Abstract
Significant tumor regressions have been observed in up to 70% of patients receiving adoptively transferred autologous melanoma-reactive tumor infiltrating lymphocytes (TIL) 1,2, and in pilot trials, 40% of treated patients experienced complete regressions of all measurable lesions for at least five years following treatment 3. To evaluate the potential association between the ability of TIL to mediate durable regressions and their ability to recognize potent antigens that presumably include mutated gene products, a novel screening approach was developed that involved mining whole exome sequence data to identify the mutated proteins that were expressed in patient tumors. Candidate mutated T cell epitopes that were identified using an MHC binding algorithm 4 were then synthesized and evaluated for recognition by TIL. Using this approach, mutated antigens expressed on autologous tumor cells were identified as targets of three TIL that were associated with objective tumor regressions following adoptive transfer. This simplified approach, which avoids the need to generate and laboriously screen cDNA libraries from tumors, may represent a generally applicable method for identifying mutated T cell antigens expressed in melanoma as well as other tumor types.
The identification of antigens associated with tumor rejection mediated by TIL has been challenging, given the diversity of these bulk populations and the relatively laborious nature of current antigen screening approaches. We have developed a novel screening method to identify mutated candidate epitopes that initially involved whole exome sequencing of tumor and matched normal cell DNA to identify somatic mutations. High-affinity candidate T cell epitopes identified in silico by scanning 19-mer polypeptides centered on mutated residues with a peptide-MHC binding algorithm 4 were then screened for recognition by TIL. We focused on identifying T cell epitopes presented in the context of HLA-A class I gene products, which were previously found to be expressed at higher levels in melanomas than HLA-B and C products 5, in three individuals with metastatic melanoma who demonstrated regression of bulky metastatic lesions following the adoptive transfer of autologous TIL.
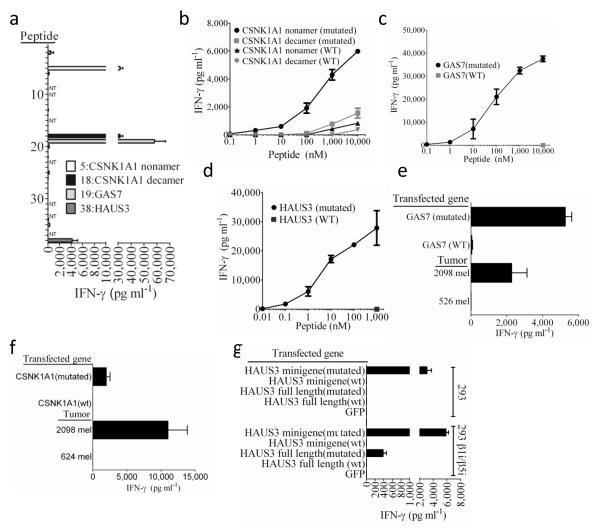
Initially, DNA isolated from the melanoma cell line 2098 mel, which was generated from a metastatic lesion an individual who was homozygous for the highly prevalent HLA class I allele HLA-A*0201, was subjected to whole exome sequencing. The top 55 mutated candidate nonamers and decamer peptides identified from 2098 mel that were predicted to bind with high affinity to HLA-A*0201 were then synthesized and evaluated for their ability to sensitize the HLA-A*0201+ cell line T2 6 for recognition by autologous TIL. The results indicated that four peptides, 5th, 18th 19th and 38th of the predicted high affinity HLA-A*02:01 binders, reproducibly stimulated high levels of IFN-γ release from the autologous TIL 2098 (Fig. 1a and Supplementary Table 1). Peptides #5 and #18 represented overlapping nonamer and decamers that corresponded to residues 26-34 and 26-35, respectively, of the casein kinase 1, alpha 1 (CSNK1A1) protein, a key regulator of the Wnt/β-catenin signaling pathway 7. A di-nucleotide substitution of TA for CC at positions 80 and 81 of the wild type CSNK1A1 coding region resulted in a serine to leucine change at position 27 in the wild type protein, .Peptide #19 was encoded by a point-mutated transcript of the growth arrest specific 7 (GAS7) gene, previously identified as a mutated target of TIL 2098 using a conventional cDNA library screening approach 8, and peptide #38 was corresponded to residues 154-162 of the HAUS augmin-like complex, subunit 3 (HAUS3) protein (Figure 1a), a molecule involved with microtubule formation within the mitotic spindle 9. Peptide titration assays demonstrated that A*0201+ target cells pulsed with a minimum of 1 nM of the mutated CSNK1A1 nonamer stimulated significant cytokine release from TIL 2098, while 100 nM of the mutated decamer stimulated a significant, albeit a significantly weaker response (Fig. 1b). The corresponding wild type CSNK1A1 peptides, which stimulated weaker responses than the mutated peptides, were not recognized at concentrations below 1 μM. The change at position two in the CSNK1A1 peptide from serine to leucine, which represents an optimal residue at one of the primary anchor positions in HLA-A*0201-binding peptides 10 enhanced the predicted HLA-A*0201 binding affinities of the CSNK1A1 peptides from > 1 μM to approximately 10 nM (Supplementary Table 1). TIL 2098 recognized targets pulsed with as little as 1 nM of the mutated GAS7 peptide and 0.1 nM of the mutated HAUS3 peptides, and failed to recognize the corresponding wild type peptides (Fig. 1c,d). The change in the HAUS3 peptide was not predicted to alter the binding affinity of the mutated peptide and the change in the GAS7 peptide was predicted to lead to a modest reduction in the binding affinity from 12 to 39 nM, indicating that these may predominantly represent T cell contact residues.
Figure 1. Response of TIL 2098 to candidate epitopes identified from autologous tumors.
(a) A screening assay was carried out to evaluate the release of IFN-γ from TIL 2098 in an overnight co-culture with peptide-pulsed T2 cells that were pulsed individually with the top 61 candidate HLA-A*0201 binding peptides (Supplementary Table 1) identified from 2098 mel, with the exception of peptides 9,10,14,23,24,31,33 and 37, which overlapped with peptides 1,4,4,25,12,15,16 and 4, respectively (NT). Peptides 39-62 (Supplementary Table 1) stimulated the release of 100 pg/ml or less of IFN-γ from TIL 2098 and are not depicted in this graph. The autologous 2098 mel stimulate the release of 10,000 pg/ml of IFN-γ from 2098 TIL in this assay. (b-d) T2 cells were pulsed with tittered doses of the indicated mutated or wild type (WT) peptides, and IFN-γ release measured in an overnight co-culture with TIL 2098. (e) Stable transfected of COS7 cells (COS-A2) expressing HLA-A*0201 were transiently transfected with the indicated transcripts and evaluated for their ability to stimulate IFN-γ release from TIL 2098 in an overnight co-culture. (f) 293 cells were transiently transfected with a construct encoding HLA-*0201 as well as the indicated transcripts and evaluated for their ability to stimulate IFN-γ release from TIL 2098 in an overnight co-culture.
We then transfected antigen negative but HLA-A*0201+ target cells with genes encoding mutated epitopes in order to evaluate their ability to be naturally processed and presented. TIL 2098 recognized HLA-A*0201-transduced COS7 cells (COS-A2) that were transiently transfected with the mutated but not wild type CSNK1A1 and GAS7 constructs (Fig 1e,f), but failed to recognize 293 cells that were transiently transfected with HLA-A*0201 (293-A2) plus the full length mutated HAUS3 construct (Fig. 1g). TIL 2098 did, however, recognize 293-A2 cells that were transfected with a mini-gene construct that encoded a methionine followed by the nine amino mutated HAUS3 epitope (Fig. 1g), indicating that this may represent an epitope that is preferentially processed either by the immunoproteasome 11,12 or recently described intermediate proteasomes that contain one or two of the three catalytic subunits of the immunoproteasome 13. Further analysis demonstrated that TIL 2098 recognized stable 293 transfectants expressing the β1i and β5i immunoproteasomal subunits that were transiently transfected with HLA-A*0201 plus either the mutated HAUS3 mini-gene or full length gene, but not the corresponding wild type HAUS3 constructs (Fig. 1g).
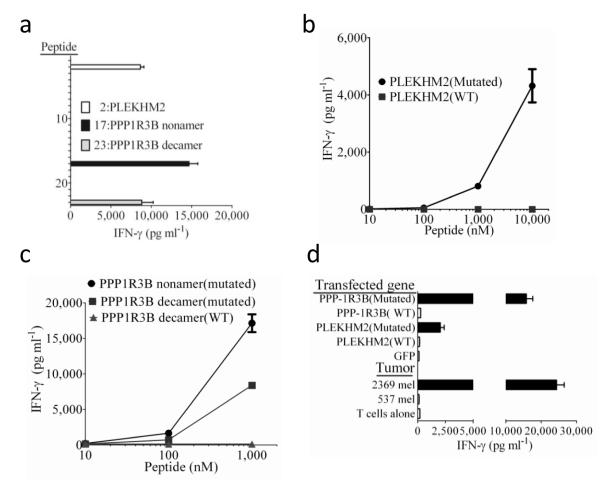
We then attempted to identify mutated antigens recognized by TIL 2369, which was derived from an individual with melanoma who expressed HLA-A*0101 and A*2601, by co-incubating TIL with HLA-A*0101+ target cells that were pulsed individually with the top 53 mutated candidate HLA-A*0101-binding peptides identified from 2369 mel (Supplementary Table 2). A mutated decamer encoded by the pleckstrin homology domain containing, family M member 2 (PLEKHM2) gene that possessed the 2nd highest affinity of the predicted HLA-A*0101-binding peptides, as well as two overlapping mutated nonamer and decamers encoded by the protein phosphatase 1, regulatory subunit 3B (PPP1R3B) gene that represented the 17th and 23rd highest predicted binders, stimulated high levels of cytokine release from TIL 2369 (Fig. 2a and Supplementary Table 2). TIL 2369 recognized target cells pulsed with a minimum of 1.0 μM of the mutated PLEKHM2 peptide (Fig. 2b) and 0.1 μM of the overlapping PPP1R3B peptides (Fig. 2c), but failed to recognize cells pulsed with 10 μM of the corresponding wild type peptides (Fig. 2b,c). TIL 2369 also recognized antigen negative HLA-A*0101+ cells transfected with cDNAs encoding the mutated but not the wild type PPP1R3B and PLEKHM2 gene products (Fig 2d). TIL 2369 failed to recognize any of the top 46 mutated candidate peptides that were predicted to bind to HLA-A*2601 with affinities ranging between 4 and 259 nM (Supplementary Table 3).
Figure 2. Response of TIL 2369 to candidate epitopes identified from autologous tumors.
(a) A screening assay was carried out to evaluate the release of IFN-γ from TIL 2369 in an overnight co-culture with peptide-pulsed 293-A1 cells that were pulsed individually with the top 56 candidate HLA-A*0101-binding peptides identified from 2369 mel (Supplementary Table 2), with the exception of peptides 35, 39 and 44, which overlapped with peptides 20, 33 and 43, respectively, and were not tested (NT). Peptides 24-56 (Supplementary Table 2) stimulated the release of less than 100 pg/ml of IFN-γ from TIL 2369 and are not depicted in this graph. The autologous 2369 mel stimulated the release of 20,838 pg/ml of IFN-γ from TIL 2369 in this assay. (b,c) 293-A1 cells were pulsed with the indicated concentrations of peptides in an overnight co-culture with TIL 2098, and IFN-γ release measured. d. 293-A1 cells were transiently transfected with the indicated constructs and evaluated for their ability to stimulate IFN-γ release from TIL 2369 in an overnight co-culture.
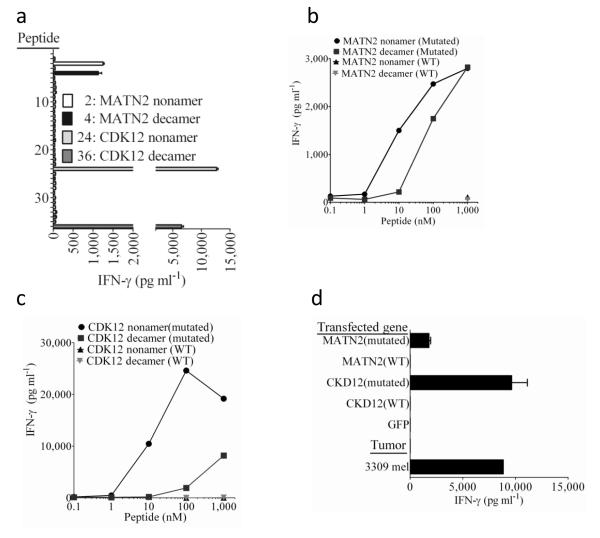
This approach was further examined by evaluating the ability of TIL isolated from a third individual with melanoma, TIL 3309, to recognize mutated candidate peptides identified from 3309 mel that were predicted to bind to either the autologous HLA-A*0101 or the A*1101 class I alleles. While none of the top 29 candidate HLA-A*0101-binding mutated peptides stimulated significant cytokine release from TIL 3309 (Supplementary Table 4), four of the top 46 candidate HLA-A*1101-binding mutated peptides, which included overlapping nonamer and decamer products of the matrilin 2 (MATN2) and cyclin-dependent kinase 12 (CDK12) genes, were recognized by TIL 3309 (Fig. 3a and Supplementary Table 5). The MATN2 protein is a member of the von Willebrand factor A domain containing family and may play a role in extracellular matrix formation 14, and CDK12 has been shown to regulate the expression of DNA damage response genes 15. The mutated MATN2 nonamer and decamer possessed the second and fourth highest predicted binding affinities, respectively, of the candidate HLA-A*1101-binding peptides, while the mutated CDK12 nonamer and decamer possessed the 24th and 36th highest predicted HLA-A*1101 binding affinities (Supplementary Table 5). The mutated MATN2 nonamer and decamer stimulated responses at minimum concentrations of 1 nM, whereas the corresponding wild type MATN2 peptides failed to stimulate significant cytokine release from this TIL at the highest concentration tested, 1 μM (Fig. 3b). TIL 3309 recognized targets pulsed with a minimum of 1 nM and 100 nM of the mutated CDK12 nonamer and decamers, respectively, but failed to recognized targets pulsed with 1 μM of the corresponding wild type peptides (Fig. 3c). In addition, HLA-A*1101+ target cells transfected with the mutated but not the corresponding wild type MATN2 and CDK12 genes stimulated significant cytokine release from TIL 3309 (Fig. 3d).
Figure 3. Response of TIL 3309 to candidate epitopes identified from autologous tumors.
(a) A screening assay was carried out to evaluate the release of IFN-γ from TIL 3309 in an overnight co-culture with COS-A11 cells that were pulsed individually with the top 46 candidate HLA-A*1101-binding peptides identified from 3309 mel (Supplementary Table 5). Peptides 37-46 (Supplementary Table 5) stimulated the release of less than 100 pg/ml of IFN-γ from TIL 3309 and are not depicted in this graph. The autologous 3309 mel stimulated the release of 4,600 pg/ml of IFN-γ from TIL 3309 in this assay. (b,c) COS7 cells stably transduced with a retroviral vector expressing HLA-A*1101 (COS-A11) were pulsed with the indicated concentrations of peptides in an overnight co-culture with TIL 3309, and IFN-γ release measured. d. COS-A11 cells were transiently transfected with the indicated constructs and evaluated for their ability to stimulate IFN-γ release from TIL 3309 in an overnight co-culture.
We then evaluated the expression of genes encoding mutated candidate epitopes by attempting to amplify partial cDNA transcripts from tumor cell mRNA using primers that flanked the putative mutation sites. In addition, amplified transcripts were subjected to Sanger sequencing in an attempt to validate whole exome sequencing results. Partial cDNA transcripts encoding 19 of the 62 candidate mutated A*0201-binding peptides identified from 2098 mel could be amplified from tumor cell RNA, while three of the amplified products appeared to exclusively encode the wild type and not the expected amino acid variant (Supplementary Table 1 and Supplementary Methods Primer List). Transcripts encoding 27 of the top 56 candidate mutated high affinity HLA-A*0101 peptides identified from 2369 mel could be amplified from this cell line, while one of the amplified products encoded the wild type, rather than the expected mutated residue (Supplementary Table 2 and Supplementary Methods Primer List). In addition, transcripts encoding 28 of the 46 candidate mutated HLA-A*1101-binding peptides evaluated for recognition by TIL 3309 could be amplified from 3309 mel, whereas one of the amplified products appeared to exclusively encode the wild type and not the expected mutated amino acid (Supplementary Table 5 and Supplementary Methods Primer List). Combining the peptide prediction results with gene expression analysis demonstrated that TIL 2098 and 2369 recognized the mutated peptides with the highest predicted binding affinity for HLA-A*0201 and *0101, respectively, and TIL 3309 recognized the peptide with the second highest predicted binding affinity for HLA-A*1101 among the proteins expressed by autologous tumor cells (Supplementary Table 6). Taken together, these results indicated that TIL may generally recognize mutated epitopes that bind with high affinity to MHC class I molecules.
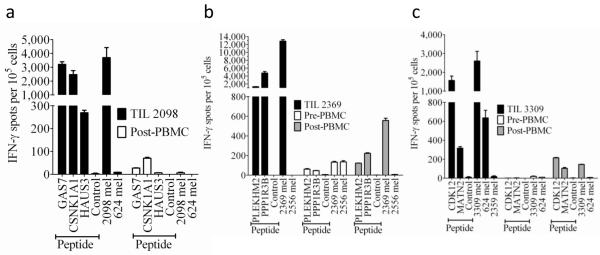
We then evaluated the contribution of T cells recognizing mutated targets to in vitro as well as in vivo anti-tumor responses by carrying out ELISPOT assays on infused TIL as well as samples of peripheral blood mononuclear cells (PBMC) obtained prior to and following therapy. The responses of TIL 2098 to HLA-A*0201+ target cells that were pulsed with either the mutated GAS7 and CSNK1A1 nonamer peptides were comparable to those directed against the autologous tumor, while lower but significant responses were directed against the mutated HAUS3 peptide (Fig. 4a). Although T cells may in some cases be stimulated more efficiently by the high levels of peptide that result from pulsing target cells than those resulting from endogenous processing, the ELISPOT response of TIL 2098 to the 2098 mel cell line that was nearly 60% of that generated in response to a polyclonal activator, indicating that 2098 mel represented a potent activator of the autologous TIL. Pre-treatment samples were not available for the first subject, but the responses against the mutated GAS7 and CSNK1A1 peptides observed in PBMC obtained approximately one month after transfer may have reflected low levels of persistence of adoptively transferred T cells recognizing these antigens (Fig. 4a). These results, taken together with the lack of response against HLA-matched melanomas (Fig. 4a and 8), indicated that TIL 2098 predominantly recognized mutated and not shared T cell epitopes.
Figure 4. IFN-γ ELISPOT responses of TIL and PBMC obtained prior to and following autologous TIL transfer.
Samples of PBMC prior to adoptive TIL transfer as well as samples of PBMC obtained approximately one month following transfer were evaluated along with samples of infused TIL. A pre-treatment sample was not available for subject 1, and the pre-treatment sample analyzed from subject 2 was obtained approximately one month following an adoptive TIL transfer that was administered in the absence of non-myeloablative chemotherapy one month prior to infusion of the TIL analyzed in (b). TIL 2098 from subject 1 was cultured with HLA-A*0201+ COS-7 cells (a), TIL 2369 from subject 2 was cultured with HLA*0101+ COS7 cells (b), and TIL 3309 from subject 3 were cultured with HLA*1101+ COS-7 cells (c) that were pulsed with candidate nonamer peptides for two hours at 370C. T cells were also cultured with autologous as well as allogeneic tumor cells, and in addition, each of the individual T cell populations were stimulated in parallel cultures with PMA plus ionomycin (PMA/I). The numbers of IFN-γ ELISPOTS per 105 T cells generated in responses to PMA/I were as follows: Subject 1 TIL 2098:7,300, subject 1 post transfer PBMC:4,700, Subject 2 TIL 2369:21,800, Subject 2 pre-transfer PBMC:21,800, Subject 2 post-transfer PBMC:11,000, Subject 3 TIL 3309:7,700, Subject 3 pre-transfer PBMC: 12,500, Subject 3 post-transfer PBMC:26,300. TIL 2098 was plated at 2,000 cells per well, TIL 2369 and 3309 were plated at 1,000 cells per well, and PBMC were plated at 100,000 cells per well, whereas all of the groups stimulated with PMA/I were plated at 1,000 cells per well. The error bars represent the mean + and − the standard error of the mean of three replicate wells per group.
Similarly, TIL 2369 responded strongly to HLA-A0101+ cells that were pulsed with the mutated PPP1R3B nonamer, which stimulated approximately 50% of the number of ELISPOTs than those observed against the autologous 2369 mel, and a lower but significant response against the mutated PLEKHM2 peptide (Fig. 4b). TIL 2369 failed to recognize allogeneic melanoma cell lines that shared expression of HLA-A*01 (Fig. 2d) or HLA-A*01 and A*26 with 2369 mel (Fig. 4b), which, taken together with the robust response against the mutated PPP1R3B and PLEKHM2 peptides, indicated that TIL 2369 may predominantly recognized mutated gene products. This individual had failed to respond clinically to treatment with a similar TIL product that was administered in the absence of a prior conditioning two months before receiving the TIL analyzed in this report. The low levels of T cells recognizing the mutated PLEKHM2 and PPP1R3B peptides and tumor cells in the earlier peripheral blood sample obtained approximately one month following the initial TIL transfer (designated Pre-PBMC) therefore presumably reflected the in vivo persistence of T cells following the initial TIL transfer. Higher levels of T cells reactive with the mutated PLEKHM2 and PPP1R3B peptides as well as 2369 mel were observed in PBMC obtained following the second TIL treatment (designated Post-PBMC), although we cannot rule out the possibility that T cells from the first treatment were present at this time.
An evaluation of ELISPOT responses in the third subject revealed that the response of autologous TIL 3309 to HLA-A*1101+ target cells that had been pulsed with the mutated CDK12 epitope was approximately 60% of that observed against the autologous 3309 mel, while the response to the MATN2 peptide was approximately 10% of that observed against the autologous mel. TIL 3309 also strongly recognized an allogeneic melanoma, 624 mel, that shared expression of HLA-C*07 with autologous cells (Fig. 4c), indicating that these T cells also recognized a shared epitope that was likely to represent a non-mutated gene product. Significant reactivity against the CDK12 and MATN2 peptides, as well as autologous 3309 mel, was observed in peripheral blood one month following autologous TIL transfer (Fig. 4c), at a time when CD8+ T cells comprised approximately 90% of the CD3+ T cells in peripheral blood. Reactivity against the CDK12 and MATN2 peptides, however, was undetectable in peripheral blood obtained prior to adoptive TIL transfer (Fig. 4c), and thus any increase in the percentage of CD8+ T cells reactive with these epitopes also reflected an increase in their absolute number.
Antigens recognized by tumor-reactive T cells have previously been identified by a variety of approaches that include the screening of antigen negative target cells that were transfected with individual cDNAs 16,17 or cDNA pools 18,19 generated from tumor cell lines, mass spectrometric analysis of peptides eluted from the surface of tumor cells 20, as well as proteomic analysis of tumor cell lysates 21. More than 400 T cell epitopes including over 50 mutated epitopes that were identified primarily from clonal or bulk populations of melanoma reactive T cells have been identified using these approaches 22. The approach described in this report complements cumbersome cDNA library cloning methodologies, which can be influenced by factors such as the size and expression levels of transcripts that encode T cell epitopes.
A mutated cancer antigen was recently identified from a methylcholanthrene-induced sarcoma that arose in a Rag2−/− mouse 23 and mutated T cell epitopes identified in B16F10 murine melanoma cells 24 from whole exome sequencing. The results of in vivo tumor model studies indicated that these mutated epitopes provided partial protection from tumor growth, supporting the potential utility of this method for identifying clinically relevant tumor antigens.
We have begun to evaluate the potential correlation between reactivity to mutated antigens and clinical response to adoptive TIL transfer. In the current study, three therapeutic TIL contained dominant populations of T cells reactive with mutated epitopes. Reactivity to mutated antigens appears to be of high importance, since only a small proportion of T cells in TIL appear to recognize melanocyte differentiation antigens and cancer germline antigens 25. Preliminary results indicate that TIL from one of two patients who failed to response to autologous TIL transfer recognized a single mutated epitope identified by exomic sequencing. Thus, this approach has lead to the identification of a total of eight mutated epitopes recognized by four of the five melanoma TIL that have been evaluated to date, demonstrating that this represents a reliable method for identifying tumor-specific T cell epitopes. This simple and rapid genomic approach should also facilitate the identification of candidate epitopes from additional tumor types and provide an opportunity to generate tumor reactive T cells by in vitro sensitization against the identified epitopes.
ONLINE METHODS
Subjects
Subject 1, a 53-year old female with extensive refractory metastatic melanoma to the lung and lower extremity received autologous TIL (#2098) in 2003 and underwent a complete regression of all metastatic melanoma that was sustained until her death from an unrelated ovarian cancer six years later. Subject 2, a 32-year old male with metastatic melanoma to the brain, liver and periportal lymph nodes, refractory to prior treatment, received autologous TIL (#2369) in 2005 and experienced a complete cancer regression that is ongoing 6-1/2 years later. Subject 3, a 27 year old female with metastatic melanoma to the brain, lung, thigh and popliteal fossa, received autologous TIL (#3309) in August of 2009 and had a nearly complete regression of all lesions but then developed recurrent thigh and chest wall lesions six months following therapy. All patients signed an informed consent form approved by the Institutional Review Board of the US National Cancer Institute.
Whole exome sequencing
Genomic DNA purification, library construction, exome capture of approximately 20,000 coding genes, and next generation sequencing of tumor and normal samples were performed at Personal Genome Diagnostics (Baltimore, MD), while bioinformatic analyses were carried out by Personal Genome Diagnostics and the Genome Technology Access Center, Genomics and Pathology Services of the Washington University School of Medicine. In brief, genomic DNA from tumor and normal samples were fragmented and used for Illumina TruSeq library construction (Illumina, San Diego, CA). Exonic regions were captured in solution using the Agilent SureSelect 50 Mb kit (version 3) according to the manufacturer’s instructions (Agilent, Santa Clara, CA). Paired-end sequencing, resulting in 100 bases from each end of the fragments, was performed using a HiSeq 2000 Genome Analyzer (Illumina, San Diego, CA). Sequence data were mapped to the reference human genome sequence and sequence alterations were determined by comparison of over 50 million bases of tumor and normal DNA. Over 8 billion bases of sequence data were obtained for each sample, with a high fraction from the captured coding regions. Over 43 million bases of target DNA were analyzed in the tumor and normal samples, and an average of 42 to 51 reads were obtained at each base in the normal and tumor DNA samples. The tags were aligned to the human genome reference sequence (hg18) using the Eland algorithm of CASAVA 1.6 software (Illumina, San Diego, CA). The chastity filter of the BaseCall software of Illumina was used to select sequence reads for subsequent analysis. The ELANDv2 algorithm of CASAVA 1.6 software (Illumina, San Diego, CA) was then a applied to identify point mutations and small insertions and deletions. Known polymorphisms recorded in dbSNP were removed from the analysis. Potential somatic mutations were filtered and visually inspected as described previously 26. The DNA isolated from the 2098 mel, 2369 mel and 3309 mel cell lines possessed 264, 574 and 278 non-synonymous single and di-nucleotide substitutions, respectively. Between 80 and 90% of the non-synonymous point mutations identified in the 2098, 2369 and 3309 tumor cell linesrepresented C/G to T/A transitions, consistent with the role of UV in generating the majority of mutations found in non-acral melanomas 27,28.
Analysis of gene expression
Oligonucleotide primers were designed to amplify fragments of gene products ranging between approximately 100 and 600 nucleotides encompassing the mutated epitopes in 2098, 2369 and 3309 mel (Supplementary Methods Primer List). These primers sets were used to carry out RT-PCRs, as previously described 19, from approximately 1 μg of RNA that was isolated from the appropriate tumor cell lines using RNAeasy (Qiagen Inc., Valencia, CA 91355). Amplified cDNA transcripts were either directly sequenced using after purification of the RT-PCR products using the PureLink PCR Purification Kit (Life Technologies, Grand Island, NY 14072) or cloned into pCDNA3.1 TOPO TA (Life Technologies) and sequenced using the T7 oligonucleotide primer 5′- TAATACGACTCACTATAGGG-3′.
Peptides
The residues surrounding amino acids resulting from non-synonymous mutations were scanned to identify candidate nonamer and decamer peptides that were predicted to bind with high affinity to individual HLA class I alleles using the NetMHCPan2.4 binding algorithm 4. Peptides were obtained from PiProteomics (Huntsville, AL, 35816) or Peptide 2.0 (Chantilly, VA 20151). Mutated peptides identified by this approach were then purified to greater than 95% homogeneity by HPLC and re-evaluated to confirm their ability to stimulate T cell responses.
Analysis of T cell responses
The TIL 2098, 2369 and 3309 were expanded from fresh tumor digests as previously described 29,30. Reactivity of TIL 2098 was evaluated by incubating HLA-A*02:01+ T2 cells that possess antigen processing defects that allow efficient loading of exogenous peptides 31 with candidate HLA-A*02:01 binding peptides at a concentration of 10 μM for two hours at 370C followed by two washing steps. Endogenous processing of the epitopes recognized by TIL 2098 was evaluated in 293 cells that were co-transfected with 200 ng of a recombinant pCDNA3.1 plasmid (Life Technologies, Grand Island, NY) encoding candidate epitopes and 50 ng of a recombinant pCDNA3.1 plasmid encoding HLA-A*02:01, and the levels of soluble IFN-γ released from TIL that were cultured overnight with either peptide pulsed target cells, transfectants or tumor cells were measured, as previously described 19. Evaluation of responses of TIL 2098 against the GAS7 and C4orf15 mutated and wild type constructs were carried out in 293 cells expressing the β1i and β5i immunoproteasomal subunits. The HLA-A*01:01+ tumor cell line 537 mel, which is not recognized by TIL 2369, and the HLA-A*02:01+ tumor cell line 624 mel, that is not recognized by TIL 2098, served as negative controls for T cell reactivity. Evaluation of peptide reactivity of TIL 2369 was carried out by pulsing 293 human embryonic kidney cells that were transduced with a recombinant retroviral expression plasmid encoding HLA-A*01 (293-A1) with candidate HLA-A*01:01 binding peptides at a concentration of 10 μM for two hours at 370C, followed by two washing steps. To determine if the epitopes identified as targets of TIL 2369 were endogenously processed and presented, 293-A1 target cells were transiently transfected with either the genes encoding the mutated candidates or the corresponding wild type genes. Evaluation of the reactivity of TIL 3309 was carried out by pulsing COS cells that had been stably transduced with retroviral constructs encoding either HLA-A*0101 or HLA-A*1101 with candidate HLA-A*01 or A*11-binding peptides at a concentration of 10 μM for two hours at 370C, followed by two washing steps. To determine if the epitopes identified as targets of TIL 3309 were endogenously processed and presented, COS-A11 target cells were transiently transfected with either the genes encoding the mutated candidates or the corresponding wild type genes, and the levels of soluble IFN-γ released from TIL that were cultured overnight with target cells were measured as previously described 19. Quantitation of responses directed against tumor cell lines and peptide pulsed target cells was evaluated in an IFN-γ ELISPOT assay using a monoclonal antibody pair obtained from Mabtech. ELISPOT. Assays were carried out by initially thawing T cells overnight in the absence of exogenous cytokines, followed by culturing with tumor cells or COS-7 cells expressing either HLA-A*0201, A*0101 or A*1101 that were pulsed with candidate peptides for two hours at 370C. Cells were also incubated in parallel overnight in complete media containing 50 ng/ml phorbol 12-myristate 13-acetate plus 1 μM ionomycin (PMA/I) to provide an estimate of the maximum number of IFN-γ-secreting cells present in these populations.
Supplementary Material
Acknowledgements
We would like to thank B. Van den Eynde, Ludwig Institute, Brussels, Belgium, for kindly providing 293 cells transfected with immunoproteasomal subunits, and to thank Ms. Susan Schwarz and Mr. Robert Fisch for assisting with experiments.
Footnotes
Author contributions P.F.R. designed and developed the experimental screening system, analyzed data and drafted the manuscript, Y.-C.L, and M.E.-G. performed experiments evaluating TIL responses against candidate mutated peptides and analyzed results, Y.L. cloned and sequenced gene products encoding candidate epitopes identified by exomic sequencing and analyzed results, J.K.T., C.G., E.T., J.C.L., and P.C. carried out bio-informatic analysis, J. G. provided advice on exomic sequencing, prepared samples for sequencing, and carried out validation studies using Sanger sequencing, Y.S. provided advice on sequencing of DNA isolated from tumor and normal cells and assisted with data analysis, and S.A.R. supervised the studies and edited the manuscript.
REFERENCES
- 1.Dudley ME, et al. Cancer Regression and Autoimmunity in Patients After Clonal Repopulation with Antitumor Lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudley ME, et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol. 2008;26:5233–5239. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marincola FM, et al. Locus-specific analysis of human leukocyte antigen class I expression in melanoma cell lines. J Immunother Emphasis Tumor Immunol. 1994;16:13–23. doi: 10.1097/00002371-199407000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salter RD, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amit S, et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Dudley ME, Rosenberg SA, Robbins PF. Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy. J Immunother. 2005;28:53–62. doi: 10.1097/00002371-200501000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 11.Morel S, et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 12.Chapiro J, et al. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J Immunol. 2006;176:1053–1061. doi: 10.4049/jimmunol.176.2.1053. [DOI] [PubMed] [Google Scholar]
- 13.Guillaume B, et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci U S A. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahle JJ, et al. Comparison of an expanded ataxia interactome with patient medical records reveals a relationship between macular degeneration and ataxia. Hum Mol Genet. 20:510–527. doi: 10.1093/hmg/ddq496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami Y, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Bruggen P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 18.Boel P, et al. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 19.Robbins PF, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox AL, et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 21.Pieper R, et al. Biochemical identification of a mutated human melanoma antigen recognized by CD4(+) T cells. J Exp Med. 1999;189:757–766. doi: 10.1084/jem.189.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Bruggen P, Stroobant V, Vigneron N, Van den Eynde B. Peptide database: T cell-defined tumor antigens. Cancer Immun. 2012 [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castle JC, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 25.Kvistborg P, et al. TIL therapy broadens the tumor-reactive CD8(+) T cell compartment in melanoma patients. Oncoimmunology. 1:409–418. doi: 10.4161/onci.18851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topalian SL, Muul LM, Solomon D, Rosenberg SA. Expansion of human tumor infiltrating lymphocytes for use in immunotherapy trials. J.Immunol.Meth. 1987;102:127–141. doi: 10.1016/s0022-1759(87)80018-2. [DOI] [PubMed] [Google Scholar]
- 30.Dudley ME, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold D, et al. Proteasome subunits encoded in the MHC are not generally required for the processing of peptides bound by MHC class I molecules. Nature. 1992;360:171–174. doi: 10.1038/360171a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.