a tool to study in vivo spinal cord regeneration. Dev. Dyn. 2003;226:418–425. doi: 10.1002/dvdy.10238. [DOI] [PubMed] [Google Scholar]
- Echeverri K., Tanaka E.M. Proximodistal patterning during limb regeneration. Dev. Biol. 2005;279:391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Echeverri K., Clarke J.D., Tanaka E.M. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol. 2001;236:151–164. doi: 10.1006/dbio.2001.0312. [DOI] [PubMed] [Google Scholar]
- García-Otín A.L., Guillou F. Mammalian genome targeting using site-specific recombinases. Front. Biosci. 2006;11:1108–1136. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- Glaser T., Perez-Bouza A., Klein K., Brustle O. Generation of purified oligodendrocyte progenitors from embryonic stem cells. FASEB J. 2005;19:112–114. doi: 10.1096/fj.04-1931fje. [DOI] [PubMed] [Google Scholar]
- Habermann B., Bebin A.G., Herklotz S., Volkmer M., Eckelt K., Pehlke K., Epperlein H.H., Schackert H.K., Wiebe G., Tanaka E.M. An Ambystoma mexicanum EST sequencing project: analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biol. 2004;5:R67. doi: 10.1186/gb-2004-5-9-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Yokotani N., Tane S., Matsumoto A., Myouga A., Okamoto M., Takeuchi T. Molecular genetic system for regenerative studies using newts. Dev. Growth Differ. 2013;55:229–236. doi: 10.1111/dgd.12019. [DOI] [PubMed] [Google Scholar]
- Israel D.I. A PCR-based method for high stringency screening of DNA libraries. Nucleic Acids Res. 1993;21:2627–2631. doi: 10.1093/nar/21.11.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iten L.E., Bryant S.V. The interaction between the blastema and stump in the establishment of the anterior—posterior and proximal—distal organization of the limb regenerate. Dev. Biol. 1975;44:119–147. doi: 10.1016/0012-1606(75)90381-4. [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez Esteban C., Raya M., Kawakami H., Marti M., Dubova I., Izpisua Belmonte J.C. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerney R., Hall B.K., Hanken J. Regulatory elements of Xenopus col2a1 drive cartilaginous gene expression in transgenic frogs. Int. J. Dev. Biol. 2010;54:141–150. doi: 10.1387/ijdb.092848rk. [DOI] [PubMed] [Google Scholar]
- Khattak S., Richter T., Tanaka E.M. Generation of transgenic axolotls (Ambystoma mexicanum) Cold Spring Harb. Protoc. 2009;2009 doi: 10.1101/pdb.prot5264. pdb prot5264. [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H.H., Tanaka E.M. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kumar A., Brockes J.P. Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 2012;35:691–699. doi: 10.1016/j.tins.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Kumar A., Godwin J.W., Gates P.B., Garza-Garcia A.A., Brockes J.P. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Pevny L., Lovell-Badge R., Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr. Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- Mallo M. Controlled gene activation and inactivation in the mouse. Front. Biosci. 2006;11:313–327. doi: 10.2741/1799. [DOI] [PubMed] [Google Scholar]
- McGuire S.E., Le P.T., Osborn A.J., Matsumoto K., Davis R.L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- McHedlishvili L., Epperlein H.H., Telzerow A., Tanaka E.M. A clonal analysis of neural progenitors during axolotl spinal cord regeneration reveals evidence for both spatially restricted and multipotent progenitors. Development. 2007;134:2083–2093. doi: 10.1242/dev.02852. [DOI] [PubMed] [Google Scholar]
- McHedlishvili L., Mazurov V., Grassme K.S., Goehler K., Robl B., Tazaki A., Roensch K., Duemmler A., Tanaka E.M. Reconstitution of the central and peripheral nervous system during salamander tail regeneration. Proc. Natl. Acad. Sci. USA. 2012;109:E2258–E2266. doi: 10.1073/pnas.1116738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N., Tanaka E.M., Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Metzger D., Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71–80. doi: 10.1006/meth.2001.1159. [DOI] [PubMed] [Google Scholar]
- Monaghan J.R., Epp L.G., Putta S., Page R.B., Walker J.A., Beachy C.K., Zhu W., Pao G.M., Verma I.M., Hunter T. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J.I., Borg P., Simon A. Plasticity and recovery of skeletal muscle satellite cells during limb regeneration. FASEB J. 2010;24:750–756. doi: 10.1096/fj.09-134825. [DOI] [PubMed] [Google Scholar]
- Mullen L.M., Bryant S.V., Torok M.A., Blumberg B., Gardiner D.M. Nerve dependency of regeneration: the role of Distal-less and FGF signaling in amphibian limb regeneration. Development. 1996;122:3487–3497. doi: 10.1242/dev.122.11.3487. [DOI] [PubMed] [Google Scholar]
- Muneoka K., Fox W.F., Bryant S.V. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol. 1986;116:256–260. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- Nacu E., Tanaka E.M. Limb regeneration: a new development? Annu. Rev. Cell Dev. Biol. 2011;27:409–440. doi: 10.1146/annurev-cellbio-092910-154115. [DOI] [PubMed] [Google Scholar]
- Nacu E., Glausch M., Le H.Q., Damanik F.F., Schuez M., Knapp D., Khattak S., Richter T., Tanaka E.M. Connective tissue cells, but not muscle cells, are involved in establishing the proximo-distal outcome of limb regeneration in the axolotl. Development. 2013;140:513–518. doi: 10.1242/dev.081752. [DOI] [PubMed] [Google Scholar]
- Ogino H., McConnell W.B., Grainger R.M. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat. Protoc. 2006;1:1703–1710. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- Pajcini K.V., Corbel S.Y., Sage J., Pomerantz J.H., Blau H.M. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A., Akam M. Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:2855–2860. doi: 10.1073/pnas.1015077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescitelli M.J., Jr., Stocum D.L. The origin of skeletal structures during intercalary regeneration of larval Ambystoma limbs. Dev. Biol. 1980;79:255–275. doi: 10.1016/0012-1606(80)90115-3. [DOI] [PubMed] [Google Scholar]
- Putta S., Smith J.J., Walker J.A., Rondet M., Weisrock D.W., Monaghan J., Samuels A.K., Kump K., King D.C., Maness N.J. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics. 2004;5:54. doi: 10.1186/1471-2164-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Gardiner D.M., Bryant S.V. Vaccinia as a tool for functional analysis in regenerating limbs: ectopic expression of Shh. Dev. Biol. 2000;218:199–205. doi: 10.1006/dbio.1999.9556. [DOI] [PubMed] [Google Scholar]
- Sarov M., Schneider S., Pozniakovski A., Roguev A., Ernst S., Zhang Y., Hyman A.A., Stewart A.F. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- Smith J.J., Putta S., Zhu W., Pao G.M., Verma I.M., Hunter T., Bryant S.V., Gardiner D.M., Harkins T.T., Voss S.R. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics. 2009;10:19. doi: 10.1186/1471-2164-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobkow L., Epperlein H.H., Herklotz S., Straube W.L., Tanaka E.M. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev. Biol. 2006;290:386–397. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Sordino P., van der Hoeven F., Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- Steen T.P. Stability of chondrocyte differentiation and contribution of muscle to cartilage during limb regeneration in the axolotl (Siredon mexicanum) J. Exp. Zool. 1968;167:49–78. doi: 10.1002/jez.1401670105. [DOI] [PubMed] [Google Scholar]
- Stocum D.L. Regulation after proximal or distal transposition of limb regeneration blastemas and determination of the proximal boundary of the regenerate. Dev. Biol. 1975;45:112–136. doi: 10.1016/0012-1606(75)90246-8. [DOI] [PubMed] [Google Scholar]
- Stocum D.L., Cameron J.A. Looking proximally and distally: 100 years of limb regeneration and beyond. Dev. Dyn. 2011;240:943–968. doi: 10.1002/dvdy.22553. [DOI] [PubMed] [Google Scholar]
- Suzuki K.T., Kashiwagi K., Ujihara M., Marukane T., Tazaki A., Watanabe K., Mizuno N., Ueda Y., Kondoh H., Kashiwagi A. Characterization of a novel type I keratin gene and generation of transgenic lines with fluorescent reporter genes driven by its promoter/enhancer in Xenopus laevis. Dev. Dyn. 2010;239:3172–3181. doi: 10.1002/dvdy.22451. [DOI] [PubMed] [Google Scholar]
- Urlinger S., Baron U., Thellmann M., Hasan M.T., Bujard H., Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrou C., Zhang Y., Zurn C., Schamel W.W., Reth M. Comparison of the tamoxifen regulated chimeric Cre recombinases MerCreMer and CreMer. Biol. Chem. 1999;380:1435–1438. doi: 10.1515/BC.1999.184. [DOI] [PubMed] [Google Scholar]
- Wallace B.M., Wallace H. Participation of grafted nerves in amphibian limb regeneration. J. Embryol. Exp. Morphol. 1973;29:559–570. [PubMed] [Google Scholar]
- Whited J.L., Lehoczky J.A., Tabin C.J. Inducible genetic system for the axolotl. Proc. Natl. Acad. Sci. USA. 2012;109:13662–13667. doi: 10.1073/pnas.1211816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited J.L., Tsai S.L., Beier K.T., White J.N., Piekarski N., Hanken J., Cepko C.L., Tabin C.J. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. 2013;140:1137–1146. doi: 10.1242/dev.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji N., Yokoyama H., Tamura K. Repatterning in amphibian limb regeneration: a model for study of genetic and epigenetic control of organ regeneration. Semin. Cell Dev. Biol. 2009;20:565–574. doi: 10.1016/j.semcdb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Zappone M.V., Galli R., Catena R., Meani N., De Biasi S., Mattei E., Tiveron C., Vescovi A.L., Lovell-Badge R., Ottolenghi S. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Riesterer C., Ayrall A.M., Sablitzky F., Littlewood T.D., Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


