Abstract
The incoherent type-1 feed-forward loop (I1-FFL) is ubiquitous in biological regulatory circuits. Although much is known about the functions of the I1-FFL motif, the energy cost incurred in the network and how it affects the performance of the network have not been investigated. Here, we study a generic I1-FFL enzymatic reaction network modelled after the GEF–GAP–Ras pathway responsible for chemosensory adaptation in eukaryotic cells. Our analysis shows that the I1-FFL network always operates out of equilibrium. Continuous energy dissipation is necessary to drive an internal phosphorylation–dephosphorylation cycle that is crucial in achieving strong short-time response and accurate long-time adaptation. In particular, we show quantitatively that the energy dissipated in the I1-FFL network is used (i) to increase the system's initial response to the input signals; (ii) to enhance the adaptation accuracy at steady state; and (iii) to expand the range of such accurate adaptation. Moreover, we find that the energy dissipation rate, the catalytic speed and the maximum adaptation accuracy in the I1-FFL network satisfy the same energy–speed–accuracy relationship as in the negative-feedback-loop (NFL) networks. Because the I1-FFL and NFL are the only two basic network motifs that enable accurate adaptation, our results suggest that a universal cost–performance trade-off principle may underlie all cellular adaptation processes independent of the detailed biochemical circuit architecture.
Keywords: incoherent type-1 feed-forward loop, non-equilibrium system, energy dissipation, sensitivity, adaptation accuracy
1. Introduction
Cells regulate their responses to environmental changes by using different intracellular biochemical circuits (networks). Despite their complexity, these regulatory networks contain many recurring interaction patterns called network motifs, each carrying out specific biological functions [1,2]. One of the most common regulatory motifs is the incoherent type-1 feed-forward loop (I1-FFL), where the input triggers the output response and also activates a negative controller, which eventually suppresses the output. Many occurrences of the I1-FFL-type circuits have been identified in bacteria, yeast, animal and human stem cells [3–8]. Experimental and theoretical studies have further discovered that the I1-FFL motif carries out many beneficial functions in various biological contexts, such as generating pulse responses upon external stimuli [9,10], accelerating gene expression [10,11], measuring fold changes of environmental signals [12–14] and adapting accurately to a broad range of chemical concentrations [15,16].
As illustrated in figure 1a, an I1-FFL network motif is composed of an input A which activates the downstream output C and at the same time turns on a negative controller R to deactivate C [10,17]. These two opposing pathways from input to output operate at different time scales, with the activation faster than the inhibition. This time-scale difference results in a fast response to an external stimulus followed by a slower recovery. An important feature of the I1-FFL network is that both the activation and inhibition effects originate from the same input. Therefore, in response to a persistent input, the inhibition and the activation effects become equal at long time scale and they cancel each other, resulting in a steady-state output that is independent of the input [16]. In other words, the I1-FFL network can achieve accurate adaptation, an important function for all sensory systems in biology.
Figure 1.
Schematic of the incoherent type-1 feed-forward loop (I1-FFL) network motif. (a) The I1-FFL motif comprises three nodes: the upstream signal and activator A, the intermediate inhibitor R and the downstream output C. (b) A realistic enzymatic reaction network that exemplifies the I1-FFL motif. The upstream signal A activates an activator B1 and an inhibitor B2 that catalyse two opposing reactions C ↔ C* through different intermediate complexes M1 and M2. The output C* is the activated form of C. External energy drives γ < 1, which results in the persistent reaction circulation illustrated by the thick arrows. (c) The dynamics of the I1-FFL motif in response to a step stimulus at time t = 0. The output C* first rises from its pre-stimulus level [C*]0 to its peak level [C*]max at time Tmax before it decreases to a steady-state level [C*]steady. The response dynamics shown here is calculated with γ = 0.
It has been well established that accurate adaptation is essential for gradient sensing in eukaryotic chemotaxis [18,19]. A recent study of Dictyostelium discoideum chemotaxis by Takeda et al. [20] provided strong evidence that the membrane-localized Ras activity central to gradient sensing in D. discoideum is controlled by an I1-FFL-type reaction network. In particular, two opposing enzymes, guanine nucleotide exchange factor (GEF) and GTPase-activating protein (GAP), activate and deactivate Ras, respectively. GEF exchanges guanosine diphosphate (GDP) with guanosine triphosphate (GTP), whereas GAP stimulates the hydrolysis of GTP to GDP [21,22]. Because these two enzymes are activated (with different rates) after receiving the same signal from the G-protein-coupled receptors (GPCRs), the activated Ras level adapts accurately to its pre-stimulus level after an initial transient response.
Although much progress has been made in understanding the functional benefits of various biochemical networks, little is known about the costs of achieving biological functions, such as gene regulation and sensory adaptation. It is not surprising that energy dissipation is required to drive biological processes, but a detailed understanding of how such energy consumption dissipation affects the functional performance of the biochemical network is still lacking. For example, in the D. discoideum chemotaxis pathway, GTP is continuously hydrolysed such as in all other GPCR signalling pathways, but we do not know whether the energy release from GTP hydrolysis is used in carrying out certain biological functions, such as response and adaptation, or whether it is merely a by-product of a futile biochemical cycle.
In this paper, we address this problem by studying the energetics of an I1-FFL-type biochemical network model motivated by the GEF–GAP–Ras pathway crucial for Dictyostelium chemotaxis. First, our analysis shows explicitly that continuous energy dissipation occurs in the I1-FFL-type reaction network for it to respond and adapt to the external stimuli. Following this insight, the main part of the paper is focused on understanding the way energy is used in enabling the sensory functions of the I1-FFL network. In particular, our analysis reveals quantitatively how continuous energy dissipation enhances both the short-time response sensitivity and the steady-state (long-time) adaptation accuracy. Furthermore, we find that the energy–performance trade-off for I1-FFL follows the same energy–speed–accuracy (ESA) relation as in negative-feedback-loop (NFL) networks studied in [23]. According to a recent study [16], the I1-FFL and NFL networks are the only two three-node network motifs that enable accurate adaptation. Therefore, our results here for the I1-FFL network, together with our earlier results for the NFL network [23], suggest that the trade-off relationship between adaptation accuracy and its minimum energy cost may be universal and independent of the detailed underlying network architecture.
2. Model
2.1. The catalytic network and the kinetic model
To analyse the properties of the abstract I1-FFL motif within a realistic biological realm, we have constructed a detailed biochemical reaction network motivated by the Ras pathway in Dictyostelium chemotaxis. In this biochemical reaction network shown in figure 1b, the upstream signal A triggers the synthesis (or activation) of two opposing enzymes B1 (activator) and B2 (inhibitor) at different rates  and
and 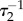 , respectively. The substrate has two conformations: the inactive conformation C and the active conformation C*. The two enzymes act in opposite directions: the activator B1 catalyses the reaction from C to C*; the inhibitor B2 catalyses the ‘reverse’ reaction from C* to C. The output of the network is defined as the level of unbound C*. C* responds to the input signal and is also subject to regulation by the network. In the case of Dictyostelium chemotaxis, A is the GPCR, B1 and B2 are GEF and GAP, respectively, and C is Ras–GDP and C* is Ras–GTP.
, respectively. The substrate has two conformations: the inactive conformation C and the active conformation C*. The two enzymes act in opposite directions: the activator B1 catalyses the reaction from C to C*; the inhibitor B2 catalyses the ‘reverse’ reaction from C* to C. The output of the network is defined as the level of unbound C*. C* responds to the input signal and is also subject to regulation by the network. In the case of Dictyostelium chemotaxis, A is the GPCR, B1 and B2 are GEF and GAP, respectively, and C is Ras–GDP and C* is Ras–GTP.
In the two catalytic reactions, the enzymes first form intermediate complexes with initial reactants (M1 for the activation complex and M2 for the deactivation complex), which then turn over into final products [24]. The kinetics within the network can be mathematically described by the rate equations
 |
2.1 |
 |
2.2 |
| 2.3 |
| 2.4 |
| 2.5 |
| 2.6 |
where f1(2) and g1(2) are forward reaction rates for enzymes B1 and B2, respectively;  are the corresponding reverse reaction rates, as denoted in figure 1b. Here, we have normalized the enzyme concentrations so that their synthesis rates (from A) equal their degradation rates, which are
are the corresponding reverse reaction rates, as denoted in figure 1b. Here, we have normalized the enzyme concentrations so that their synthesis rates (from A) equal their degradation rates, which are  and
and  for B1 and B2, respectively.
for B1 and B2, respectively.
2.2. Model parameters and characteristics
To distinguish the equilibrium systems from the non-equilibrium ones, we define a parameter  . In equilibrium systems, detailed balance holds, so γ = 1. If the enzymatic reactions operate out of equilibrium, then γ is different from unity [25]. Note that the Michaelis–Menten catalytic mechanism assumes
. In equilibrium systems, detailed balance holds, so γ = 1. If the enzymatic reactions operate out of equilibrium, then γ is different from unity [25]. Note that the Michaelis–Menten catalytic mechanism assumes  (i.e. γ = 0), which corresponds to the extreme non-equilibrium case of forbidding the reverse reaction from the products to the intermediate complexes. Here, we use the parameter 0 ≤ γ ≤ 1 to characterize the non-equilibriumness of the network. The activation of B1 by A is assumed to be much faster than all the other processes,
(i.e. γ = 0), which corresponds to the extreme non-equilibrium case of forbidding the reverse reaction from the products to the intermediate complexes. Here, we use the parameter 0 ≤ γ ≤ 1 to characterize the non-equilibriumness of the network. The activation of B1 by A is assumed to be much faster than all the other processes,  ,
,  and
and  , so that we can take the approximation
, so that we can take the approximation  and set [B1] = [A]. We denote the other time scale τ2 = 1 to set the time scale in our model. For convenience of analytical analysis, we also consider the ‘symmetric’ case where the catalytic rates are identical for the two enzymes (f1(2) = g1(2),
and set [B1] = [A]. We denote the other time scale τ2 = 1 to set the time scale in our model. For convenience of analytical analysis, we also consider the ‘symmetric’ case where the catalytic rates are identical for the two enzymes (f1(2) = g1(2),  ) so that
) so that  . The total substrate concentration [C]0 = [C] + [C]* + [M1] + [M2] is set to be unity to normalize the scale for chemical concentrations in our model. After these rescalings, our model is described by five dimensionless independent parameters: f1,
. The total substrate concentration [C]0 = [C] + [C]* + [M1] + [M2] is set to be unity to normalize the scale for chemical concentrations in our model. After these rescalings, our model is described by five dimensionless independent parameters: f1,  , f2, the input signal strength [A] and the non-equilibrium factor γ. We have tested the generality of the conclusions derived from this simplified symmetric case by testing two representative asymmetric cases: the two enzymes have (i) the same Michaelis constants but different turnover rates or (ii) the same turnover rates but different Michaelis constants (see the electronic supplementary material for details). Both scenarios lead to qualitatively the same conclusions that are presented in the rest of this paper.
, f2, the input signal strength [A] and the non-equilibrium factor γ. We have tested the generality of the conclusions derived from this simplified symmetric case by testing two representative asymmetric cases: the two enzymes have (i) the same Michaelis constants but different turnover rates or (ii) the same turnover rates but different Michaelis constants (see the electronic supplementary material for details). Both scenarios lead to qualitatively the same conclusions that are presented in the rest of this paper.
A response of the system to an input change at time t = 0 is shown in figure 1c for γ = 0. Because of the fast synthesis of B1, [C*] first increases to its peak level [C*]max at time Tmax. Then, as the inhibitor B2 is slowly turned on, [C*] reduces back to a steady-state level [C*]steady that is close to the pre-stimulus value [C*]0. The response dynamics of the system is characterized by [C*]max and Tmax, and its adaptation accuracy is characterized by  . In the following, we study how these characteristics of the network performance depend on the pathway parameters; in particular, the non-equilibrium parameter γ, which determines the energy cost for operating the biochemical circuit.
. In the following, we study how these characteristics of the network performance depend on the pathway parameters; in particular, the non-equilibrium parameter γ, which determines the energy cost for operating the biochemical circuit.
3. Results
3.1. The dynamics and energy dissipation of the network
We first simulated the dynamics of the reaction network after addition of A for different γ values (unless otherwise stated, the rate constants used in this paper are f1 = 10,  , f2 = 100 and
, f2 = 100 and  ). The results are shown in figure 2a. For any finite γ > 0, right after the stimulus, the output C* first quickly dips to a lower level owing to a temporary accumulation at the intermediate state (see appendix A for a detailed explanation). Following this initial ‘dipping’, the C* concentration increases to its peak level [C*]max and then decreases back to a new steady-state level [C*]steady. The non-equilibrium parameter γ affects both the response magnitude
). The results are shown in figure 2a. For any finite γ > 0, right after the stimulus, the output C* first quickly dips to a lower level owing to a temporary accumulation at the intermediate state (see appendix A for a detailed explanation). Following this initial ‘dipping’, the C* concentration increases to its peak level [C*]max and then decreases back to a new steady-state level [C*]steady. The non-equilibrium parameter γ affects both the response magnitude  and the adaptation accuracy (how close [C*]steady is to the pre-stimulus level [C*]0). For the equilibrium case when γ = 1, the system has weak (or even negative) response and poor adaptation accuracy. As γ decreases from 1, the system operates out of equilibrium with a smaller initial activity dip and enhanced performance in both response strength and adaptation accuracy.
and the adaptation accuracy (how close [C*]steady is to the pre-stimulus level [C*]0). For the equilibrium case when γ = 1, the system has weak (or even negative) response and poor adaptation accuracy. As γ decreases from 1, the system operates out of equilibrium with a smaller initial activity dip and enhanced performance in both response strength and adaptation accuracy.
Figure 2.

The response dynamics and energy cost of the I1-FFL network. (a) Response of the I1-FFL network to a step increase of the input ([A] = 0.1 used here) for different γ (represented by different coloured lines). As highlighted by the black and red dotted arrows, both the maximum response and the adaptation accuracy decrease as γ increases from 0 (Michaelis–Menten-type reactions) to 1 (equilibrium reactions). The blue dotted arrow indicates the initial activity dip, which disappears only when γ = 0. (b)  versus γ for different values of [A]. ΔW increases with 1/γ logarithmically. The results are from direct simulations of equations (2.1)–(2.6) and equation (3.6). Unless otherwise stated, the rate constants used in this paper are: f1 = 10,
versus γ for different values of [A]. ΔW increases with 1/γ logarithmically. The results are from direct simulations of equations (2.1)–(2.6) and equation (3.6). Unless otherwise stated, the rate constants used in this paper are: f1 = 10,  and f2 = 100.
and f2 = 100.
When γ < 1, the reaction network shown in figure 1b operates out of equilibrium, and free energy needs to be dissipated to maintain its function. More specifically, substrate C is continuously activated to C* by B1 and, at the same time, C* is continuously deactivated to C by B2. This generates a circulating flux in the network (thick arrows in figure 1b), and it consumes free energy to perform the (chemical) work of maintaining this persistent flux. Quantitatively, the energy dissipation rate can be determined as [26]
| 3.1 |
where Ji± are the transition fluxes in the forward and backward directions for the ith reaction. The energy dissipation rate is in unit of KBT, with KB the Boltzmann constant and T the temperature of the environment. We have calculated W in the I1-FFL network (figure 1b) for different values of γ.
The reaction cycle, the most distinctive feature of a non-equilibrium system, can be characterized by a time scale τcyc, defined as the average time for a C molecule to complete the cycle C → M1 → C* → M2 → C. In the limit  , the expression for τcyc can be obtained by a first-passage-time analysis (see appendix A for details): τcyc = 2(1 + KM/[A])/f2 where
, the expression for τcyc can be obtained by a first-passage-time analysis (see appendix A for details): τcyc = 2(1 + KM/[A])/f2 where  is the Michaelis constant. By using W and τcyc, we can define
is the Michaelis constant. By using W and τcyc, we can define  as the average thermodynamic work per cycle. As shown in figure 2b, ΔW is zero for the equilibrium case γ = 1, and it depends on 1/γ logarithmically for
as the average thermodynamic work per cycle. As shown in figure 2b, ΔW is zero for the equilibrium case γ = 1, and it depends on 1/γ logarithmically for  . Thus, from figure 2a,b, it is clear that energy dissipation (or equivalently the value of γ) strongly affects the I1-FFL network behaviours, which we study in the following sections.
. Thus, from figure 2a,b, it is clear that energy dissipation (or equivalently the value of γ) strongly affects the I1-FFL network behaviours, which we study in the following sections.
3.2. Energy dissipation enhances response sensitivity
The initial response to a sudden change of input from 0 to [A] is characterized by the response amplitude Δ[C*]max and the response time Tmax. In figure 3a, we show that the response amplitude Δ[C*]max increases with 1/γ (or equivalently the energy dissipation) for different values of [A]. The expression of the response amplitude can be derived analytically when [A] is small and  (see the electronic supplementary material, for the detailed derivation),
(see the electronic supplementary material, for the detailed derivation),
| 3.2 |
which shows that the maximum response increases as γ decreases. It is also clear from equation (3.2) that there exists a critical value  and the response is negative for γ > γc owing to the initial ‘dipping’ effect (figure 2a).
and the response is negative for γ > γc owing to the initial ‘dipping’ effect (figure 2a).
Figure 3.

Energy dissipation enhances response sensitivity. (a) Larger 1/γ (i.e. larger energy dissipation) leads to higher sensitivity Δ[C*]max, which saturates to an intrinsic upper limit determined by the catalytic rates of the network. (b) The response time Tmax increases as 1/[A] increases, but only weakly depends on γ.
The increase in the response with 1/γ can be understood intuitively: during the initial response time before the reverse enzyme B2 is synthesized, we can take the approximation [B1] = [A] and [B2] = 0, and γ determines the population distribution among the three states C, M1 and C*. A larger value of 1/γ leads to a more stable and therefore more populated C* state, which results in a bigger initial response. As γ approaches 0, Δ[C*]max saturates to a value dependent on the signal [A] and other intrinsic parameters of the network.
The response time Tmax is not strongly affected by γ (figure 2a), it mainly depends on the input strength [A] (figure 3b). The response time is controlled by the catalytic reaction rates which are larger when signal strength is higher, because the amount of enzymes [B1] and [B2] is proportional to [A] as shown in equations (2.1) and (2.2).
The dependence of Δ[C*]max on [A] is more complex, as can be seen in equation (3.2). From direct simulation of the model, we find that Δ[C*]max depends non-monotonically on [A]. It first increases with [A] to a maximum value Δ[C*]c when [A] = [A]c and then decreases as [A] > [A]c. Both [A]c and Δ[C*]c increase with 1/γ (see the electronic supplementary material, figure S1). One reason for this behaviour is that a stronger input signal leads to the faster synthesis of inhibitor B2, which results in a higher effective inhibition rate of C*. When the production rate of B2 is comparable to or larger than the catalytic rates f1 and f2, the maximum response can decrease with [A].
3.3. Energy dissipation improves adaptation accuracy
Living organisms need to adapt to their environment and maintain the steady-state output [C*]steady near its set level [C*]0. For the simplified symmetric I1-FFL-type reaction network studied here, we have [C*]0 = 1/2 and we can calculate [C*]steady analytically (see appendix A for details),
| 3.3 |
from which the relative adaptation error ε can be determined,
 |
3.4 |
Equation (3.4) shows that a smaller γ or equivalently higher energy dissipation rate (figure 2b) leads to smaller error ε, with its upper and lower limits achieved at γ = 1 (equilibrium) and γ = 0 (the Michaelis–Menten kinetics), respectively,
| 3.5 |
The energy dissipation rate  of the I1-FFL network can be calculated by using equation (3.1). For the symmetric network considered here,
of the I1-FFL network can be calculated by using equation (3.1). For the symmetric network considered here,  can also be obtained analytically (see appendix A for details):
can also be obtained analytically (see appendix A for details):
| 3.6 |
For the equilibrium case γ = 1,  ;
;  diverges for the extreme non-equilibrium (Michaelis–Menten) case with γ = 0. In figure 4, ε/ε1 is plotted as a function of
diverges for the extreme non-equilibrium (Michaelis–Menten) case with γ = 0. In figure 4, ε/ε1 is plotted as a function of  for various γ values. The results show that higher energy dissipation improves adaptation accuracy (i.e. smaller ε/ε1). However, stronger signal strength [A] leads to larger error ε/ε1. As shown in figure 4, at a moderate level of signal strength [A] ≤ 1, the adaptation error εγ can be reduced to 10% of its equilibrium level ε1 for ΔW ∼ 5−6 KBT.
for various γ values. The results show that higher energy dissipation improves adaptation accuracy (i.e. smaller ε/ε1). However, stronger signal strength [A] leads to larger error ε/ε1. As shown in figure 4, at a moderate level of signal strength [A] ≤ 1, the adaptation error εγ can be reduced to 10% of its equilibrium level ε1 for ΔW ∼ 5−6 KBT.
Figure 4.

Relative error versus ΔW for different signal strength [A]. Increasing energy dissipation reduces adaptation error. For a moderate level of [A] ≤ 1, it takes ΔW ∼ 5−6 KBT to reduce the adaptation error to 10% of its equilibrium level.
From equations (3.4) and (3.5), we obtain an analytical expression for the relative error ε/ε1,
 |
3.7 |
The dependence of ε/ε1 on γ and [A] is bounded by two limits,
These limits show that the intrinsic catalytic properties of the participating enzymes, such as the value of KM, determine the highest adaptation accuracy in an I1-FFL-type reaction network, even with unlimited energy consumption (γ = 0). Larger KM leads to a lower minimum error ε0, as also shown in equation (3.5).
3.4. Energy dissipation expands the dynamic range of accurate adaptation
To survive in a changing environment, living organisms have to adapt in a wide range of backgrounds. In figure 5a, we show the dependence of the steady-state output [C*] on the background level [A] for different values of γ. For a given γ, the adapted (steady state) value of [C*] can remain around its target value [C*]0 for a range of input [A] before it decreases for a stronger external signal [A]. We define the dynamic range of adaptation A1/2 as the signal strength at which Csteady is half of its targeted value [C*]0 = 0.5. Thus, A1/2 characterizes the width of the region in which high adaptation accuracy can be achieved.
Figure 5.

Energy dissipation expands the accurate adaptation regime. (a) Steady-state dose–activity curves for different values of γ (as labelled). The curves shift towards higher signal strength as γ → 0, indicating that dissipated energy helps expand the accurate adaptation regime, characterized by A1/2. (b) A1/2 versus 1/γ for different KM. Larger energy dissipation leads to larger A1/2, which saturates to KM (shown as the dotted lines), as γ → 0. In this set of simulations,  f2 = 500 and f1 is determined from
f2 = 500 and f1 is determined from  .
.
From equation (3.3), we derive the analytical expression for A1/2,
| 3.8 |
Equation (3.8) shows that larger 1/γ expands the range of signal to which the system can adapt accurately, as illustrated in figure 5a. Our results also indicate that the maximum range that an I1-FFL-type reaction network can maintain its accurate adaptation is limited by the Michaelis constant KM of the participating enzymes; a larger KM leads to a greater expansion of the accurate adaptation regime (figure 5b).
Overall, our analysis (equations (3.2)–(3.8)) shows that the performance of the I1-FFL network is enhanced by having higher energy dissipation (smaller γ), but the best-achievable performance (when γ → 0) is determined by the intrinsic properties of the participating enzymes. For the GEF–GAP–Ras pathway in Dictyostelium chemotaxis, the kinetic constants of the enzymes have not been measured. However, accurate adaptation with better than 90% accuracy to cyclic adenosine monophosphate (cAMP) levels as high as 100 nM–1 µM was reported in [20]. Assuming the input in our model is in the same range as the cAMP level, our analysis (equation (3.5)) allows us to estimate the value of the underlying Michaelis constant KM ≈ 1–10 µM. For a cAMP level of 100 nM, the adaptation time was measured to be approximately 20 s [20], which determines the average cycle time τcyc ∼ 20 s in our model. Together with the measured adaptation error ε0 ∼ 1–4%, we can estimate for the catalytic rate f2 = 2/(ε0τcyc) ∼ 2.5−10 s−1. Interestingly, both these estimated values, KM and f2, fall into the range of typical kinetic constants according to a recent systematic study on enzyme parameters [27]. To estimate the energy dissipation rate in the GEF–GAP–Ras pathway in a single Dictyostelium cell, we need to know the copy number of the Ras protein, which has not been measured. If we assume that the Ras copy number is comparable to that of the upstream receptor cAR1, which is estimated to be approximately 80 000 [28], we can obtain the dissipation rate to be approximately 4000 GTP s−1 in the presence of 100 nM stimulus (cAMP).
4. Discussion
In summary, we have constructed a realistic I1-FFL reaction network ubiquitous in biochemical circuits. We have investigated the energetic costs for the key functions of this network. Our analysis demonstrates how the energy dissipated in the I1-FFL network is used (i) to enhance the system's sensitivity in its initial response to an external stimulus; (ii) to achieve high accuracy in adapting to persistent stimuli; and (iii) to expand the range of stimuli over which such accurate adaptation occurs.
In biochemical networks, there are many so-called ‘futile cycles’, in which two metabolic pathways run simultaneously in opposite directions with no apparent function other than dissipating chemical energy in the form of heat [29]. The phosphorylation and dephosphorylation cycle in the I1-FFL network shown in figure 1b exemplifies such a futile cycle. However, contrary to what the name might suggest, we show here that the energy dissipated in the futile cycle driven by continuous hydrolysis of the high-energy fuel molecules, such as adenosine triphosphate (ATP) and GTP, enables critical regulatory functions, for example high sensitivity and accurate adaptation. In the following, we discuss the possible microscopic mechanism for such a cost–performance trade-off in the I1-FFL network. We also compare I1-FFL with other important regulatory networks to suggest a possible universal relationship among accuracy, speed and the energy cost of all regulatory functions.
4.1. Origin of the adaptation error in an incoherent type-1 feed-forward loop and its suppression by energy dissipation
One key function of the I1-FFL motif (figure 1a) is accurate adaptation, which is the ability to keep the activated substrate [C*]steady in steady state around the targeted level [C*]0 (=1/2) independent of the input [A]. From mass conservation, the deviation of [C*]steady from 1/2 equals the population of C in the intermediate complexes M1 and M2, so that the relative adaptation error ε = |1–2[C*]steady| = 2 × [M1]. Therefore, the origin of this error is the population of substrate trapped in the enzyme-bound intermediate complexes.
In an equilibrium system (γ = 1) where detailed balance is satisfied in all individual reactions throughout the network, the population in each chemical state is entirely determined by the free energy landscape. However, for a non-equilibrium reaction system (γ < 1) studied in this paper, the reverse reactions from the final products back to the intermediate complexes are suppressed. Effectively, this corresponds to decreasing the free energy of the C*-state to disfavour the intermediate complexes to reduce adaptation error. From equation (3.3), the effective energy difference between the intermediate state (M1) and the C*-state can be derived as
| 4.1 |
where the first term is the equilibrium free energy difference at γ = 1, and the second term
| 4.2 |
defines the free energy shift owing to energy dissipation (γ < 1).
Equation (4.2) shows the dependence of Hγ on γ and the turnover ratio  . At any given value of r, the magnitude of Hγ increases with 1/γ and saturates at ln(1 + r) as 1/γ → ∞. For a given γ < 1, Hγ increases with r and saturates to −0.5 ln γ when r → ∞. The saturation of Hγ shows that the maximum ‘useful’ energy contribution from the non-equilibrium effect is determined by intrinsic kinetic rates of the catalytic reactions. This is consistent with equation (3.5), which shows that larger KM leads to smaller adaptation error because larger KM disfavours the intermediate states.
. At any given value of r, the magnitude of Hγ increases with 1/γ and saturates at ln(1 + r) as 1/γ → ∞. For a given γ < 1, Hγ increases with r and saturates to −0.5 ln γ when r → ∞. The saturation of Hγ shows that the maximum ‘useful’ energy contribution from the non-equilibrium effect is determined by intrinsic kinetic rates of the catalytic reactions. This is consistent with equation (3.5), which shows that larger KM leads to smaller adaptation error because larger KM disfavours the intermediate states.
4.2. Energy dissipation enhances adaptation accuracy and response sensitivity simultaneously
In an equilibrium network with γ = 1, accurate adaptation is possible but it comes with the price of a small or even negative response. This can be seen by taking the following transformation:
 |
This new network with transformed parameters  operates at equilibrium because
operates at equilibrium because 
 . From equation (3.4), this equilibrium network has the same steady-state adaptation accuracy (ε) and the range of accurate adaptation (A1/2) as the
. From equation (3.4), this equilibrium network has the same steady-state adaptation accuracy (ε) and the range of accurate adaptation (A1/2) as the  non-equilibrium network. However, from equation (3.2) and figure 2a, it is clear that the response sensitivity of the equilibrium network is much smaller than that of the non-equilibrium network with γ < 1, which is confirmed by direct simulations (see the electronic supplementary material, figure S2).
non-equilibrium network. However, from equation (3.2) and figure 2a, it is clear that the response sensitivity of the equilibrium network is much smaller than that of the non-equilibrium network with γ < 1, which is confirmed by direct simulations (see the electronic supplementary material, figure S2).
To fully appreciate the effects of energy dissipation, we calculated the system's response dynamics to a series of stepwise stimuli (figure 6a) and computed the response-to-signal gain (figure 6b) for different γ. Our results clearly show that higher energy consumption, i.e. smaller γ, leads to both enhanced response gain (sensitivity) and more accurate adaptation simultaneously.
Figure 6.
The dependence of adaptation accuracy and responsive sensitivity on γ. (a) Response and adaptation for a stepwise increase of the input [A] (shown at the top) for different γ. Signal increases from 0 to 1 with step size Δ[A] = 0.1. Adaptation is more accurate for smaller γ. (b) The gain  as a function of background [A] for different γ. Smaller γ leads to higher gain.
as a function of background [A] for different γ. Smaller γ leads to higher gain.
4.3. A universal energy–speed–accuracy trade-off relation for adaptation
Other than the I1-FFL motif, there is another simple network motif, namely the NFL motif (figure 7a), which is used in many different sensory systems (e.g. Escherichia coli chemotaxis) to achieve accurate adaptation. A recent exhaustive study of all three-node regulatory networks has shown that I1-FFL- and NFL-type topologies are the only two three-node networks that could provide robust accurate adaptation [16]. For the NFL network, an ESA trade-off relation was found recently among the speed and accuracy of adaptation and the required energy dissipation rate [23]. Here, for the I1-FFL network, when γ is small 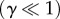 and signal is weak
and signal is weak  , a similar exponential relation between adaptation error and the energy dissipation rate can also be derived from equations (3.4) and (3.6) (see appendix A for details),
, a similar exponential relation between adaptation error and the energy dissipation rate can also be derived from equations (3.4) and (3.6) (see appendix A for details),
 |
4.3 |
It can easily be shown (for the same limit  ) that the cycle time τcyc is the same as the relaxation time of the adapted state, and can thus be considered as the adaptation time of the system.
) that the cycle time τcyc is the same as the relaxation time of the adapted state, and can thus be considered as the adaptation time of the system.
Figure 7.
The universal ESA relationship for the two basic adaptation networks. (a,b) Three-node adaptive motifs NFL and I1-FFL are shown in the upper panels. The lower panels illustrate the circulatory reactions driven by external energy inputs (the red arrows) in these two systems. For NFL networks, external energy (W) is consumed to push the system uphill (red arrows) to maintain the adaptation state at the cross-over point of the active and inactive states [23]. For the I1-FFL network, the facilitating energy Hγ pumps C and C* from their lower energy states to their higher energy states to drive the circulatory reactions within the network. (c) These two adaptive systems share the same ESA relationship.
The relationship among the energy dissipation rate  , the adaptation speed
, the adaptation speed  and the adaptation accuracy (1/ε), as given in equation (4.3), is essentially the same as the recently derived ESA trade-off relation for the NFL network [23]. Furthermore, the adaptation errors in both networks all originate from the probabilities in the undesired chemical states (the two intermediate states for I1-FFL and the two boundary states for NFL), and the intrinsic properties of the catalytic reactions determine the maximum performance of both networks.
and the adaptation accuracy (1/ε), as given in equation (4.3), is essentially the same as the recently derived ESA trade-off relation for the NFL network [23]. Furthermore, the adaptation errors in both networks all originate from the probabilities in the undesired chemical states (the two intermediate states for I1-FFL and the two boundary states for NFL), and the intrinsic properties of the catalytic reactions determine the maximum performance of both networks.
The similarities between NFL and I1-FFL as shown here are quite remarkable given their different topologies (e.g. one feed-forward, one feedback). Because NFL and I1-FFL are the two basic network motifs for accurate adaptation [16], the results presented here for I1-FFL, together with our previous work [23] on NFL, strongly suggest that the ESA relationship is universal for all regulatory systems that enable accurate adaptation (figure 7).
4.4. The difference between a negative-feedback loop and an incoherent type-1 feed-forward loop
Despite the same ESA relation shared by the NFL and I1-FFL networks, however, they have several notable differences. For NFL (figure 7a), the dissipated energy is used to stabilize the targeted adaptation state that is unstable in the equilibrium model (see [23] for details), whereas for I1-FFL (figure 7b), external energy is used to drive C and C* to higher energy states to maintain the continuous reaction cycle C → M1 → C* → M2 → C, which leads to large responses upon input changes. The energy dissipation in I1-FFL also increases the steady-state probability in the ‘correct’ (desired) state C* relative to that of the ‘error’ state M1. In this sense, I1-FFL is similar to the case of kinetic proofreading, where energy is used to further decrease the probability in the error state which is already less favoured [26,30,31].
These two networks also have different dynamic properties. The time scales for both the response and the adaptation processes in I1-FFL depend on the input signal, whereas in NFL they do not. Relatedly, in I1-FFL, the continuous turnovers of the two opposing enzymes are directly controlled by the input signal (with different time scales) and is necessary for response and adaptation. In NFL, however, the two opposing enzymes (such as CheR and CheB in E. coli chemotaxis) for the NFL are not directly controlled by the input signal and their turnovers are not required for adaptation. The implications of these differences and the advantages of each of the two adaptation schemes over the other in different biological systems are interesting subjects for future investigations.
Acknowledgements
We thank Pablo Sartori, Bill Loomis, Wouter Rappel and Herbert Levine for discussions and comments. Y.T. initiated the work; G.L. and Y.T. carried out the work and wrote the paper.
Appendix A
A.1. Mathematical explanation of the initial ‘dipping’ effect after stimulation
In figure 2a, responding trajectories of the I1-FFL motif (figure 1b) after stimulation are shown for different γ values. For any finite γ > 0, right after the stimulus, the output C* first rapidly dips to a lower level. This initial decrease in C* is due to the fast synthesis of activator B1 and resulting transient back-flow from C* into the M1 state. Immediately after the A concentration is increased from 0 to [A], only the B1 concentration changes, so, for  , equation (2.6) becomes
, equation (2.6) becomes
 |
A1 |
which leads to the initial ‘dipping’ for any non-zero γ. The ‘dipping’ behaviour becomes more pronounced as the reaction network approaches equilibrium when γ → 1 as shown in figure 2a (blue arrow).
A.2. The cycle time (τcyc)
We define a cycle time (τcyc) as the average time for a molecule C to complete a cycle C → M1 → C* → M2 → C. Here, we calculate the cycle time by using a first-passage-time analysis. Owing to the symmetry between the two halves of the cycle, we can consider half of the cycle C → M1 → C*. Let P1(t) and P2(t) be the probabilities in states C and M1, respectively. The dynamics of P1, P2 can be written as
| A2 |
and
| A3 |
Here, we consider the case  , where the reverse reaction rate
, where the reverse reaction rate  can be neglected.
can be neglected.
Equations (A 2) and (A 3) can be solved analytically with the initial condition P1(t = 0) = 1, P2(t = 0) = 0,
| A4 |
and
| A5 |
where σ1 and σ2 are
 |
A6 |
which are the two roots of the quadratic equation
| A7 |
The probability of arriving at state C* during time interval (t → t + dt) is f2P2(t)dt. Therefore, the average arrival time is
 |
A8 |
In the above derivation, we have used 
 and σ1σ2 = f1f2[A].
and σ1σ2 = f1f2[A].
Finally, we have the expression for the cycle time τcyc (for  ),
),
| A9 |
A.3. The steady-state output level [C*]steady
Owing to the symmetry assumption about the catalytic rates of two enzymes, the following relations hold at steady state: [C*] = [C], [M1] = [M2], [B1] = [B2] – [A], so that the steady-state solution of equations (2.1)–(3.1) satisfies
| A10 |
and
| A11 |
which lead us to the steady-state output level
 |
A12 |
as well as the steady-state level of the intermediate complex
| A13 |
A.4. The steady-state energy dissipation rate Ẇ
We can compute the energy dissipation using equation (3.1),
 |
From equations (A 12) and (A 13), the forward and backward fluxes in each reaction step can be evaluated at steady state:
 |
A14 |
 |
A15 |
 |
A16 |
and
 |
A17 |
Therefore, the energy dissipation rates in reactions  and
and  can be calculated,
can be calculated,
 |
A18 |
and
 |
A19 |
which leads to the total energy dissipation
 |
A20 |
A.5. Derivation of the energy–speed–accuracy relation for incoherent type-1 feed-forward loop networks
At the limit of small γ (i.e. γ → 0), equation (3.4) can be linearized as
 |
A21 |
Meanwhile, equation (3.6) can also be simplified,
 |
A22 |
| A23 |
Equations (A 21) and (A 23) lead to the ESA relation for I1-FFL networks,
 |
A24 |
as shown in equation (4.3), where  is used in the exponential.
is used in the exponential.
Funding statement
This work is partially supported by an NIH grant (no. R01GM081747 to Y.T.).
References
- 1.Alon U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461. ( 10.1038/nrg2102) [DOI] [PubMed] [Google Scholar]
- 2.Alon U. 2007. An introduction to systems biology: design principles of biological circuits, vol. 10 New York, NY: CRC Press. [Google Scholar]
- 3.Eichenberger P, et al. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2, e328 ( 10.1371/journal.pbio.0020328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TI, et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804. ( 10.1126/science.1075090) [DOI] [PubMed] [Google Scholar]
- 5.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298, 824–827. ( 10.1126/science.298.5594.824) [DOI] [PubMed] [Google Scholar]
- 6.Odom DT, et al. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303, 1378–1381. ( 10.1126/science.1089769) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer LA, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. ( 10.1016/j.cell.2005.08.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swiers G, Patient R, Loose M. 2006. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev. Biol. 294, 525–540. ( 10.1016/j.ydbio.2006.02.051) [DOI] [PubMed] [Google Scholar]
- 9.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. 2004. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA 101, 6355–6360. ( 10.1073/pnas.0307571101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangan S, Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11 980–11 985. ( 10.1073/pnas.2133841100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangan S, Itzkovitz S, Zaslaver A, Alon U. 2006. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J. Mol. Biol. 356, 1073–1081. ( 10.1016/j.jmb.2005.12.003) [DOI] [PubMed] [Google Scholar]
- 12.Goentoro L, Shoval O, Kirschner MW, Alon U. 2009. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 36, 894–899. ( 10.1016/j.molcel.2009.11.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goentoro L, Kirschner MW. 2009. Evidence that fold-change, and not absolute level, of β-catenin dictates Wnt signaling. Mol. Cell 36, 872–884. ( 10.1016/j.molcel.2009.11.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoval O, Goentoro L, Hart Y, Mayo A, Sontag E, Alon U. 2010. Fold-change detection and scalar symmetry of sensory input fields. Proc. Natl Acad. Sci. USA 107, 15 995–16 000. ( 10.1073/pnas.1002352107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behar M, Hao N, Dohlman HG, Elston TC. 2007. Mathematical and computational analysis of adaptation via feedback inhibition in signal transduction pathways. Biophys. J. 93, 806–821. ( 10.1529/biophysj.107.107516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. 2009. Defining network topologies that can achieve biochemical adaptation. Cell 138, 760–773. ( 10.1016/j.cell.2009.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen-Orr SS, Milo R, Mangan S, Alon U. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31, 64–68. ( 10.1038/ng881) [DOI] [PubMed] [Google Scholar]
- 18.Parent CA, Devreotes PN. 1999. A cell's sense of direction. Science 284, 765–770. ( 10.1126/science.284.5415.765) [DOI] [PubMed] [Google Scholar]
- 19.Levchenko A, Iglesias PA. 2002. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys. J. 82, 50–62. ( 10.1016/S0006-3495(02)75373-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeda K, Shao D, Adler M, Charest PG, Loomis WF, Levine H, Groisman A, Rappel WJ, Firtel RA. 2012. Incoherent feedforward control governs adaptation of activated Ras in a eukaryotic chemotaxis pathway. Sci. Signal. 5, p. ra2 ( 10.1126/scisignal.2002413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. 2000. Molecular cell biology. New York, NY: W. H. Freeman. [Google Scholar]
- 22.Vigil D, Cherfils J, Rossman KL, Der CJ. 2010. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857. ( 10.1038/nrc2960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan G, Sartori P, Neumann S, Sourjik V, Tu Y. 2012. The energy–speed–accuracy tradeoff in sensory adaptation. Nat. Phys. 8, 422–428. ( 10.1038/nphys2276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaelis L, Menten ML. 1913. Die Kinetik der Invertinwirkung. Biochem. Z 49, 333–369. [Google Scholar]
- 25.Van Kampen NG. 1981. Stochastic processes in physics and chemistry. Amsterdam, The Netherlands: North-Holland Physics Publishing. [Google Scholar]
- 26.Qian H. 2007. Phosphorylation energy hypothesis: open chemical systems and their biological functions. Annu. Rev. Phys. Chem. 58, 113–142. ( 10.1146/annurev.physchem.58.032806.104550) [DOI] [PubMed] [Google Scholar]
- 27.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. 2011. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410. ( 10.1021/bi2002289) [DOI] [PubMed] [Google Scholar]
- 28.Ueda M, Shibata T. 2007. Stochastic signal processing and transduction in chemotactic response of eukaryotic cells. Biophys. J. 93, 11–20. ( 10.1529/biophysj.106.100263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voet D, Voet JG, Pratt CW. 1999. Fundamentals of biochemistry. New York, NY: Wiley. [Google Scholar]
- 30.Hopfield JJ. 1974. Kinetics proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl Acad. Sci. USA 71, 4135–4139. ( 10.1073/pnas.71.10.4135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian H. 2006. Reducing intrinsic biochemical noise in cells and its thermodynamic limit. J. Mol. Biol. 362, 387–392. ( 10.1016/j.jmb.2006.07.068) [DOI] [PubMed] [Google Scholar]





