Abstract
The impact of biodiversity on the stability of ecological communities has been debated among biologists for more than a century. Recently summarized empirical evidence suggests that biodiversity tends to enhance the temporal stability of community-level properties such as biomass; however, the underlying mechanisms driving this relationship remain poorly understood. Here, we report the results of a microcosm study in which we used simplified systems of freshwater microalgae to explore how the phylogenetic relatedness of species influences the temporal stability of community biomass by altering the nature of their competitive interactions. We show that combinations of two species that are more evolutionarily divergent tend to have lower temporal stability of biomass. In part, this is due to negative ‘selection effects’ in which bicultures composed of distantly related species are more likely to contain strong competitors that achieve low biomass. In addition, bicultures of distantly related species had on average weaker competitive interactions, which reduced compensatory dynamics and decreased the stability of community biomass. Our results demonstrate that evolutionary history plays a key role in controlling the mechanisms, which give rise to diversity–stability relationships. As such, patterns of shared ancestry may help us predict the ecosystem-level consequences of biodiversity loss.
Keywords: community ecology, temporal stability, biodiversity, ecosystem functioning, phylogenetic distance, compensatory dynamics
1. Introduction
Biologists since Darwin have argued that biodiversity is a major determinant of the stability of ecological systems in face of environmental fluctuations [1,2], but this often-cited influence of biodiversity continues to be hotly debated among biologists [3]. The persistence of this debate can be attributed to at least three non-mutually exclusive factors. First, researchers have loosely used the term ‘stability’ to refer to a wide variety of ecological concepts (resistance, resilience, temporal variation, ecological thresholds, potential for alternative states, etc.); yet theory has shown that diversity should not be expected to influence all these forms of ecological stability in the same way [4]. Second, stability has been measured at a variety of levels of biological organization (e.g. genes, populations and communities), and the stability of those different levels can be influenced by diversity differently [5–7]. Lastly, empirical studies rarely go beyond documentation of simple diversity–stability relationships to identify the underlying causal mechanisms [7–9], which has left the interpretation of results open to speculation and debate.
Despite continued controversy, one of the few diversity–stability relationships that appears to be robust is the positive influence of species richness on the temporal stability of community biomass [7,10–13]. Over 20 studies have now experimentally manipulated the number of species of bacteria, aquatic invertebrates, algae or plants growing in some experimental unit (field plots, greenhouse pots and laboratory microcosms), and examined how a change in species richness influences fluctuations in the summed biomass of species through time. The most commonly used measure of stability in these studies has been the coefficient of variation, which is the standard deviation of community biomass through time scaled to the mean biomass of a community. Several experiments have shown that species richness tends to increase stability by increasing mean community biomass, leading to lower coefficients of variation [12,14–16]. Fewer studies have documented impacts of diversity on the variance component of stability [7], and for those that have, the mechanisms have been a source of debate [6,8,17]. Theory suggests that species richness can reduce temporal variation in community biomass by (i) reducing the summed variances of species biomasses if, for example, diversity increases the chances of including more stable species or by (ii) reducing the summed covariances in species biomasses, such as occurs when species exhibit compensatory dynamics [8,17]. Compensatory dynamics can be driven by negative species interactions such as competition that cause populations to exhibit negative covariance, or by species displaying independent, but asynchronous responses to environmental fluctuations [8,9,17]. Separating these possibilities has proved difficult for two reasons. First, past experiments have seldom been designed to identify the causes of compensatory dynamics; Second, the statistical metrics historically used to quantify covariance in population dynamics have been shown to be biased and inaccurate when quantifying the synchrony of fluctuations among more than two populations [8,9]. Improved metrics for estimating variances and covariances in communities containing more than or equal to three species are being developed [8,18]; but at present, there are few interpretable measurements of how biodiversity influences population covariances.
To better understand what contributes to biomass stability, researchers have recently begun to explore how the evolutionary relationships among species might influence the ecological functions performed by a community [19–23]. This focus parallels a broader trend among ecologists to consider how patterns of shared ancestry moderate the distribution of biological traits among species—traits that influence species' abilities to coexist and to perform ecological functions [24–26]. Interest in the ecosystem-level impacts of evolution has also been driven by practical constraints. Namely, functional differentiation among species is often difficult to quantify directly, since the functions performed by organisms are controlled by a wide variety of biological traits, many of which are hard to identify and measure [23]. If genetic divergence corresponds to ecological divergence as is often assumed (i.e. phylogenetic niche conservatism; [25,27] but see [28]), then ecological differentiation may be measured more easily and holistically using molecular phylogenies that quantify evolutionary divergence among species [26].
Assuming that closely related species are more ecologically similar than less related species, evolutionary divergence among species has the potential to influence the stability of community biomass in several ways. First, a select group of studies have shown that evolutionarily divergent species tend to produce more biomass when together than do closely related species [20,21]. We might, therefore, expect these communities to have lower coefficients of variation through time (higher stability). Phylogenetic divergence might also influence community stability by altering the strength of species interactions and/or the responses to environmental fluctuations that control compensatory dynamics. One prediction is that genetically and ecologically similar species are more likely to respond similarly to environmental fluctuations in a similar way, thereby decreasing compensatory dynamics [23]. An alternative prediction is that more similar species might compete more strongly for shared resources, leading to more negative covariances that increase compensatory dynamics [29,30]. The key point here is that evolutionary divergence has potential to influence community stability in contrasting ways that have yet to be disentangled. To understand the influence of evolutionary divergence on community stability, we need data showing how phylogenetic relationships influence both species interactions as well as similarities in species environmental responses, as both of these influence compensatory dynamics.
To explore how evolutionary divergence impacts the temporal stability of community biomass, we constructed a molecular phylogeny for 37 of the most widespread and abundant species of North American green algae. We then selected nine species for use in our experiments, with taxa chosen to maximize variation in evolutionary relatedness (as measured by phylogenetic distance (PD) among species pairs). All species were grown alone in monoculture, and together in all two-species bicultures, in each of two treatments—a temporally constant environment and an environment with random fluctuations in water temperature. These treatments allowed us to assess how evolutionary relatedness (PD) influences each facet of biomass stability (mean biomass, variances and covariances among species biomasses). In addition, the designs allowed us to test whether covariances were influenced by species interactions (in bicultures) or were instead the result of independent, asynchronous responses of species to a fluctuating environment (correlated dynamics of monocultures). While our laboratory microcosms and simplified algal assemblages are not intended to mimic natural lake ecosystems and should not be over-extrapolated, these model systems allow us, for the first time, to demonstrate how each of the mechanisms that contribute to stability vary as a function of the shared ancestry of species.
2. Material and methods
(a). Species pool
This study focused on nine species of freshwater green algae (Ankistrodesmus falcatus, Chlorella sorokiniana, Coelastrum microporum, Cosmarium turpinii, Elakatothrix obtusata, Scenedesmus acuminatus, Selenastrum capricornotum, Staurastrum punctulatum and Tetraedron minimum), all of which were obtained from the culture collections at the University of Texas at Austin or the University of Gottingen (Germany). Based on a US Environmental Protection Agency's 2007 survey of more than 1200 lakes in North America (http://water.epa.gov/type/lakes/), all nine genera are widespread, being present in at least 17% of all lakes sampled. In addition, the taxa were all relatively abundant in lakes for which they were found, ranking in the top 50% of species densities (for over 400 taxa in total). Aside from being widespread and abundant, all of these species grow well under laboratory conditions using common growth media and are relatively easy to distinguish morphologically, allowing for reliable estimates of population dynamics from cell counts (described more later). Lastly, and importantly, these taxa represent a relatively even frequency distribution of PDs among species spanning a more widely sampled phylogenetic tree of North American freshwater algae, which we describe in the electronic supplementary material.
(b). Phylogenetic distance
Phylogenetic diversity is defined as the total PD among species in a community [31] and is jointly influenced by both the number of species (richness) and the evolutionary (phylogenetic) relatedness among species in a community. By keeping constant the number of species in culture (richness = 2), we obtained a measure of PD that depends only on relatedness among species and is independent from species richness. Using the generated phylogenetic tree (see the electronic supplementary material), we calculated the PD between each pair of species as the sum of tree branch lengths connecting them [19]. A number of additional metrics have recently been proposed for measuring phylogenetic relatedness, but many of these have become sufficiently convoluted that they suffer from lack of clear interpretation [32]. We calculated PD using a custom Bioperl [33] script from mean branch lengths (of all bootstrap pseudoreplicates for maximum likelihood and for all tress retained from the MCMC search for Bayesian analyses) connecting each species pair, and ignoring the root branch. In two cases (C. turpinii and A. falcatus), sequences were not available for our experimental species. We therefore used distances for the genus rather than the species by including two representative species per genus and calculating distances from the genus. Results did not depend on whether or not we assumed a relaxed molecular clock to ‘smooth’ branch lengths to make them ultrametric. If fact, smoothed and unsmoothed branch length estimates resulted in highly correlated estimates of PD (ρ = 0.973, p < 0.0001).
(c). Experimental design
We grew four replicates of all nine species in monoculture and four replicates of all 36 pairwise species combinations in 2 ml 48 well microplates (hereafter plates), with each well filled with 1.5 ml of COMBO growth medium [34]. To reduce the risk of cross-contamination among wells, we interposed at least one empty well between each culture. To reduce the effect of evapotraspiration, we did not culture algae in any edge well and filled eight of them with sterile water, increasing relative humidity in the plate. The different monocultures and bicultures were randomly distributed in different plates, with each plate holding 10 cultures for a total of 18 plates. Initial biovolumes inoculated in both mono and bicultures were 0.1 mm3 ml−1, with the latter evenly divided among species assigned to a biculture. Algae were cultured for 35 days under a 16 L : 8 D cycle at a light intensity of ca 110 µmol m−2 s−1 in temperature controlled Thermo-Scientific BOD growth chambers. Fresh COMBO growth medium was replaced in well plates daily at a 20% exchange rate (300 µl), and during exchanges, cultures were resuspended with a pipette to initiate daily mixing.
Over the first two weeks of the study, we measured daily fluorescence of chlorophyll-a, which is a widely used proxy for algal biomass, to track growth dynamics of the cultures (Fluorometer, Synergy H1 Hybrid Reader, Biotek). We allowed all cultures to reach steady-state biomass (ca after 13 days), so that we could exclude the transient phase of population dynamics that occur during early phases of logistic growth, which have potential to bias measures of temporal stability. Once all cultures approached steady state (end of exponential growth phase), we divided the replicates in half and imposed two experimental treatments. Two replicates of all nine monocultures and each of the 36 bicultures (randomly distributed over nine plates) were kept in a growth chamber with a constant 18°C temperature (constant treatment), whereas the other two replicates of monocultures and bicultures (another set of nine plates) were transferred to a chamber with daily random temperature fluctuations (fluctuating treatment). In the chamber with the fluctuating treatment, temperatures ranged between 13°C and 23°C (average 18°C) and were selected from a uniform distribution within this range. This range of variation was chosen to mimic a scenario that algae encounter in natural lakes where daily temperature fluctuations depend on weather conditions (sun, wind) and water depth. To avoid any influence on growth depending on culture positions within the growth chambers, we randomly changed the position of each plate inside the room every other day over the duration of the experiment. The average biomass did not vary among plates, indicating homogeneous growing conditions inside both growth chambers (constant temperature: F = 0.42, p = 0.85, n = 10; fluctuating temperature: F = 0.34, p = 0.92, n = 10).
(d). Sampling and data collection
On days 14, 16, 19, 21, 23, 26, 28, 30, 33 and 35 of the experiment, the 300 µl of algal suspension removed from every well for medium exchange (see above) was fixed by adding 60 µl of formalin (to keep algal morphology intact) and stored in the dark for further analysis. From these samples, we estimated the abundances of each species in culture by analysing 100 µl of fixated algal suspension in a FlowCam (Fluid Imaging Technologies, Inc. with 10× magnification lens), resulting in an analysis of approximately 7% of the entire culture volume and a number of cells per sample ranging from 103 to 108 depending on species' cell sizes. Mean cell biovolume of each algal species was estimated from measures of 200 individual cells per species grown in monoculture at the end of the experiment. Estimates of community-level biovolume (summed biovolumes used as a measure of total community biomass) were made by multiplying cell densities by mean cell biovolumes. We used data from the 10 sampling dates to estimate the stability of total biomass in the algal cultures over the 21-day sampling period. All monocultures were included in our final analyses, as all species grew and persisted to the end of the experiment in both the constant and fluctuating temperature treatments. We excluded 11 bicultures from the final analyses: nine of these were excluded because one of the species went extinct in both replicates prior the end of sampling. It is impossible to calculate summed variances or covariances from replicates that collapsed to only one species. Two other bicultures were excluded because of cross-contamination of both replicates with algae that were not assigned to the treatment (Chlorella sorokiniana). At present, the analytical methods do not exist to correctly estimate summed variances and covariances for communities with more than two species (see [8] for a mathematical explanation of the bias that results). In total, 25 different species combinations were retained for our analysis.
(e). Data analysis
The temporal stability of total biomass in each replicated biculture was measured as the inverse of the temporal coefficient of variation
| 2.1 |
with 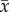 being the community biomass averaged over all time points, s.d. the standard deviation of community biomass over all time points, var the summed variance of population biomasses over time and cov the covariance in species biomasses over time. Thus, the temporal stability of community biomass can be influenced both by changes in mean biomass (numerator) or by changes in any of the components of the temporal standard deviation (denominator), which include the sum of the individual species variances and the covariance of individual species biomasses over time.
being the community biomass averaged over all time points, s.d. the standard deviation of community biomass over all time points, var the summed variance of population biomasses over time and cov the covariance in species biomasses over time. Thus, the temporal stability of community biomass can be influenced both by changes in mean biomass (numerator) or by changes in any of the components of the temporal standard deviation (denominator), which include the sum of the individual species variances and the covariance of individual species biomasses over time.
We used linear regressions to assess the effect of PD on each component of stability (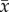 , var and cov). Two non-mutually exclusive factors can influence the covariance component of species fluctuations through time, including species interactions (e.g. competition that leads to compensatory responses) and species-specific responses to a fluctuating environment (e.g. resulting in synchrony or asynchrony of their biomasses over time). Our experimental design allowed us to further tease apart how each of these contributed to covariance (see the electronic supplementary material). By performing separate regressions on bicultures that were exposed to a constant environment, we were able to isolate how species interactions contributed to covariance, as independent responses to a fluctuating environment were not possible. By examining monocultures that were grown in a fluctuating environment, we were able to isolate species independent responses to a fluctuating environment, as there were no interspecific interactions. Each data point used in these regressions represented the mean value of two independently cultured replicates having the same species composition, with each summarizing species biomasses over 10 time points.
, var and cov). Two non-mutually exclusive factors can influence the covariance component of species fluctuations through time, including species interactions (e.g. competition that leads to compensatory responses) and species-specific responses to a fluctuating environment (e.g. resulting in synchrony or asynchrony of their biomasses over time). Our experimental design allowed us to further tease apart how each of these contributed to covariance (see the electronic supplementary material). By performing separate regressions on bicultures that were exposed to a constant environment, we were able to isolate how species interactions contributed to covariance, as independent responses to a fluctuating environment were not possible. By examining monocultures that were grown in a fluctuating environment, we were able to isolate species independent responses to a fluctuating environment, as there were no interspecific interactions. Each data point used in these regressions represented the mean value of two independently cultured replicates having the same species composition, with each summarizing species biomasses over 10 time points.
To examine how species interactions influenced the performance of each species, we estimated two different relative yields (RYs). An RY is the ratio of the biomass of a species in polyculture to its biomass in monoculture and is the most widely used metric of competition [35]. First, we estimated RY(x,i) which is the mean effect of competitors on a particular focal species (equivalent to ‘competitive response’ or ‘tolerance’ (sensu [36]); e.g. species A in monoculture compared with species A in presence of species B, to species A in presence of C, to species A in presence of D, etc.). When RY(x,i) < 1, this indicates that the biomass of the species decreases in the presence of another species (e.g. interference, exploitative competition). Second, we estimated for each species RY(i,x), which is the mean effect of a focal species on other competitors (equivalent to ‘competitive effect’ (sensu [36]); e.g. species B in monoculture compared with B in presence of species A, species C in monoculture compared with C in presence of species A, species D in monoculture compared with D in presence of species A, etc.). When RY(i,x) < 1, this indicates that the focal species decreases the biomass of its competitors. Thus, RY(i,x) (competition effect) and RY(x,i) (competitive response) are two complementary measures of the average competitive abilities of each species. To establish how the presence of each species in a culture influenced community biomass, we also calculated the mean selection effect of diversity. The selection effect is estimated as the covariance between the biomass of species in monoculture and their biomass in biculture. A negative value of the selection effect suggests that the most productive species in monoculture became the less productive in biculture and vice versa (for a more detailed description of this method, see the electronic supplementary material).
To quantify the strength of interactions among species and determine how these varied with relatedness, we first estimated for each biculture the mean RY (RYcom) as the ratio of the biomass a species achieves when grown in polyculture to its biomass in monoculture. RYcom is an average of two different RY(x,i) values previously described (e.g. for the community composed species A and B RYcom is the average of RY(A,B) and RY(B,A)). Then, for each biculture, we took the opposite of the mean RYcom as a measure of the strength of interspecific competition. When 1 – RYcom is close to 1 (RYcom close to 0) this means that species achieved much lower biomasses in polycultures than in monoculture. As 1 – RYcom diminished (RYcom increases), the biomasses achieved in polyculture were closer to the biomasses in monoculture.
To further quantify the similarity in species' responses to environmental variation, we performed a supplemental experiment in which we measured species' responses to temperature fluctuations along a gradient from 13°C to 23°C. For this, we first allowed the nine monocultures to reach steady state at 18°C. Then, for the next 6 days, cultures were subjected to daily media exchanges (20% replacement) and temperature fluctuations. Each monoculture was replicated twice and encountered the same random fluctuations at six different temperature levels (15°C, 23°C, 19°C, 13°C, 17°C and 21°C). This set of temperatures was also encountered by algae during the main experiment. Every day, after growing in a particular temperature, we measured their immediate response to temperature fluctuations between two media exchanges as ln(Nt+1/Nt), with Nt+1/Nt being the ratio of abundances over a period of 24 h. The compiled environmental responses were used to calculate a temperature response ‘similarity’ among species, which was measured as the correlation coefficient between any two-species growth rates along the temperature gradient.
3. Results
The temporal stability of total biomass in the algal bicultures decreased as the PD among species increased (linear fit: stability = 8.63 – 12.28 × PD, r2 = 0.21, p = 0.02 or exponential decay fit: stability = 5.52 + 27.01 × exp(−29.538 × PD), r2 = 0.45, p < 0.01, n = 25; figure 1). This trend was partially driven by the fact that bicultures characterized by higher values of PD (less related species) also tended to have a reduced mean biomass (r2 = 0.22, p = 0.02, n = 25; figure 2a). The two additional components of temporal stability (summed variances and covariance) were not influenced by the PD among species (r2 < 0.01, p = 0.75, n = 25 for both, figure 2b,c, respectively).
Figure 1.
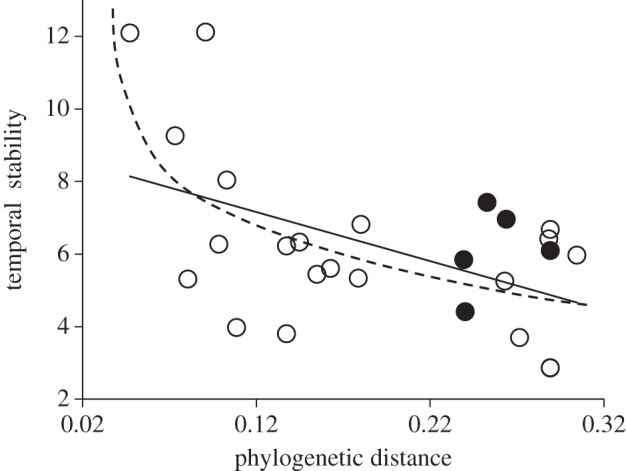
Effect of PD on the stability of total biomass of algal bicultures. Temporal stability was measured as the inverse of the temporal coefficient of variation. Each dot represents one of the 25 bicultures. Black dots are bicultures that included the low productivity-superior competitor C. turpinii. The solid line represents the general linear relationship, whereas the broken line represents an exponential decay fit. Both fits were significant regardless of whether C. turpinii is included or excluded from the analyses (see main text).
Figure 2.
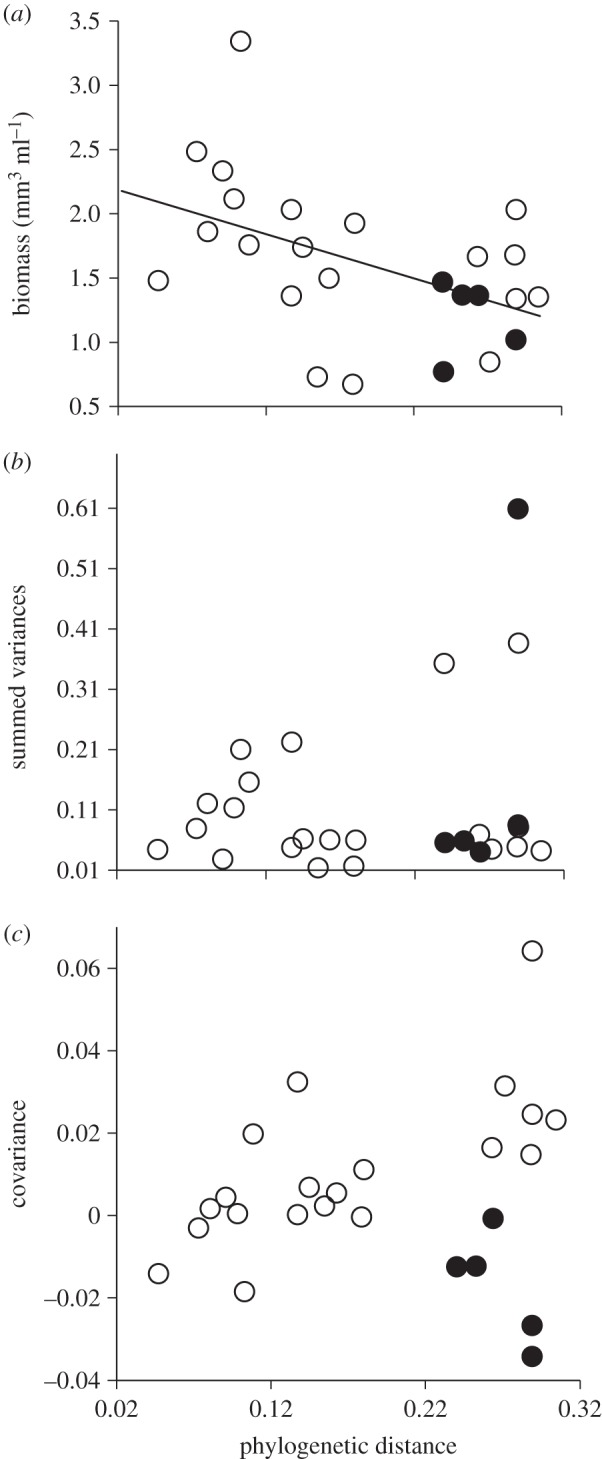
Effects of PD on the three components of stability. (a) Effect of PD on mean temporal biomass of algal bicultures. The solid line represents the general linear relationship, which is only significant when C. turpinii is included (see main text). (b) Effect of PD on the summed variances. (c) Effect of PD on the temporal covariance. In (a–c), each dot represents a single biculture, with black dots showing those bicultures that include the low productivity-superior competitor C. turpinii.
To better understand why bicultures composed of less related species achieved lower total biomass (figure 2a), we examined the yields and RYs (relative to monocultures) of all the species. We found that species differed considerably in their biomass achieved at steady state (figure 3a). For instance, Chlorella sorokiniana, C. turpinii and Elakatrothrix obtusta achieved the lowest biomasses both in mono- and biculture. We also found evidence for large differences in the competitive abilities of the different species (figure 3b). Notably, the biomasses of two species: C. turpinii and Coelastrum microporum were unaffected by the presence of other species (RYx.i, or competitive response not different from 1); yet, they considerably reduced the biomass achieved by their competitors (RYi.x or competitive effect less than 1). However, only communities that contained C. turpinii showed a selection effect that was significantly less than 0 (see Material and methods; figure 3c). Taken together, these results suggest that the presence of superior competitor with low biomass production (C. turpinii) led to a negative selection effect that decreased productivity in bicultures with high PD (figure 2a).
Figure 3.
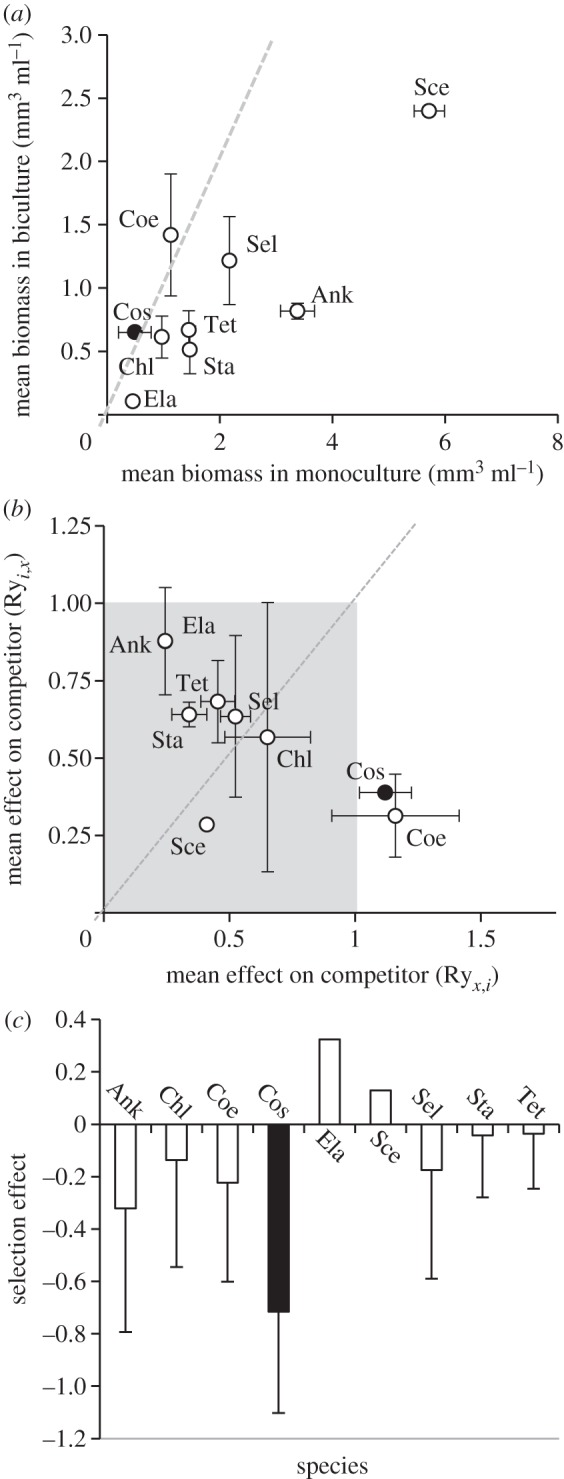
Species biomasses, competitive capacities and selection effects. (a) The nine species differed in their mean temporal biomasses achieved in monoculture (x-axis) and in their mean biomasses in biculture in the fluctuating treatment (y-axis); black dot shows C. turpinii; (b) Species also differed in their competitive abilities as indicated by differences in RYs. The grey zone represents RY values less than 1. (c) Mean selection effect of bicultures including each of the nine species used in this experiment. (Ank, Ankistrodesmus; Chl, Chlorella; Coe, Coelastrum; Cos, Cosmarium; Ela, Elakatrothrix; Sce, Scenedesmus; Sel, Selenastrum; Sta, Staurastrum and Tet, Tetraedron). Error bars represent standard errors among bicultures containing the species. Ela and Sce have no error bars because only present in one biculture. Dashed lines in (a,b) represent the 1 : 1 isocline.
When we excluded the cultures containing C. turpinii from analyses of temporal stability, we were surprised to find that the negative relationship between PD and temporal stability remained significant (r2 = 0.26, p = 0.02, n = 20). This suggests that, while the negative selection effect was contributing to the inverse relationship between PD and stability, it was not the only causal factor. For the subset of data that excluded bicultures containing C. turpinii, we found that the negative relationship between PD and stability was instead driven by an increase in the covariance component of stability (r2 = 0.48, p < 0.001, n = 20; figure 4a), whereas the mean biovolume was no longer affected by PD (r2 = 0.104, p = 0.222, n = 20).
Figure 4.
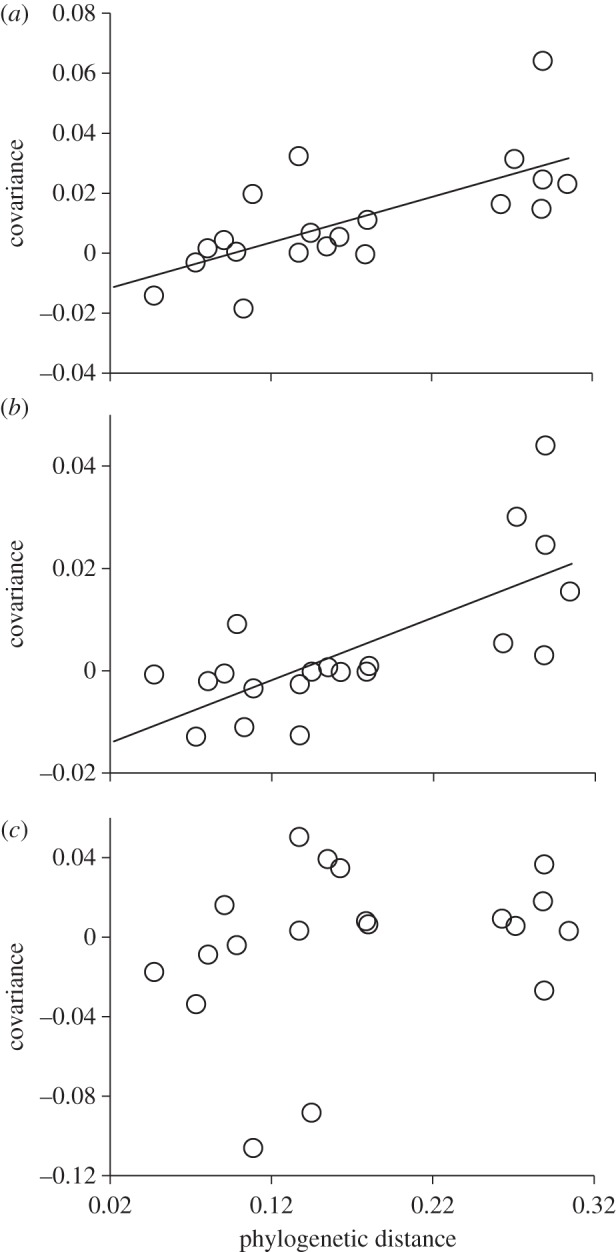
Effect of PD on the covariance component of stability. Three different culture scenarios (see text and electronic supplementary material for more details). In (a), the covariance is measured in bicultures in a fluctuating environment, representing the joint effect of species interactions and independent responses to a fluctuating environment. Note that (a) is similar to figure 2c, but does not include bicultures containing C. turpinii. In (b), the covariance is measured in bicultures grown in a constant environment, thus representing only the effects of species interactions. In (c), the covariance is measured by comparing the dynamics of independent monocultures of each species exposed to the same temperature fluctuations. Hence, the covariance results from their independent responses to the environment and is not influenced by species interactions. Each dot represents one of the 20 bicultures (the five bicultures, including C. turpinii were not included here). Linear regressions are only present when significant.
To determine which factors were contributing to covariance, we analysed the two experimental treatments allowing us to separate the role of competitive interactions from the roles played by species independent responses to a fluctuating environment (see the electronic supplementary material). Analyses of bicultures that were grown at a uniform temperature, which reveals the covariance component that was driven solely by species interactions, showed that the covariance was positively related to PD (r2 = 0.52, p < 0.001, n = 20; figure 4b). Analyses of species grown as monocultures that experienced temperature fluctuations, which reveals the covariance component that was driven by species' independent responses to environmental fluctuations (in the absence of interactions), showed that the covariance was not related to the PD among species (r2 = 0.009, p = 0.69, n = 20; figure 4c). Thus, for the subset of data that excluded the superior competitor C. turpinii, competition among species appeared to be the primary contributor to stability, and the influence of competition on stability tended to decrease as species became more evolutionary dissimilar.
Consistent with this interpretation, we found that the strength of competition—measured as 1 – RYcom of the two interacting species—decreased as PD increased (r2 = 0.27, p = 0.02, n = 20; figure 5a). Thus, closely related species competed more strongly and reduced each other's yields more than distantly related species. By contrast, the similarity in species growth responses to the temperature gradient, measured as the correlation coefficient relating species growth rates along the temperature gradient, was not related to PD (r2 = 0.0077, p = 0.712, n = 20; figure 5b). Thus, closely related species showed stronger compensatory dynamics than less related species, resulting solely from strong competitive interactions among similar species rather than any type of temporal asynchrony in their response to environmental fluctuations.
Figure 5.
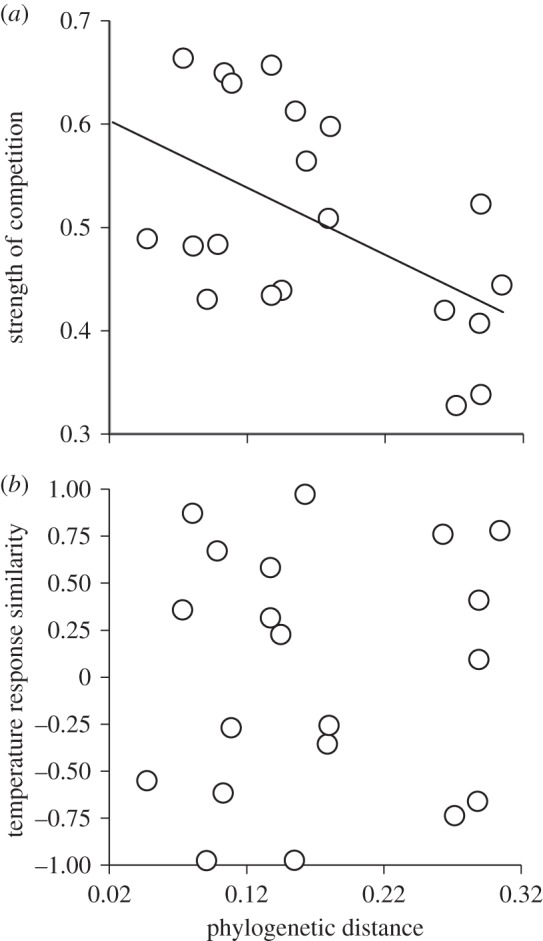
Effect of PD on competition and environmental responses' similarity. (a) The strength of interspecific competition measured as the opposite of the mean RY of the two interacting species (1 – RYcom). (b) Similarity in specific responses to a temperature gradient from 13°C to 23°C. Each dot represents one of the 20 bicultures. Bicultures including C. turpinii were not included in this analysis. The solid line in (a) represents a significant linear relationship.
4. Discussion
Identifying the mechanism by which biodiversity influences the stability of community and ecosystem processes is crucial for understanding the ecological consequences of diversity loss, and for developing appropriate conservation strategies. Here, we have shown that the biomass of algal cultures composed of closely related species was more stable in the face of daily temperature fluctuations than the biomass of cultures composed of distantly related species. Our experiment helped quantify two independent mechanisms that contribute to the relationship between biodiversity and the temporal stability of biomass production. First, evolutionarily diverse assemblages had a greater chance that a competitively superior species would dominate biomass of bicultures. In our case, the competitively superior taxon had low biomass production, which reduced the mean biomass of bicultures, lowering the coefficient of variation and reducing stability. This so-called ‘negative selection effect’ occurs in roughly 40% of studies that have attempted to link the diversity of primary producers' to production of biomass [37]. Because the presence of positive selection effects—where the competitively superior species increases rather than decreases community biomass—tends to be more common [37,38], one might expect that phylogenetic divergence would frequently lead to increased overyielding, as has been shown in other studies [19,20]. Regardless of the direction of the effect, one key point is that the evolutionary relatedness of species correlates with the probability of including a competitively superior taxon in a community and this will either increase or decrease stability depending on the productivity of the dominant species.
The second mechanism by which evolutionary relatedness impacted the temporal stability of community biomass was by influencing the strength of compensatory dynamics that produce negative covariance in species populations. There has been much speculation about the underlying cause of compensatory dynamics in ecological communities [6,9,10,39–42]. It is generally thought that two distinct mechanisms produce compensatory dynamics: competition among species that lead to negative covariance in population dynamics, and asynchrony in the independent responses of species to environmental fluctuations. To our knowledge, our study is the first to disentangle the relative contributions of these two mechanisms to stability. We found no evidence for an effect of evolutionary relatedness on the asynchrony in species responses to temperature fluctuations. However, analyses of bicultures grown at a uniform temperature revealed compensatory dynamics driven solely by species interactions. Strong interactions led to greater compensatory dynamics and negative covariance when species had high levels of evolutionary relatedness. But the strength of competition decreased as PDs among species increased, and this tended to reduce the negative covariances among species populations that are needed to maintain stability. A similar negative influence of evolutionary divergence on the strength of species interactions has been observed in protists [28] and terrestrial plant systems [29]. If, as many have assumed, evolutionary relatedness is negatively associated with the degree of ecological divergence among species [43–45], then PD could generally be associated with a decline in the strength of competitive interactions and a reduction in the compensatory dynamics that maintain community stability. See the electronic supplementary material for a brief discussion of possible traits influencing competitive interactions in our study.
It is important to note that our experiment was never intended to represent the complexity of real lakes, which have much higher environmental variation than considered here, and where levels of diversity are typically far greater (although mono- and bicultures do exist). The value of our study, and the benefit of using simplified experimental systems, is that we were able to explicitly assess the influence of phylogenetic relatedness on each facet contributing to biomass of stability—something that would be difficult, if not impossible, in more natural environments or with higher levels of diversity. If the mechanisms documented in our laboratory study ultimately prove to be common in more natural systems, then our work would have several important implications for conservation and management. The stabilizing effect of diversity on community-level processes like biomass production is often used to argue that conservation will provide ‘insurance’ that minimizes change in ecosystems exposed to environmental fluctuations or stress [46–49]. To maximize this insurance, it is often argued that we should maximize the evolutionary diversity of species to ensure adaptability of communities and future ‘options’ [33], and/or maximize species diversity to ensure communities are composed of taxa that will exhibit differing, but complementary, responses to environmental fluctuations [50–52]. Our results run counter to these suggestions. We found no evidence that species exhibit complementary responses to environmental fluctuations, and we showed that the compensatory dynamics which maintain community stability are strongest in systems that have the lowest (not highest) evolutionary diversity. Therefore, the assumption that species and evolutionary diversity provide insurance against changes in community-level processes may need to be re-evaluated. As the field of diversity–stability continues to amass data, it is worth considering whether management goals that focus on community stability might be better achieved by conservation of ecologically redundant species or overlapping levels of evolutionary relatedness.
Acknowledgements
We thank Keith Fritschie and Charlotte Wilson for technical assistance during the experiment.
Funding statement
This work was supported by the U.S. National Science Foundation's DIMENSIONS of Biodiversity program in a grant to B.J.C. and T.H.O. (DEB-1046121).
References
- 1.Elton CS. 1958. The ecology of invasions by animals and plants. London, UK: Methuen [Google Scholar]
- 2.May RM. 1973. Stability and complexity in model ecosystems. Princeton, NJ: Princeton University Press [Google Scholar]
- 3.McCann KS. 2000. The diversity–stability debate. Nature 405, 228–233 (doi:10.1038/35012234) [DOI] [PubMed] [Google Scholar]
- 4.Ives AR, Carpenter SR. 2007. Stability and diversity of ecosystems. Science 317, 58–62 (doi:10.1126/science.1133258) [DOI] [PubMed] [Google Scholar]
- 5.Tilman D. 1996. Biodiversity: population versus ecosystem stability. Ecology 77, 350–363 (doi:10.2307/2265614) [Google Scholar]
- 6.Doak DF, Bigger D, Harding EK, Marvier MA, O'Malley RE, Thomson D. 1998. The statistical inevitability of stability–diversity relationships in community ecology. Am. Nat. 151, 264–276 (doi:10.1086/286117) [DOI] [PubMed] [Google Scholar]
- 7.Jiang L, Pu Z. 2009. Effects of species diversity on temporal stability in single-trophic and multitrophic communities. Am. Nat. 174, 651–659 (doi:10.1086/605961) [DOI] [PubMed] [Google Scholar]
- 8.Loreau M, de Mazancourt C. 2008. Species synchrony and its drivers: neutral and nonneutral community dynamics in fluctuating environments. Am. Nat. 172, E48–E66 (doi:10.1086/589746) [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez A, Loreau M. 2009. The causes and consequences of compensatory dynamics in ecological communities. Annu. Rev. Ecol. Evol. Syst. 40, 393–414 (doi:10.1146/annurev.ecolsys.39.110707.173349) [Google Scholar]
- 10.Cottingham KJ, Brown BL, Lennon JT. 2001. Biodiversity may regulate the temporal variability of ecological systems. Ecol. Lett. 4, 72–85 (doi:10.1046/j.1461-0248.2001.00189.x) [Google Scholar]
- 11.Griffin JN, O'Gorman EJ, Emmerson MC, Jenkins SR, Klein AM, Loreau M, Symstad AJ. 2009. Biodiversity and the stability of ecosystem functioning. In Biodiversity, ecosystem functioning, and human wellbeing: an ecological and economic perspective (eds Naeem S, Bunker D, Hector A, Loreau M, Perrings C.), pp. 78–93 Oxford, UK: Oxford University Press [Google Scholar]
- 12.Hector A, et al. 2010. General stabilizing effects of plant diversity on grassland productivity through population asynchrony and overyielding. Ecology 91, 2213–2220 (doi:10.1890/09-1162.1) [DOI] [PubMed] [Google Scholar]
- 13.Campbell V, Murphy G, Romanuk TN. 2011. Experimental design and the outcome and interpretation of diversity-stability relations. Oikos 120, 399–408 (doi:10.1111/j.1600-0706.2010.18768.x) [Google Scholar]
- 14.Steiner CF, Long ZT, Krumins JA, Morin PJ. 2005. Temporal stability of aquatic food webs: partitioning the effects of species diversity, species composition and enrichment. Ecol. Lett. 8, 819–828 (doi:10.1111/j.1461-0248.2005.00785.x) [Google Scholar]
- 15.Vogt RJ, Romanuk TN, Kolasa J. 2006. Species richness–variability relationships in multi-trophic aquatic microcosms. Oikos 113, 55–66 (doi:10.1111/j.0030-1299.2006.14494.x) [Google Scholar]
- 16.Jiang L, Joshi H, Patel SN. 2009. Predation alters relationships between biodiversity and temporal stability. Am. Nat. 173, 389–399 (doi:10.1086/596540) [DOI] [PubMed] [Google Scholar]
- 17.Tilman D, Lehman CL, Bristow CE. 1998. Diversity–stability relationships: statistical inevitability or ecological consequence? Am. Nat. 151, 277–282 (doi:10.1086/286118) [DOI] [PubMed] [Google Scholar]
- 18.Gross K, Cardinale BJ, Fox J, Gonzalez A, Loreau M, Polley HW, van Ruijven J, Reich P. In press. Species richness and the temporal stability of biomass production: an analysis of recent biodiversity experiments. Am. Nat. [DOI] [PubMed] [Google Scholar]
- 19.Maherali H, Klironomos JN. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748 (doi:10.1126/science.1143082) [DOI] [PubMed] [Google Scholar]
- 20.Cadotte MW, Cardinale BJ, Oakley TH. 2008. Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017 (doi:10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn DFB, Mirotchnick N, Jain M, Palmer MI, Naeem S. 2011. Functional and phylogenetic diversity as predictors of biodiversity–ecosystem–function relationships. Ecology 92, 1573–1581 (doi:10.1890/10-1245.1) [DOI] [PubMed] [Google Scholar]
- 22.Jousset A, Schmid B, Scheu S, Eisenhauer N. 2011. Genotypic richness and dissimilarity opposingly affect ecosystem functioning. Ecol. Lett. 14, 537–545 (doi:10.1111/j.1461-0248.2011.01613.x) [DOI] [PubMed] [Google Scholar]
- 23.Cadotte MW, Dinnage R, Tilman D. 2012. Phylogenetic diversity promotes ecosystem stability. Ecology 93, S223–S233 (doi:10.1890/11-0426.1) [Google Scholar]
- 24.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505 (doi:10.1146/annurev.ecolsys.33.010802.150448) [Google Scholar]
- 25.Wiens JJ, et al. 2010. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324 (doi:10.1111/j.1461-0248.2010.01515.x) [DOI] [PubMed] [Google Scholar]
- 26.Srivastava DS, Cadotte MW, MacDonald AM, Marushia RG, Mirotchnick N. 2012. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 15, 637–648 (doi:10.1111/j.1461-0248.2012.01795.x) [DOI] [PubMed] [Google Scholar]
- 27.Johnson MTJ, Stinchcombe JR. 2007. An emerging synthesis between community ecology and evolutionary biology. Trends Ecol. Evol. 22, 250–257 (doi:10.1016/j.tree.2007.01.014) [DOI] [PubMed] [Google Scholar]
- 28.Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1007 (doi:10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 29.Violle C, Nemergut DR, Pu Z, Jiang L. 2011. Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett. 14, 782–787 (doi:10.1111/j.1461-0248.2011.01644.x) [DOI] [PubMed] [Google Scholar]
- 30.Verdu M, Gomez-Aparicio L, Valiente-banuet A. 2012. Phylogentic relatedness as a tool in restoration ecology: a meta-analysis. Proc. R. Soc. B 279, 1761–1767 (doi:10.1098/rspb.2011.2268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Cons. 61, 1–10 (doi:10.1016/0006-3207(92)91201-3) [Google Scholar]
- 32.Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH. 2009. Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE 4, e5695 (doi:10.1371/journal.pone.0005695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stajich JE, et al. 2002. The bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618 (doi:10.1101/gr.361602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L. 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377, 147–159 (doi:10.1023/A:1003231628456) [Google Scholar]
- 35.Austin MP, Fresco LFM, Nicholls AO, Groves RH, Kaye PE. 1988. Competition and relative yield: estimation and interpretation at different densities and under various nutrient concentrations using Silybum marianum and Cirsium vulgare. J. Ecol. 76, 157–171 (doi:10.2307/2260460) [Google Scholar]
- 36.Goldberg DE, Fleetwood L. 1987. Competitive effect and response in four annual plants. J. Ecol. 75, 1131–1143 (doi:10.2307/2260318) [Google Scholar]
- 37.Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L, Balvanera P, O'Connor MI, Gonzalez A. 2011. The functional role of producer diversity in ecosystems. Am. J. Bot. 98, 1–21 (doi:10.3732/ajb.1000364) [DOI] [PubMed] [Google Scholar]
- 38.Jiang L, Pu Z, Nemergut DR. 2008. On the importance of the negative selection effect for the relationship between biodiversity and ecosystem functioning. Oikos 117, 488–493 (doi:10.1111/j.0030-1299.2008.16401.x) [Google Scholar]
- 39.Tilman D. 1999. The ecological consequences of changes in biodiversity: a search for general principles. Ecology 80, 1455–1474 (doi:10.2307/176540) [Google Scholar]
- 40.Houlahan JE, et al. 2007. Compensatory dynamics are rare in natural ecological communities. Proc. Natl Acad. Sci. USA 104, 3273–3277 (doi:10.1073/pnas.0603798104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranta E, Kaitala V, Fowler MS, Laakso J, Ruokolainen L, O'Hara R. 2008. Detecting compensatory dynamics in competitive communities under environmental forcing. Oikos 117, 1907–1911 (doi:10.1111/j.1600-0706.2008.16614.x) [Google Scholar]
- 42.Valone TJ, Barber NA. 2008. An empirical evaluation of the insurance hypothesis in diversity–stability models. Ecology 89, 522–531 (doi:10.1890/07-0153.1) [DOI] [PubMed] [Google Scholar]
- 43.Blomberg SP, Garland T., Jr 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods . J. Evol. Biol. 15, 899–910 (doi:10.1046/j.1420-9101.2002.00472.x) [Google Scholar]
- 44.Cahill JF, Jr, Kembel SW, Lamb EG, Keddy PA. 2008. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspect. Plant Ecol. Evol. Syst. 10, 41–50 (doi:10.1016/j.ppees.2007.10.001) [Google Scholar]
- 45.Mayfield MM, Levine JM. 2010. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13, 1085–1093 (doi:10.1111/j.1461-0248.2010.01509.x) [DOI] [PubMed] [Google Scholar]
- 46.Faith DP. 2002. Quantifying biodiversity: a phylogenetic perspective. Conserv. Biol. 16, 248–252 (doi:10.1046/j.1523-1739.2002.00503.x) [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues ASL, Gaston KJ. 2002. Maximizing phylogenetic diversity in the selection of networks of conservation areas. Biol. Conserv. 105, 103–111 (doi:10.1016/S0006-3207(01)00208-7) [Google Scholar]
- 48.Mouquet N, et al. 2012. Ecophylogenetics : advances and perspectives. Biol. Rev. 87, 769–785 (doi:10.1111/j.1469-185X.2012.00224.x) [DOI] [PubMed] [Google Scholar]
- 49.Srivastava DS, Vellend M. 2005. Biodiversity-ecosystem function research: is it relevant to conservation?. Annu. Rev. Ecol. Evol. Syst. 36, 267–294 (doi:10.1146/annurev.ecolsys.36.102003.152636) [Google Scholar]
- 50.Tilman D, Downing JA. 1994. Biodiversity and stability in grasslands. Nature 367, 363–365 (doi:10.1038/367363a0) [Google Scholar]
- 51.Ives AR, Klug JL, Gross K. 2000. Stability and species richness in complex communities. Ecol. Lett. 3, 399–411 (doi:10.1046/j.1461-0248.2000.00144.x) [Google Scholar]
- 52.Yachi S, Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl Acad. Sci. USA 96, 1463–1468 (doi:10.1073/pnas.96.4.1463) [DOI] [PMC free article] [PubMed] [Google Scholar]


