Abstract
Mammalian primordial germ cells (PGCs) are unipotent progenitors of the gametes. Nonetheless, they can give rise directly to pluripotent stem cells in vitro or during teratocarcinogenesis. This conversion is inconsistent, however, and has been difficult to study. Here, we delineate requirements for efficient resetting of pluripotency in culture. We demonstrate that in defined conditions, routinely 20% of PGCs become EG cells. Conversion can occur from the earliest specified PGCs. The entire process can be tracked from single cells. It is driven by leukemia inhibitory factor (LIF) and the downstream transcription factor STAT3. In contrast, LIF signaling is not required during germ cell ontogeny. We surmise that ectopic LIF/STAT3 stimulation reconstructs latent pluripotency and self-renewal. Notably, STAT3 targets are significantly upregulated in germ cell tumors, suggesting that dysregulation of this pathway may underlie teratocarcinogenesis. These findings demonstrate that EG cell formation is a robust experimental system for exploring mechanisms involved in reprogramming and cancer.
Graphical Abstract
Highlights
-
•
A defined system for generation of pluripotent EG cells at high efficiency
-
•
20% of single primordial germ cells become EG cells
-
•
Stimulation with LIF but not FGF drives conversion to pluripotency
-
•
LIF/STAT3 targets are upregulated in pluripotent germ cell tumors
Introduction
In sexually reproducing organisms germ cells provide the continuous link between the generations, delivering the genetic and epigenetic information required to construct a new organism (Surani, 2007). Primordial germ cells (PGCs) represent the founder cells of the germline lineage. In mice, they are induced from Oct4- (also known as Pou5f1) positive pluripotent epiblast cells at the onset of gastrulation. By E7.5, PGCs are said to be specified, coincident with the expression of Stella (also known as Dppa3, Pgc7) (McLaren and Lawson, 2005; Saitou et al., 2002). During normal development, PGCs behave as unipotent progenitors and produce only germ cells. Yet, they express pluripotency genes until after colonization of the genital ridges (Surani et al., 2007). Significantly, PGCs can give rise to pluripotent tumors in ectopic sites and they can serve as the cell of origin of testicular teratocarcinomas (Stevens, 1967). Ex vivo PGCs can directly give rise to pluripotent stem cell lines known as embryonic germ (EG) cells (Matsui et al., 1992; Resnick et al., 1992). Like embryonic stem (ES) cells, EG cells are genetically normal and are capable of contributing to chimeras (Labosky et al., 1994; Stewart et al., 1994).
The process by which PGCs convert to pluripotency is erratic and poorly characterized. Three growth factors are reported to play key roles; stem cell factor (SCF), leukemia inhibitory factor (LIF), and basic fibroblast growth factor (bFGF). Individually, each factor positively influences PGC proliferation and/or survival (Dolci et al., 1991; Godin et al., 1991; Matsui et al., 1991), but in combination they facilitate conversion to EG cells. Only LIF plays a role in subsequent self-renewal of EG cells. bFGF is important during the first day of culture but not thereafter, suggesting it may trigger the conversion process (Durcova-Hills et al., 2006). bFGF appears to act though the PI3K/AKT pathway because it is not required for EG cell formation from Pten deletion mutants (Kimura et al., 2003) or if AKT is hyperactivated (Kimura et al., 2008). Retinoic acid (RA) and forskolin (FK), two potent PGC mitogens, can substitute for bFGF in EG cell derivation (Koshimizu et al., 1996), as can the histone deacetylase inhibitor trichostatin A (Durcova-Hills et al., 2008). However, whether the activity of these factors is direct or mediated through induction of FGFs or other factors remains unclear due to the complex culture conditions, which include serum, feeders, and heterogeneous somatic cells. Previously, we showed that addition of two small molecule inhibitors of mitogen-activated protein kinase (MAPK) signaling and glycogen synthase kinase 3 (GSK3) (2i) (Ying et al., 2008) enables reliable generation of EG cells from mouse and rat PGCs (Leitch et al., 2010; Blair et al., 2012). However, undefined components should be eliminated to delineate the individual contributions of signaling molecules and pathways that mediate the derestriction of PGCs to pluripotency. Here, we develop a defined culture system and exploit this to clarify pathway requirements and in addition to track the PGC to EG cell conversion at the single cell level.
Results
EG Cell Derivation Does Not Require Serum or Feeders
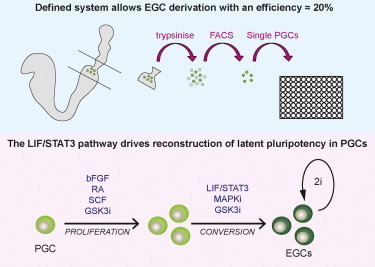
EG cells can be obtained after plating PGCs directly in 2i/LIF on feeders (Leitch et al., 2010). Past attempts to culture PGCs without feeders have resulted in rapid cell death within 24 hr (De Felici et al., 1998). We therefore investigated whether addition of known PGC-supportive factors might increase proliferation and viability. Posterior regions of mouse E8.5 embryos were trypsinized and plated in 2i/LIF, with the addition of bFGF, SCF, RA, and FK (henceforth referred to collectively as four factors—4Fs) for the first 2 days only. In these feeder-free conditions, EG cell lines were readily obtained. However the addition of the 4Fs resulted in substantial growth of somatic cells (data not shown) calling into the question the cell-autonomous ability of PGCs to produce EG cells. Therefore, we used flow cytometry to obtain a pure population of PGCs (Figures S1A and S1B available online). This approach enabled accurate calculation of derivation efficiency (percentage of PGCs forming colonies), which on fibronectin approached 4% (Figure S1C.) Previously, it has been suggested that inhibition of MAPK has a negative effect on PGC proliferation (De Miguel et al., 2002). Therefore, we plated equal numbers of flow-sorted PGCs on fibronectin in either 2i/LIF or GSK3 inhibitor plus LIF (CH/LIF). Over the first 72 hr, many more PGCs were evident per cluster in the CH/LIF cultures; however, many, although not all, of the cells downregulated the Oct4-ΔPE-GFP reporter that is active in both PGCs and EG cells. By 7 days, only a small number of EG cell colonies were present in CH/LIF compared with 2i/LIF (Figure 1A). These colonies in CH/LIF were partially differentiated, but they had a tightly packed core with EG cell morphology and after picking cells could be expanded in 2i/LIF indistinguishably from other EG cells.
Figure 1.
Efficient EG Cell Derivation in Defined Conditions without Serum or Feeders
(A) Histogram showing EG-cell-colony frequency in 2i/LIF versus CH/LIF. The four factors (4Fs), bFGF, SCF, FK, and RA, were added for the first 48 hr of culture. Error bars denote SE of two biological replicates. **p < 0.01, Student’s t test.
(B) Schematic of derivation protocol. PGCs were plated in CH/LIF plus 4Fs, with or without PD for the first 48 hr. All cultures were subsequently fed with 2i/LIF.
(C) Quantitation of EG-cell-colony frequency in each condition.
(D) Histogram showing EG-cell-colony formation from E11.5 PGCs in 2i/LIF and CH/LIF. The four factors (4Fs), bFGF, SCF, FK, and RA, were added for the first 48 hr of culture. All cultures were subsequently fed with 2i/LIF. Error bars denote SE of three biological replicates. Phase and fluorescence images show a primary E11.5 EG cell colony.
(E–G) FACS plot showing gated GFP-positive E8.5 PGCs and phase-contrast and fluorescence images of primary EG cell colonies derived from (E) Oct4-ΔPE-GFP embryos, (F) Blimp1-GFP embryos, and (G) Stella-GFP embryos.
(H) Summary of E7.5 EG-cell-derivation experiments.
(I) Bright-field and fluorescence images of E11.5 chimeric embryos made from aggregations of E7.5 EG cells carrying a constitutively active DsRed reporter transgene.
(J) Coat color chimeras generated with agouti E7.5 EG cells injected into C57BL/6 blastocysts (upper panel) and an adult chimera with C57BL/6 mate and brown pup, indicating transmission of the EG cell genome (lower panel).
These results indicate that while MAPK inhibition contributes substantially to the production of EG cells, it may impair the initial viability of PGCs. We therefore investigated whether delayed inhibition of MAPK may reduce early cell death and improve overall conversion efficiency. We plated 250 flow-sorted PGCs on fibronectin in CH/LIF plus 4Fs, with or without PD, for the first 48 hr and thereafter transitioned to 2i/LIF by half-medium changes (Figure 1B). After 12 days, 72 Oct4-ΔPE-GFP colonies were obtained in cultures initiated in CH, compared with only six from 2i (Figure 1C). In the course of scoring these plates, we also noted that some EG cell colonies were clustered together, raising the possibility that single PGCs may produce more than one colony (see later). However, even when colony clusters are scored as single conversion events, deferring addition of PD for 48 hr leads to a 10-fold increase in yield (Figure 1C). All subsequent experiments were thus performed using these conditions unless otherwise stated.
We investigated formation of EG cells from gonadal PGCs at E11.5. From 2,000 PGCs per well, we recovered a maximum of seven EG cells colonies per well (Figure 1D). The conversion frequency of 1/286 is some 50-fold lower than for E8.5 PGCs. This is consistent with previous reports of increasing refractoriness as development progresses (Durcova-Hills and Capel, 2008; Labosky et al., 1994). EG cells have never been derived from before E8.0 even though PGCs are specified at E7.5 (Labosky et al., 1994; McLaren and Lawson, 2005). To test whether early PGCs are competent to produce EG cells, we collected embryos at the early/midallantoic bud stage, excluding late head-fold stage embryos. We dissected the posterior section of 23 embryos carrying the Oct4-ΔPE-GFP transgene and were able to recover 98 GFP-positive cells by flow cytometry (Figure 1H). After 10 days of culture, we obtained a total of 13 EG cell colonies. Three colonies appeared close together and another two were doublets (Figures 1E and 1H). Assuming clustered colonies derive from one starting cell gives a corrected conversion frequency of 10/98. Importantly, GFP-negative cells did not yield any colonies. However, the expression of the Oct4-ΔPE-GFP transgene is not completely restricted to PGCs at this time point (Yoshimizu et al., 1999). To confirm that the colonies obtained were derived from PGCs rather than late epiblast or other cells, we repeated the experiment using Blimp1-GFP or Stella-GFP reporters. From five E7.5 Blimp1-GFP embryos, we obtained 2,000 GFP-positive cells. As only 20–40 PGCs are present at this stage, the majority of these cells were presumably visceral endodermal where Blimp1 is also expressed (Ohinata et al., 2005). Indeed, we observed many patches of endodermal-like cells growing in the cultures (Figure 1F). However, we also obtained 15 EG cell colonies in eight distinct clusters (Figures 1F and 1H). These colonies were Blimp1-GFP negative at the end of the experiment, which is expected because BLIMP1 is rapidly downregulated during EG derivation (Durcova-Hills, et al., 2008), and the expression of Blimp1 in ES cells in 2i/LIF is negligible (Figure 1F; Marks et al., 2012). Stella-GFP is upregulated around E7.5 specifically in PGCs (Payer et al., 2006). In two separate experiments, we were able to isolate 45 and 47 Stella-GFP-positive PGCs that produced eight and ten colony clusters, respectively, representing an EG-cell-derivation efficiency of approximately 20% (Figures 1G and 1H). Overall, these findings are comparable to EG cell generation from purified E8.5 PGCs. To confirm the identity and potency of E7.5 EG cells, we produced chimeras. We first introduced a DsRed reporter transgene to Stella-GFP-derived EG cells using the PiggyBac system. EG cells stably transfected with a DsRed expression construct (Guo et al., 2009) were injected into blastocysts and transferred to recipient pseudopregnant hosts. Pregnant females were sacrificed at midgestation, and four out of nine embryos exhibited widespread chimerism (Figure 1I). Unlabeled EG cells were also injected into blastocysts, transferred to pseudopregnant hosts, and left to term. Coat color chimerism was evident in 5/15 pups (Figure 1J; Table S1). One of these chimeras was test mated and gave germline transmission (Figure 1J). We conclude that from specification PGCs have the capacity to form pluripotent EG cell lines.
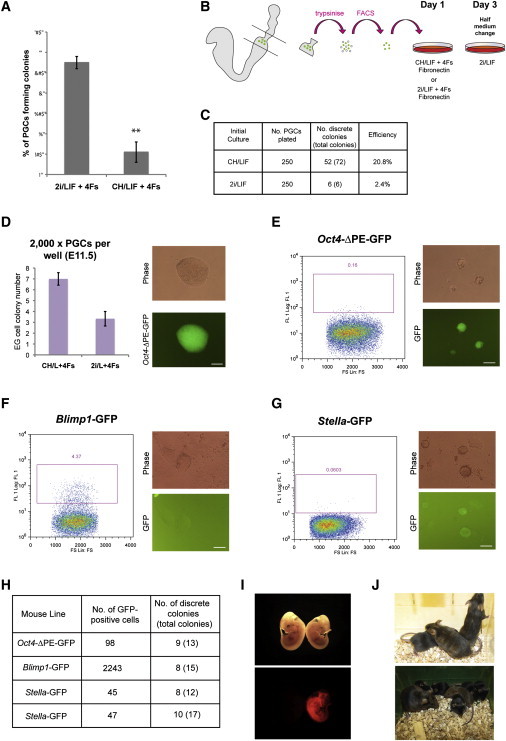
Single PGCs Efficiently Give Rise to EG Cells
To establish if isolated PGCs are capable of forming EG cells, individual Oct4-ΔPE-GFP-positive PGCs from E8.5 embryos were flow-sorted into 96-well plates containing CH/LIF plus 4Fs with or without PD (Figure 2A). Each well was examined by microscopy 4 hr after deposition, and none was found to contain more than one cell, although some mitotic figures were observed. Twenty-four hours after deposition, each well was again examined, and cells were present in 56/96 and 61/96 wells with or without PD, respectively (Figure 2B). As above, cultures were transitioned to 2i/LIF by daily half-medium changes from 48 hr. After 12 days, each well was assessed for the presence of EG cells (Figure 2B). In the plate initiated in 2i, there were three positive wells, one containing two colonies. In the CH/LIF plate, 12 cells (18%) yielded EG colonies and five contained more than one colony. We note that if total colonies are divided by the number of PGCs plated, then the calculated derivation efficiency would be 31.1%. Thus, the enumeration method routinely used for bulk culture experiments overestimates the actual number of starting PGCs that convert.
Figure 2.
EG Cell Derivation from Single PGCs
(A) Schematic of derivation protocol. Oct4-ΔPE-GFP-positive PGCs were deposited singly into each well of a 96-well plate, in medium containing CH/LIF+4Fs with or without PD. After 48 hr, cultures were transitioned to 2i/LIF by daily half-medium changes.
(B) Summary of results obtained from each 96-well plate.
(C) Single frames from time-lapse movie of EG cell derivation. Arrow denotes PGC that produces all nine EG cells colonies. Cells with PGC-like morphology proliferate up until approximately 85 hr. By 112.5 hr, extensive cell death is evident. At 154.5 hr, small colonies are evident and have expanded by 179 hr. The final image shows Oct4-ΔPE-GFP expression in EG cell colonies at 179 hr. See also Movie S1.
(D) Coat color chimera (generated with agouti EG cells derived from a colony in C) with C57BL/6 mate and all brown litter, indicating successful germline transmission.
We carried out time-lapse analyses to visualize the process of EG cell generation and observe how multiple colonies may be produced. Ten PGCs were plated in each well of a 96-well plate and the progeny of single PGCs tracked over 179 hr. We observed that in clones that generated EG cells many sister cells died. We also found several instances of single PGCs giving rise to multiple EG cell colonies. These generally arose independently from separate daughter cells rather than by colony splitting (Figure 2C; Movie S1). We picked ten colonies originating from different PGCs, and five derived from the same PGC. All could readily be expanded as EG cell lines. One was injected into blastocysts and gave rise to coat color chimeras (Figure 2D; Table S1). A chimera was test mated and exhibited germline transmission (Figure 2E), confirming naive pluripotent identity and functionality.
Signaling Requirements for EG Cell Formation
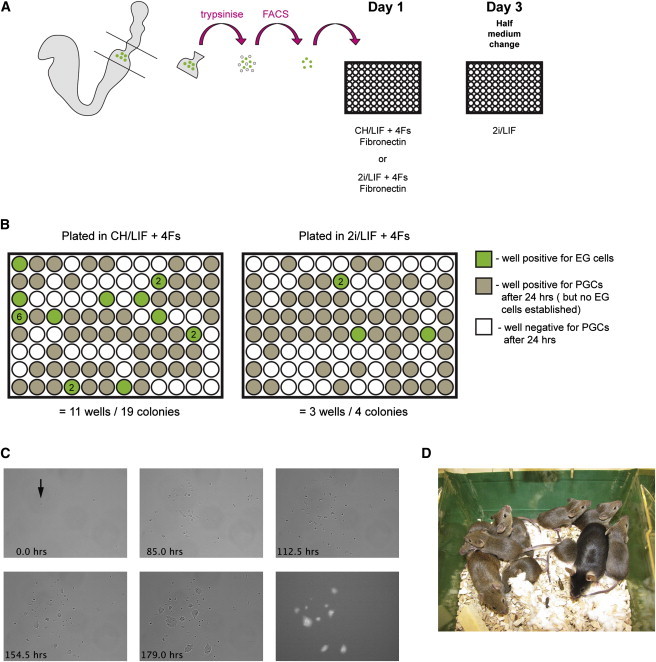
Next, we investigated the stimuli required to enable conversion of PGCs to EG cells. Sorted PGCs were cultured as above but with individual factors removed. Withdrawal of forskolin had no effect, but without initial exposure to SCF or RA there was a significant reduction in colony numbers (Figure 3A). Fewer colonies were also obtained without bFGF. Thus, all three of these factors contribute to the starting period of EG cell derivation though none is essential. Interestingly, although LIF has been reported to increase the survival and proliferation of PGCs (Matsui et al., 1991), we did not observe an overt effect on PGC survival or proliferation during the early stages of culture (Figure 3B), and EG-cell-colony formation is not significantly affected by the absence of LIF during the first 48 hr of culture (Figure 3C). However, colony number is reduced if LIF is withheld for a further 24 hr, and most significantly, in the continuous absence of LIF no colonies are obtained (Figure 3A).
Figure 3.
LIF/STAT3 Drives Acquisition of Pluripotency
(A) Histogram showing EG-cell-colony numbers obtained from flow-sorted PGCs plated either in CH/LIF+4Fs (control) or with the indicated factor removed. After 48 hr, cultures were transitioned to 2i/LIF (or 2i only in the –LIF condition) by daily half-medium changes (mean ± SEM, *p < 0.05, **p < 0.01, and ***p < 0.001, ANOVA with Dunnett’s post hoc, here and for all other figures, unless otherwise stated) (n = 12).
(B) Phase-contrast image after 65.2 hr in minus LIF and plus LIF conditions, showing the daughter cells derived from single PGCs.
(C) Histogram showing EG-cell-colony numbers obtained from flow-sorted PGCs plated in CH+4Fs. After 48 hr, cultures were transitioned to 2i by daily half-medium changes. LIF was added either from the start of the experiment (0 hr) or at the time indicated, and maintained thereafter (n = 12).
(D) Histogram showing EG-cell-colony numbers obtained from flow-sorted PGCs. At the times indicated, either LIF was removed by changing the medium (gray bars) or LIF was removed and a JAK inhibitor added (blue bars). In the control, LIF was present throughout (n = 8).
(E) Mean reads per million (RPM) of STAT3 target genes in Oct4-positive ES cells (Tang et al., 2010) and E8.5 PGCs (Hackett et al., 2012) reveals a higher expression in ES cells than PGCs.
(F) OCT4 and KLF4 immunostaining of an established EG cell line and nascent EG cell colonies after 96 or 120 hr in culture. At 120 hr KLF4 staining exhibits either heterogenous (i) or homogenous (ii) staining patterns. All cells in field were OCT4 positive; DAPI omitted for clarity. Scale bar 100 μm.
(G) Histogram indicating EG-cell-derivation efficiency, relative to an untransfected control, following transfection with two independent siRNAs targeting STAT3 and a negative control siRNA (siGFP) (error bars denote SEM, ***p < 0.001, ANOVA with Newman-Keul’s post hoc, n = 2).
We assessed the duration of LIF stimulation required to enable conversion. When LIF is present from the start and removed by medium washout after 72 hr, a small number of colonies are obtained. The yield increases after 96 hr in LIF and further after 120 hr (Figure 3D). However, medium change may not be sufficient to completely eliminate the LIF signal. Therefore, we combined LIF removal with addition of an inhibitor of Janus-associated kinases (JAK) to block ongoing signaling (Figure 3D). Under these conditions, no colonies are recovered unless prior LIF stimulation has been maintained for 144 and 192 hr exposure is needed to reach control efficiency (Figure 3D). These findings indicate that prolonged LIF stimulation between 48 and 192 hr is required to maximize EG cell formation.
STAT3 is the key mediator of LIF effects both on ES cell self-renewal (Matsuda et al., 1999; Niwa et al., 1998) and in EpiSC and somatic cell reprogramming (Bao et al., 2009; van Oosten et al., 2012; Yang et al., 2010). We compared the expression pattern of LIF/STAT3 targets (Bourillot et al., 2009) in single-cell RNA-seq data sets from E8.5 PGCs and ES cells (Hackett et al., 2012; Tang et al., 2010). The expression of 37 annotated STAT3 target genes was significantly enriched in ES cells (Welch’s t test, p < 0.01) (Figure 3E). This is consistent with a requirement to activate LIF signaling and targets for PGC conversion. We used immunostaining to detect the emergence of KLF4, a validated LIF/STAT3 target and pluripotency factor (Hall et al., 2009; Li et al., 2005; Niwa et al., 2009), which has previously been shown to be upregulated during EG cell derivation (Durcova-Hills et al., 2008; Nagamatsu et al., 2012) (Figure 3F). Positive cells were first detected at 96 hr, but, in contrast to established EG cell cultures in 2i/LIF, KLF4 expression is mosaic within colonies (Figure 3F). This heterogeneity is manifest even after 120 hr, although colonies with a more homogenous KLF4 staining pattern can also be observed by this stage (Figure 3F). These observations indicate that KLF4 expression develops asynchronously and is progressively consolidated during EG cell formation.
To establish whether STAT3 function is in fact required for EG cell derivation, we performed knockdown experiments during the conversion process using small interfering RNA (siRNA). The efficiency and specificity of siRNAs was confirmed in ES cells (Figure S2). PGCs were plated in CH plus 4Fs in the absence of LIF for 30 hr prior to siRNA transfection. Following transfection, culture medium was changed to 2i/LIF and colonies were counted after 12 days. Transfection was associated with some cellular toxicity, reducing the colony yield from control siGFP-transfected cells by approximately 50% (Figure 3G). However, over and above this effect, STAT3 knockdown abolished EG-cell-colony formation completely in each of several independent experiments (Figure 3G). We conclude that STAT3 is required to mediate conversion of PGCs to EG cells.
STAT3 Targets Are Upregulated in Germ Cell Tumors
The preceding results suggest that signaling through the LIF/STAT3 pathway is low or absent in PGCs and that activation of STAT3 targets drives regeneration of pluripotency during EG cell derivation. PGCs can be the cells of origin during teratocarcinogenesis, and many pluripotency genes are found to be upregulated in human germ cell tumors (Oosterhuis and Looijenga, 2005). We therefore investigated the expression of the STAT3 targets in a human germ cell tumor (GCT) microarray data set (Korkola et al., 2006; West et al., 2009). We found widespread upregulation of these target genes in GCTs compared with low expression in normal testes (Figure 4A). We used these STAT3 target genes to construct a KEGG pathway (“Savatier STAT3 targets”) and found this to be the fifth most upregulated pathway when comparing all GCTs with normal testes (Figure 4B). However, only a subset of human GCTs is thought to undergo teratocarcinogenesis (Oosterhuis and Looijenga, 2005). Of these, embryonal carcinomas contain a pluripotent cell compartment and exhibit a gene expression profile similar to human ES cells (Sperger et al., 2003). Notably, STAT3 target genes are the second most upregulated KEGG pathway in embryonal carcinoma (EC) samples, second only to “ribosome” (Figure S3A). Mixed GCTs often contain an EC component and STAT3 targets are ranked fourth of the upregulated pathways in this group (Figure S3B). Teratomas consist only of differentiated cell types and have low expression of pluripotency-associated genes (Figure 4A) but are believed to pass through a pluripotent cell intermediate. Intriguingly, they still exhibit a strong enrichment for STAT3 target genes (Figure S3C). Reactivation of pluripotency does not occur in seminomas (Oosterhuis and Looijenga, 2005), which retain expression of germline genes (Figure 4A). Consistently, STAT3 targets are less enriched in the seminoma samples, and also in yolk sac tumors, the etiology of which remains unclear (Figures S3D and S3E). These data suggest that STAT3 activation may be a primary lesion in germ cell tumors that transit through pluripotency.
Figure 4.
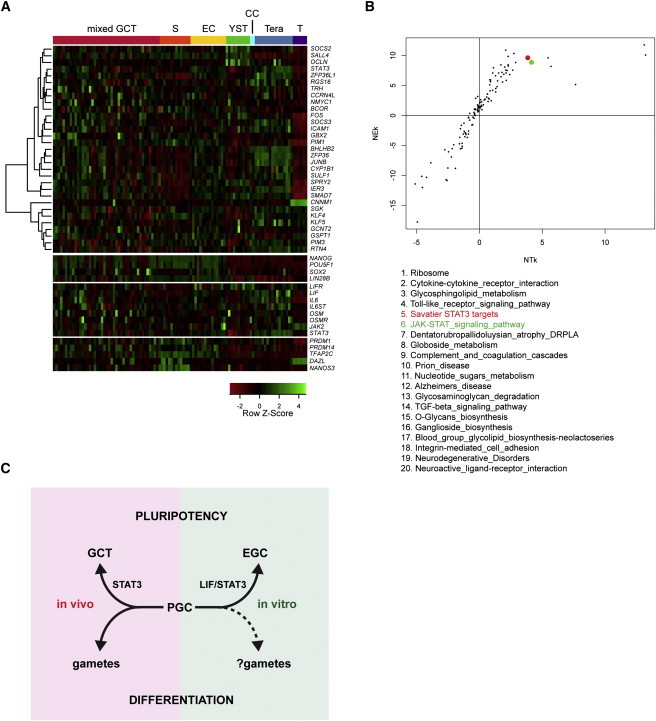
STAT3 Targets Are Upregulated in Germ Cell Tumors
(A) Heatmap of STAT3 target gene expression in adult germ cell tumors. Primary data are from Korkola et al. (2006). Samples were clustered according to the prevalent histology. Additional genes involved in pluripotency, LIF-signaling pathway, and PGC development are provided as a reference. mixed GCT, mixed germ cell tumors; S, seminoma; EC, embryonal carcinoma; YST, yolk-sac tumor; CC, choriocarcinoma; Tera, teratoma; T, normal testis.
(B) NTk and NEk values for KEGG pathways enriched in tumor samples over normal testis. STAT3 target genes were fed into the algorithm as a KEGG pathway (Savatier STAT3 targets, colored in red) for unbiased analysis of enrichment over other pathways. The standard JAK-STAT KEGG pathway is shown in green. The top 20 most enriched pathways are shown below order by average rank.
(C) Model depicting PGC conversion to pluripotency in two different contexts. In vitro, the LIF/STAT3 pathway is the key requirement for generation of EG cells. We further propose that in vivo activation of STAT3 drives the formation of the pluripotent cells that constitute embryonal carcinoma or give rise to teratomas, the “pluripotent GCTs.”
See also Figure S3.
Discussion
Mammalian primordial germ cells are considered unipotent, giving rise only to the gametes. Indeed, the sperm and egg represent two of the most overtly differentiated cell phenotypes. Yet, these two specialized cells regain access to the entire embryonic and extraembryonic differentiation programs following fertilization and zygotic reprogramming. Immature cells of the germline can also acquire pluripotency through nonphysiological routes, ex vivo formation of EG cells and multipotent germline stem cells (Kanatsu-Shinohara et al., 2004), or in vivo teratocarcinogenesis. However, those events have previously been obtained at low frequency in complex environments. Here, we demonstrate that 10%–30% of single mouse PGCs can convert to pluripotent EG cells in well-defined conditions. This is comparable to the efficiency of ES cell derivations reported from single epiblast cells (Brook and Gardner, 1997; Rugg-Gunn et al., 2012) and challenges the notion that PGCs are an intrinsically committed unipotent lineage.
PGC identity depends on the activity of determinants such as Blimp1 (Ohinata et al., 2005), Prdm14 (Yamaji et al., 2008), and Tcfap2c (Weber et al., 2010). However, PGC specification is also associated with reexpression or upregulation of core pluripotency transcription factors including Nanog, Sox2, and Klf2 (Kurimoto et al., 2008). These factors are thought to be essential in PGCs (Chambers et al., 2007; Kehler et al., 2004; Yamaguchi et al., 2009), although their role remains unclear. Their presence may mean that pluripotency is not extinguished in PGCs as in other postgastrulation lineages (Osorno et al., 2012), but could instead lie dormant. Derivation of EG cell lines from the first specified PGCs at E7.5 is consistent with the idea that latent pluripotency may be a necessary feature of the germline. The efficiency of EG cell generation does decrease during PGC development however, falling by more than an order of magnitude at E11.5. Interestingly, this coincides temporally with widespread epigenome modifications (Hajkova et al., 2008). Nonetheless, a rudiment of pluripotency is retained in later germline development as evidenced by the ability to derive a type of pluripotent stem cell from spermatogonial stem cells (Kanatsu-Shinohara et al., 2004; Ko et al., 2009).
In feeder-free cultures with 4Fs and GSK3 inhibition, we observed that PGCs exhibit features of locomotor cells, such as cell extension and lamellopodia (Movie S1). This motile phenotype persists throughout the early proliferative phase of culture for approximately 72 hr. After this time, cell death is progressive, and only cells undergoing conversion to EG cells continue to proliferate extensively. However, cell loss is heterogeneous and occasional cells with PGC morphology survive until much later time points. This raises the intriguing possibility that it may be feasible to sustain PGC proliferation and survival without EG cell formation. In this context, it might be productive to omit LIF while employing GSK3 inhibition.
LIF does not appear important for initial PGC culture but is specifically required to drive EG cell conversion. Inhibition of MAPK signaling is also not required for the initial 48 hr of PGC culture, in fact, is deleterious during that period. Our observations suggest that EG cell formation can be divided into two discrete phases: an initial 48 hr period of PGC adaptation to culture that is promoted by bFGF, RA, SCF, and GSK3 inhibition and a subsequent period of fate conversion over 6 days. The second phase is driven by LIF stimulation and MAPK inhibition, which is augmented by inhibition of GSK3. A key goal for future studies will be to elucidate the temporal pattern of STAT3 target gene induction and delineate the synergy with MAPK inhibition that reconstructs the full pluripotency and self-renewal circuit (Nichols and Smith, 2012). The intersection between these two pathways also appears crucial to achieve authentic induced pluripotency by somatic cell reprogramming (Silva et al., 2008; Sridharan et al., 2009; van Oosten et al., 2012; Yang et al., 2010). Elucidating the process of EG cell formation may therefore illuminate generally the acquisition of pluripotency.
Given the proven ability of transcription factors to artificially induce pluripotency in somatic cells (Takahashi and Yamanaka, 2006), the high expression of these factors in the germline (Kurimoto et al., 2008) raises the question of how PGCs are constrained from becoming pluripotent and thereby tumorigenic in vivo. Our findings point to the primacy of LIF/STAT3 signaling in driving fate conversion. We propose that activation of the STAT3 pathway in PGCs can result in reacquisition of pluripotency in two contexts—in vitro enabling the derivation of EG cells and in vivo allowing the formation of pluripotent GCTs (Figure 4C). The observation that STAT3 targets are underrepresented in PGCs suggests that the pathway is normally either silent or is antagonized. Indeed the LIF receptor gp130 is not required during PGC development (Molyneaux et al., 2003). This may be an important safeguard against acquisition of ectopic pluripotency.
STAT3 targets are upregulated in those GCTs that have a pluripotent compartment, or that have transited through a pluripotent state, suggesting that this pathway may play a previously unappreciated role in teratocarcinogenesis. This may merit further investigation, notably because inhibitors of the Jak/Stat pathway are being developed as chemotherapeutic agents against hematological and solid tumors (Liu et al., 2012; O’Shea et al., 2013; Quintás-Cardama et al., 2011). Although, GCTs are generally responsive to cisplatin therapy, resistance does occur particularly in teratomas (Oosterhuis and Looijenga, 2005) and conceivably might be reduced by targeting the STAT3 pathway in combination therapy.
Finally, re-evaluation of PGCs as a robust source of pluripotent stem cells and the pivotal role played by LIF in the conversion process raises the possibility that the early human germline might be a promising source of LIF-responsive pluripotent stem cells. We speculate that rebuilding pluripotency directly from in vivo or in vitro derived human PGCs may allow capture of the hypothetical human naive state (De Los Angeles et al., 2012), which has so far proved elusive starting from preimplantation embryos.
Experimental Procedures
Animal studies were authorized by a UK Home Office Project License and carried out in a Home Office-designated facility.
EG Cell Culture
For routine culture, EG cells were maintained in 2i/LIF medium on laminin (10 μg/ml, Sigma) or gelatin-coated plates. 2i/LIF medium comprise the MEK inhibitor PD0325901 (PD) 1 μM, the GSK3 inhibitor CHIR99021 (CH) 3 μM, and mouse LIF 10 ng/ml (prepared in house) in N2B27 medium (Ying et al., 2003) (Stem Cells, SCS-SF-NB-02). Cells were expanded by dissociation with trypsin and replating every 2–3 days.
EG-Cell-Derivation Medium
Derivations were performed in N2B27 (Ying et al., 2003) with the following additives as indicated: LIF, CH, PD (all as above), bFGF 25 nM (prepared in house), retinoic acid (RA) 2 μM (Invitrogen), forskolin (FK) 10 μM (Sigma), stem cell factor (SCF) 100 ng/ml (R&D), and JAK inhibitor 1 1 μM (Calbiochem). N2B27 was batch tested for EG cell derivation. Plates were coated with human plasma fibronectin (Millipore, 15–20 μg/ml in PBS, for 1 hr at 37°C) or precoated collagen IV plates (BD, Biocoat) were used, as indicated.
EG Cell Derivation
E7.5, E8.5, and E11.5 EG cell lines were established from embryos produced by crossing mixed background Oct4-ΔPE-GFP transgenic males (Yoshimizu et al., 1999) with strain 129 female mice. EG cell derivations from E8.5 embryos were performed essentially as described previously (Leitch et al., 2010). In brief, the posterior fragment of the embryo containing PGCs was dissected free of the extraembryonic membranes and trypsinized to a single cell suspension. Cells were collected by centrifugation. Cells were resuspended in derivation medium and either plated in two 2 cm2 wells per starting embryo or filtered and sorted using a MoFlo high-speed cell sorter (Dako Cytomation). Sorted PGCs were deposited directly into cell culture plates precoated with the indicated ECM protein (as above) and containing derivation medium. PGCs were plated at a density of approximately 12.5 cell per cm2, or as indicated. In all experiments, sorted GFP-negative somatic cells were cultured in parallel, and pluripotent cell colonies were not observed. E7.5 EG cell lines were derived from the above cross, as well as embryos produced by crossing mixed background Blimp1-GFP (Ohinata et al., 2005) or Stella-GFP BAC (Payer et al., 2006) transgenic males with strain 129 female mice. Embryos were dissected at 10 a.m., and embryos more advanced than the midallantoic bud stage (Downs and Davies, 1993) were excluded. The posterior fragment containing PGCs was dissected and processed as above for fluorescence activated cell sorting (FACS). GFP-positive cells were sorted as above and plated at a density of 12.5 cells per cm2 on fibronectin-coated dishes in CH/LIF plus bFGF, RA, SCF, and FK (4Fs). After 48 hr cultures were transitioned to 2i/LIF medium by daily half-medium changes. E11.5 EG cell colonies were obtained by dissecting E11.5 gonads and purifying PGCs by FACS, as above. Sorted PGCs were plated at a density of 1,000 cells per cm2, and EG cell derivation was performed as for E7.5. Time-lapse imaging was performed using a Leica DMI7000.
Piggybac Transposition
E7.5 Stella-GFP EG cells (1 × 106) were transfected using Lipofectamine 2000 (Invitrogen) with 4 μg of the PiggyBac vector pCAG-DsRed-IRES-Zeocin plus 2 μg of pCAGPBase (Wang et al., 2008). The Lipofectamine/DNA complex was applied to the cells in 2i/LIF for 7 hr and then removed and replaced with fresh medium. After 48 hr, Zeocin was added at a concentration of 100 μg/ml to select for stable transfectants. The cells were passaged three times over 7 days and Zeocin-resistant cells were flow-sorted. The brightest 20% DSRED-expressing cells were passaged as a stable pool for morula aggregation.
RNAi Experiments
siRNAs were transfected at a final concentration of 40 nM using Lipofectamine RNAiMAX (Invitrogen). For a 24-well plate (2 cm2), we used 1 μl of transfection reagent, 1 μl of 20 μM siRNA solution, and 400 μl of N2B27 medium. PGCs were sorted and plated on fibronectin-coated plates 30 hr before transfection. The medium was changed after 10 hr incubation. STAT3 siRNAs were purchased from QIAGEN (catalog numbers are indicated in parentheses): siSTAT3_1 (SI01435287), siSTAT3_2 (SI01435294), siSTAT3_3 (SI01435301), and siSTAT3_4 (SI01435308). GFP siRNAs were custom made (target sequence: GCAAGCTGACCTGAAGTTCA).
Chimera Production
Midgestation chimeras were generated by morula aggregation of fluorescent E7.5 EG cells with E2.5 embryos (Nagy et al., 2003). Term chimeras were produced by microinjection of E7.5 or E8.5 EG cells (agouti) into C57Bl/6 blastocysts (Nagy et al., 2003).
Immunostaining
Immunostaining was performed using standard protocols. Briefly, cells were fixed in 4% paraformaldehyde for 10 min, blocked, and permeabilized in PBS, 0.1% Triton X-100, and 1% BSA. Primary antibodies were incubated in the same buffer overnight at 4°C. Secondary antibodies were incubated for 1 hr at room temperature. Plates were washed 3 × 15 min in PBS after primary and secondary antibody incubations. Nuclei were stained with DAPI. Primary antibodies were: OCT4 (BD, 1:200) and KLF4 (R&D Systems, 3:500). Nuclei were stained with DAPI. Alexa Fluor secondary antibodies (Invitrogen) were used at 1:500 dilution.
Reanalysis of Published RNA-seq and Microarray Data
STAT3 targets (Bourillot et al., 2009) were compared between RNA-seq data sets for Oct4-positive ES cells (Tang et al., 2010) and E8.5 PGCs (Hackett et al., 2012) and mean reads per million (RPM) depicted as a scatterplot. Previously published microarray data of adult germ cell tumors (Korkola et al., 2006) were obtained from GEO (accession number GSE3218). Raw data files were analyzed in R/Bioconductor with the affy package and RMA normalized. To construct the heatmap, the normalized values of STAT3 target gene expression were log2 transformed and centered around the median. Pathway analysis was performed using the sigPathway package (http://bioconductor.org/packages/release/bioc/html/sigPathway.html). To create a customized gene set object, the G file specific for the human microarray used in this study, hgu133a (Tian et al., 2005), was modified to include a Savatier STAT3 targets category, containing the probes for the reported STAT3 target genes. The NTk and NEk enrichment values obtained for KEGG pathways were plotted against each other, and the pathways were ordered by average rank.
Acknowledgments
We thank Sam Jameson, Keith Savill, and staff for excellent animal husbandry; Nigel Miller (Department of Pathology, University of Cambridge) for flow cytometry support; and Bill Mansfield and Charles-Étienne Dumeau for chimera generation. We thank Petra Hajkova, Kirsten McEwen, and Ian Adams for helpful discussions. We thank Marko Hyvonen for production of recombinant LIF. This study was funded by the Biotechnology and Biological Sciences Research Council and the Medical Research Council of the United Kingdom and by the Swiss National Science Foundation Sinergia program. A.S. is a Medical Research Council Professor.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., Surani M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair K., Leitch H.G., Mansfield W., Dumeau C.-É., Humphreys P., Smith A.G. Culture parameters for stable expansion, genetic modification and germline transmission of rat pluripotent stem cells. Biology Open. 2012;1:58–65. doi: 10.1242/bio.2011029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourillot P.-Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N., Savatier P. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- Brook F.A., Gardner R.L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- De Felici M., Pesce M., Giustiniani Q., Di Carlo A. In vitro adhesiveness of mouse primordial germ cells to cellular and extracellular matrix component substrata. Microsc. Res. Tech. 1998;43:258–264. doi: 10.1002/(SICI)1097-0029(19981101)43:3<258::AID-JEMT8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Loh Y.-H., Tesar P.J., Daley G.Q. Accessing naïve human pluripotency. Curr. Opin. Genet. Dev. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miguel M.P., Cheng L., Holland E.C., Federspiel M.J., Donovan P.J. Dissection of the c-Kit signaling pathway in mouse primordial germ cells by retroviral-mediated gene transfer. Proc. Natl. Acad. Sci. USA. 2002;99:10458–10463. doi: 10.1073/pnas.122249399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolci S., Williams D.E., Ernst M.K., Resnick J.L., Brannan C.I., Lock L.F., Lyman S.D., Boswell H.S., Donovan P.J. Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature. 1991;352:809–811. doi: 10.1038/352809a0. [DOI] [PubMed] [Google Scholar]
- Downs K.M., Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Capel B. Development of germ cells in the mouse. Curr. Top. Dev. Biol. 2008;83:185–212. doi: 10.1016/S0070-2153(08)00406-7. [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Adams I.R., Barton S.C., Surani M.A., McLaren A. The role of exogenous fibroblast growth factor-2 on the reprogramming of primordial germ cells into pluripotent stem cells. Stem Cells. 2006;24:1441–1449. doi: 10.1634/stemcells.2005-0424. [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Tang F., Doody G., Tooze R., Surani M.A. Reprogramming primordial germ cells into pluripotent stem cells. PLoS ONE. 2008;3:e3531. doi: 10.1371/journal.pone.0003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin I., Deed R., Cooke J., Zsebo K., Dexter M., Wylie C.C. Effects of the steel gene product on mouse primordial germ cells in culture. Nature. 1991;352:807–809. doi: 10.1038/352807a0. [DOI] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2012;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P., Ancelin K., Waldmann T., Lacoste N., Lange U.C., Cesari F., Lee C., Almouzni G., Schneider R., Surani M.A. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomelí H., Nagy A., McLaughlin K.J., Schöler H.R., Tomilin A. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Suzuki A., Fujita Y., Yomogida K., Lomelí H., Asada N., Ikeuchi M., Nagy A., Mak T.W., Nakano T. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development. 2003;130:1691–1700. doi: 10.1242/dev.00392. [DOI] [PubMed] [Google Scholar]
- Kimura T., Tomooka M., Yamano N., Murayama K., Matoba S., Umehara H., Kanai Y., Nakano T. AKT signaling promotes derivation of embryonic germ cells from primordial germ cells. Development. 2008;135:869–879. doi: 10.1242/dev.013474. [DOI] [PubMed] [Google Scholar]
- Ko K., Tapia N., Wu G., Kim J.B., Bravo M.J.A., Sasse P., Glaser T., Ruau D., Han D.W., Greber B. Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell. 2009;5:87–96. doi: 10.1016/j.stem.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Korkola J.E., Houldsworth J., Chadalavada R.S., Olshen A.B., Dobrzynski D., Reuter V.E., Bosl G.J., Chaganti R.S. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- Koshimizu U., Taga T., Watanabe M., Saito M., Shirayoshi Y., Kishimoto T., Nakatsuji N. Functional requirement of gp130-mediated signaling for growth and survival of mouse primordial germ cells in vitro and derivation of embryonic germ (EG) cells. Development. 1996;122:1235–1242. doi: 10.1242/dev.122.4.1235. [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labosky P.A., Barlow D.P., Hogan B.L. Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development. 1994;120:3197–3204. doi: 10.1242/dev.120.11.3197. [DOI] [PubMed] [Google Scholar]
- Leitch H.G., Blair K., Mansfield W., Ayetey H., Humphreys P., Nichols J., Surani M.A., Smith A. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development. 2010;137:2279–2287. doi: 10.1242/dev.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., McClintick J., Zhong L., Edenberg H.J., Yoder M.C., Chan R.J. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
- Liu X., Guo W., Wu S., Wang L., Wang J., Dai B., Kim E.S., Heymach J.V., Wang M., Girard L. Antitumor activity of a novel STAT3 inhibitor and redox modulator in non-small cell lung cancer cells. Biochem. Pharmacol. 2012;83:1456–1464. doi: 10.1016/j.bcp.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T., Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Toksoz D., Nishikawa S., Nishikawa S., Williams D., Zsebo K., Hogan B.L. Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature. 1991;353:750–752. doi: 10.1038/353750a0. [DOI] [PubMed] [Google Scholar]
- Matsui Y., Zsebo K., Hogan B.L. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- McLaren A., Lawson K.A. How is the mouse germ-cell lineage established? Differentiation. 2005;73:435–437. doi: 10.1111/j.1432-0436.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- Molyneaux K.A., Schaible K., Wylie C. GP130, the shared receptor for the LIF/IL6 cytokine family in the mouse, is not required for early germ cell differentiation, but is required cell-autonomously in oocytes for ovulation. Development. 2003;130:4287–4294. doi: 10.1242/dev.00650. [DOI] [PubMed] [Google Scholar]
- Nagamatsu G., Kosaka T., Saito S., Takubo K., Akiyama H., Sudo T., Horimoto K., Oya M., Suda T. Tracing the conversion process from primordial germ cells to pluripotent stem cells in mice. Biol. Reprod. 2012;86:182. doi: 10.1095/biolreprod.111.096792. [DOI] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. Cold Spring Harbor Laboratory Press; New York: 2003. Manipulating the Mouse Embryo. [Google Scholar]
- Nichols J., Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol. 2012;4:a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- O’Shea J.J., Holland S.M., Staudt L.M. JAKs and STATs in immunity, immunodeficiency, and cancer. N. Engl. J. Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O’Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Oosterhuis J.W., Looijenga L.H.J. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- Osorno R., Tsakiridis A., Wong F., Cambray N., Economou C., Wilkie R., Blin G., Scotting P.J., Chambers I., Wilson V. The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development. 2012;139:2288–2298. doi: 10.1242/dev.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B., Chuva de Sousa Lopes S.M., Barton S.C., Lee C., Saitou M., Surani M.A. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis. 2006;44:75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- Quintás-Cardama A., Kantarjian H., Cortes J., Verstovsek S. Janus kinase inhibitors for the treatment of myeloproliferative neoplasias and beyond. Nat. Rev. Drug Discov. 2011;10:127–140. doi: 10.1038/nrd3264. [DOI] [PubMed] [Google Scholar]
- Resnick J.L., Bixler L.S., Cheng L., Donovan P.J. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Cox B.J., Lanner F., Sharma P., Ignatchenko V., McDonald A.C.H., Garner J., Gramolini A.O., Rossant J., Kislinger T. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev. Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger J.M., Chen X., Draper J.S., Antosiewicz J.E., Chon C.H., Jones S.B., Brooks J.D., Andrews P.W., Brown P.O., Thomson J.A. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L.C. Origin of testicular teratomas from primordial germ cells in mice. J. Natl. Cancer Inst. 1967;38:549–552. [PubMed] [Google Scholar]
- Stewart C.L., Gadi I., Bhatt H. Stem cells from primordial germ cells can reenter the germ line. Dev. Biol. 1994;161:626–628. doi: 10.1006/dbio.1994.1058. [DOI] [PubMed] [Google Scholar]
- Surani M.A. Germ cells: the eternal link between generations. C. R. Biol. 2007;330:474–478. doi: 10.1016/j.crvi.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Greenberg S.A., Kong S.W., Altschuler J., Kohane I.S., Park P.J. Discovering statistically significant pathways in expression profiling studies. Proc. Natl. Acad. Sci. USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten A.L., Costa Y., Smith A., Silva J.C. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat. Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin C., Lu D., Ning Z., Cox T., Melvin D., Wang X., Bradley A., Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Eckert D., Nettersheim D., Gillis A.J.M., Schäfer S., Kuckenberg P., Ehlermann J., Werling U., Biermann K., Looijenga L.H.J., Schorle H. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol. Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- West J.A., Viswanathan S.R., Yabuuchi A., Cunniff K., Takeuchi A., Park I.-H., Sero J.E., Zhu H., Perez-Atayde A., Frazier A.L. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Kurimoto K., Yabuta Y., Sasaki H., Nakatsuji N., Saitou M., Tada T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development. 2009;136:4011–4020. doi: 10.1242/dev.041160. [DOI] [PubMed] [Google Scholar]
- Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y., Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yang J., van Oosten A.L., Theunissen T.W., Guo G., Silva J.C.R., Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T., Sugiyama N., De Felice M., Yeom Y.I., Ohbo K., Masuko K., Obinata M., Abe K., Schöler H.R., Matsui Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


