Abstract
De novo synthetic biological design has the potential to significantly impact upon applications such as energy generation and nanofabrication. Current designs for constructing organisms from component parts are typically limited in scope, as they utilize a cut-and-paste ideology to create simple stepwise engineered protein-signalling pathways. We propose the addition of a new design element that segregates components into lipid-bound ‘proto-organelles’, which are interfaced with response elements and housed within a synthetic protocell. This design is inspired by living cells, which utilize multiple types of signalling molecules to facilitate communication between isolated compartments. This paper presents our design and validation of the components required for a simple multi-compartment protocell machine, for coupling a light transducer to a gene expression system. This represents a general design concept for the compartmentalization of different types of artificial cellular machinery and the utilization of non-protein signal molecules for signal transduction.
Keywords: synthetic biology, compartmentalized bioreactors, signal transduction
Impressive feats of synthetic biological engineering have been demonstrated by modifying the metabolism of living organisms [1–6], altering enzyme localization in metabolic pathways to improve efficiency [7,8] or sensory component addition [9]. Such designs, however, build upon existing organisms, rather than engineering from component parts. As a consequence, they often suffer from inherent complexities, such as the required regulation of the new metabolic processes [10]. Novel, simpler machines therefore offer an attractive alternative: the bottom-up approach enabling construction of self-regulating synthetic machines [11–13] utilizing repositories of parts such as ‘biobricks’ [14] in the absence of an existing biological system. This imparts advantages including elimination of unpredictable interference by the host organism metabolism [15], removal of host transcriptional gene regulation complexity [16] and the potential addition of regulatory elements or external controls that switch on the activity of the protocell machine [17]. Additionally, there is potential for addition of componentry using soluble nanolipoproteins [18] that allow the incorporation of incompatible systems without the requirement for spatial segregation. Despite the advantages of de novo synthetic biological design, the development of such systems has been limited: benefits of reduced complexity come with disadvantages including loss of control fidelity and component incompatibility. We present an experimental design that could illustrate a more general strategy for incorporating control elements without significantly increasing design complexity, which involves linking modular compartmentalized elements (proto-organelles) via non-protein-signalling molecules. This mimics the utilization of multiple signalling modes in living cells and allows for the incorporation of numerous potential subsystems, potentially including apparatus for protocell budding [19], leading the way towards protocell replication [20], liposomal biomachinery encapsulation [21], protein–protein communication that is either membrane mediated [22] or mediated through direct interaction, energy production modules [23] and genome replication modelled after retroviral systems [24].
Figure 1a presents our modular compartmentalized lipid-stabilized protocell design including: (i) a light-sensing lipid-stabilized proto-organelle, which in response to light generates a H+ gradient and triggers lactose release, and (ii) a fluorescent molecular readout module, which is a molecular signal transducer that links the output signal (the presence of lactose analogues) to protein expression. Our design segregates protocell components into subsystems, thus allowing additional levels of complexity and control to be realized, and highlights future opportunities for facilitating the development of semi-synthetic molecular machines.
Figure 1.
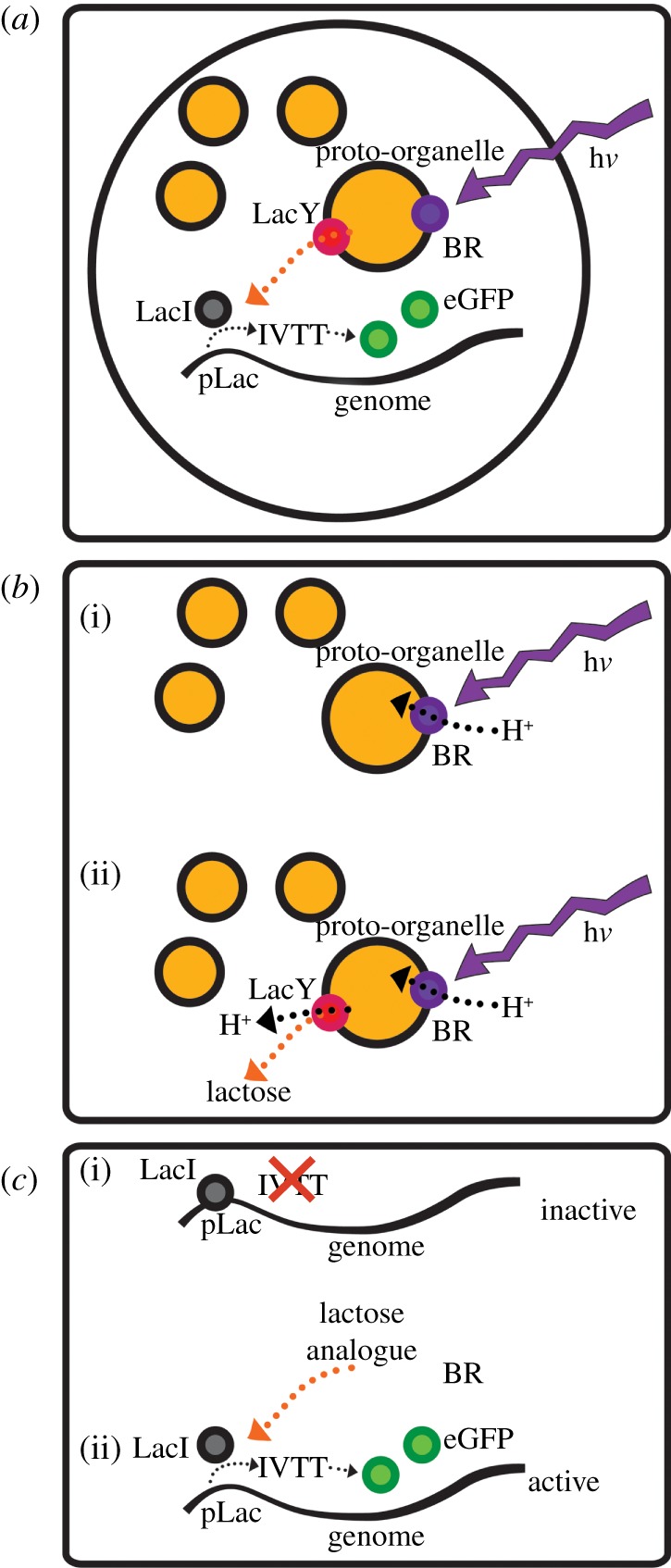
(a) Design and components of the light-sensing protocell featuring the light-sensing proto-organelle subsystem and the fluorescent molecular readout module. The entire complex is encapsulated and stabilized by a lipid monolayer of diphytanoyl phosphatidylcholine in hexadecane for measurement. (b) The light-sensing proto-organelle subsystem design. (i) hν powers acidification of proto-organelles by BR. (ii) The resulting H+ gradient provides energy for H+/lactose symport via LacY coupling lactose release to light sensing. (c) The fluorescence molecular readout module. Upper: the inactive state—transcription and production of eGFP is prevented by association of the repressor LacI to the pLac control region of the artificial genome. Lower: the active state—the lactose analogue (IPTG) results in release of LacI from the pLac region and removal of transcriptional repression resulting in eGFP production.
The first subsystem is a light-sensing proto-organelle comprising a bacteriorhodopsin (BR) light sensor and a lactose permease (LacY) signal transducer incorporated into liposomes, constructed from purified 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) lipids, that exports a control molecule (lactose) in the presence of a light signal. Illumination of BR acidifies the interior of the proto-organelle by light-driven pumping of H+ across the lipid membrane. The H+ gradient leads to LacY-driven co-transport of H+ and lactose from the proto-organelle interior to the exterior (figure 1b). This proto-organelle is housed within the second subsystem: the fluorescent molecular readout module. This module consists of a simple ‘genome’ containing enhanced green fluorescent protein (eGFP) with engineered control sequences that are repressed by the presence of the transcriptional repressor, LacI. Increasing the concentration of a lactose analogue signal molecule transduces the incoming light signal to this subsystem by repression removal. In combination with all the components required for in vitro transcription and translation (IVTT), this forms a complete system for synthesis of mRNA and consequent eGFP production (figure 1c).
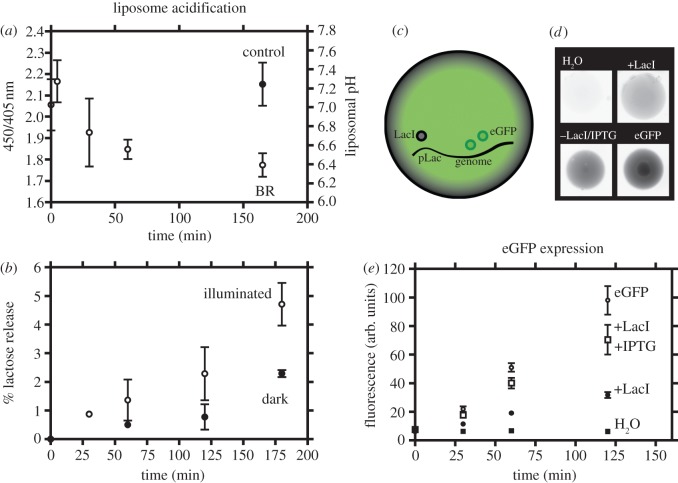
Figure 2 shows the measured activity of our protocell modules. Light energy drives H+ pumping into the liposome interior using the light-powered proton pump, BR. We monitored the resulting acidification of the liposome interior via changes in fluorescence excitation of the pH-sensitive dye pyranine (8-hydroxypyrene-1,3,6-trisulfonic acid) [25,26]. The observed changes in the ratio of the 450/405 nm fluorescence excitation bands are indicative of liposomal pH changes (see the electronic supplementary material, figure S1) and therefore showed that we could create a H+ electrochemical gradient in our proto-organelles. This was demonstrated by the acidification of liposomes containing functional BR following illumination (figure 2a). We added capability for signal molecule (lactose) release from the interior of the organelle to the external solution: coupling BR activity to signal release through LacY. Utilizing lactose as the signal molecule allowed very sensitive detection via a linked chemiluminescent assay. Figure 2b shows the result of illumination of the BR and LacY proto-organelles and the light-driven lactose release over time. Illumination doubles the concentration of lactose released from proto-organelles compared with non-illuminated under identical conditions. These observations validate our initial subsystem design (the proto-organelle light sensor), which consists of only five different molecules (BR, LacY, DOPE, DOPC and lactose), excluding the buffer components.
Figure 2.
Activity of the individual protocell components. (a) Acidification of the proto-organelle subsystem (BR liposomes) following illumination, as determined by pyranine fluorescence. Calibration for the liposomal pH is shown in the electronic supplementary material, figure S1. (b) Release of lactose from LacY and BR containing proto-organelles illuminated (open circles) and non-illuminated (filled circles). (c) The fluorescence molecular readout module. (d) Representative fluorescence images of eGFP expression in lipid-stabilized protocell drops (inverted colour for clarity). (e) Quantification of eGFP expression over time. All samples contain the eGFP ‘genome’ with the exception of the H2O control; +LacI contains LacI; +LacI/IPTG contains both LacI and IPTG; and eGFP contains neither LacI nor IPTG. Errors are calculated as s.d. from three repeat experiments. In cases where error bars are not shown, this is because they are smaller than the symbols.
The readout module required the development of a control mechanism capable of responding to the incoming signal molecule (lactose or lactose analogue) from the light-sensing proto-organelle. We designed a controllable switch that is able to turn on mRNA production to express eGFP in the presence of a signal molecule (figure 1c) monitored by fluorescent readout. We used the lac operon to repress expression, specifically mRNA production in the presence of the LacI repressor protein that tightly associates with a pLac control region, which we designed into our linear eGFP ‘genome’. This repression only occurs in the absence of a chemical control signal (in our case, the lactose analogue isopropyl β-D-1-thiogalactopyranoside (IPTG): a non-metabolizable lactose analogue that unlike lactose does not require an additional enzymatic isomerization step for LacI interaction: this efficiently binds to LacI and removes repression).
The design of our fluorescent molecular readout module subsystem is shown in figure 2c, and the resulting protocell solutions and eGFP expression in the presence and absence of the lactose analogue signal molecule and the LacI repressor in figure 2d. We measured the fluorescence of our protocells as sub-microlitre lipid-stabilized protocell drops in hexadecane allowing for the isolation of individual protocell drops. Fluorescence was negligible in the absence of an eGFP genome (see examples of raw data in figure 2d and quantified fluorescence in figure 2e). In the presence of the eGFP genome, eGFP fluorescence increased over time, consistent with efficient induction of expression. We verified that LacI could turn off this expression with eGFP production, determined by the fluorescence, decreasing to approximately 30% of that observed without repression (figure 2e). We then determined whether we could switch expression back on with the addition of the lactose analogue signal molecule (IPTG). Upon inclusion of the signal molecule in the protocells, repression by LacI was relieved, resulting in efficient eGFP production up to approximately 75% of that observed in the absence of any repression. This therefore demonstrated our ability to construct a system that can be readily switched, from an ‘off’ to an ‘on’ state, in the presence of the lactose analogue and the repressor LacI. This validates our fluorescent molecular readout module subsystem design and demonstrates the feasibility of the components used here for signal transduction between modules via a small molecule and associated protein componentry.
Designing protocell organisms from relatively simple components has potential advantages over the modification of biological systems, in particular the ability to predict the behaviour of the system under design a priori. The development of these systems will require both the segregation of components and the addition of external regulatory control elements, for example by isolating control elements in lipid-stabilized proto-organelles that can then be integrated into a signal-response element via non-protein signal molecules. This ideology allows the creation of simple synthetic systems, albeit with complex behaviour, utilizing a minimal component count. The resulting designs offer system complexity while avoiding the need to engineer existing life forms, which has inherent disadvantages such as unwanted side reactions and unanticipated changes to host metabolism [27], inhibitory regulatory control mechanisms that limit function and inherent genetic noise present in natural systems [28,29]. We have validated each of these subsystems, such that the modules can be combined as outlined in this design brief or inserted into other protocell frameworks leading to new synthetic biology machines that are controllable and will respond to environmental conditions. Future design iterations will build upon our development platform, incorporating otherwise incompatible elements together and using a high fidelity of regulation through the addition of further control elements. Such designs, where individual components can be segregated but linked by signal molecules, will provide additional control complexity to synthetic biological designs with potential applications in the construction of synthetic bioreactor machines that respond to external switches to control internal processing. Although not an issue with the protocell volumes we have utilized, in small volume systems, some consideration has to be made for the stochastic distribution of componentry, which can influence system behaviour [30]. The compartmentalized and modular design template reported here will act as the foundation for the further sequential addition of independent modules, with multiple independent input and output signals, resulting in a rapid increase in the complexity of future protocells that can be engineered.
Acknowledgements
We would like to thank Dr Kalypso Charalambous (Bristol University), Dr Lok Hang Mak (Imperial College) and Dr Andre Studer (Imperial College) for useful discussions, and Dr Heather Findlay (Bristol University), Nicola Harris (Bristol University) and Dr Paul Curnow (Bristol University) for providing protein samples.
Funding statement
This work was supported by the ESPRC (EP/G00465X and EP/H024425). L.M.C.B. was supported by a Royal Society University Fellowship.
References
- 1.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21, 796–802. ( 10.1038/nbt833) [DOI] [PubMed] [Google Scholar]
- 2.Jiang Y, Morgan-Kiss RM, Campbell JW, Chan CH, Cronan JE. 2010. Expression of Vibrio harveyi acyl-ACP synthetase allows efficient entry of exogenous fatty acids into the Escherichia coli fatty acid and lipid A synthetic pathways. Biochemistry 49, 718–726. ( 10.1021/bi901890a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ro D-K, et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. ( 10.1038/nature04640) [DOI] [PubMed] [Google Scholar]
- 4.Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD. 2008. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 7, 36 ( 10.1186/1475-2859-7-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562. ( 10.1038/nature08721) [DOI] [PubMed] [Google Scholar]
- 6.Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. ( 10.1038/nature06450) [DOI] [PubMed] [Google Scholar]
- 7.Conrado RJ, et al. 2012. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 40, 1879–1889. ( 10.1093/nar/gkr888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delebecque CJ, Lindner AB, Silver Pa, Aldaye FA. 2011. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474. ( 10.1126/science.1206938) [DOI] [PubMed] [Google Scholar]
- 9.Levskaya A, et al. 2005. Synthetic biology: engineering Escherichia coli to see light. Nature 438, 441–442. ( 10.1038/nature04405) [DOI] [PubMed] [Google Scholar]
- 10.Callura JM, Cantor CR, Collins JJ. 2012. Genetic switchboard for synthetic biology applications. Proc. Natl Acad. Sci. USA 109, 5850–5855. ( 10.1073/pnas.1203808109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stano P, Rampioni G, Carrara P, Damiano L, Leoni L, Luisi PL. 2012. Semi-synthetic minimal cells as a tool for biochemical ICT. Biosystems 109, 24–34. ( 10.1016/j.biosystems.2012.01.002) [DOI] [PubMed] [Google Scholar]
- 12.Noireaux V, Maeda YT, Libchaber A. 2011. Inaugural article: development of an artificial cell, from self-organization to computation and self-reproduction. Proc. Natl Acad. Sci. USA 108, 3473–3480. ( 10.1073/pnas.1017075108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stano P, Luisi PL. 2010. Achievements and open questions in the self-reproduction of vesicles and synthetic minimal cells. Chem. Commun. 46, 3639–3653. ( 10.1039/b913997d) [DOI] [PubMed] [Google Scholar]
- 14.Knight T. 2012. Idempotent vector design for standard assembly of biobricks. See http://hdl.handle.net/1721.1/21168.
- 15.Tan C, Marguet P, You L. 2009. Emergent bistability by a growth-modulating positive feedback circuit. Nat. Chem. Biol. 5, 842–848. ( 10.1038/nchembio.218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee TI, et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804. ( 10.1126/science.1075090) [DOI] [PubMed] [Google Scholar]
- 17.Schwille P. 2011. Bottom-up synthetic biology: engineering in a tinkerer's world. Science 333, 1252–1254. ( 10.1126/science.1211701) [DOI] [PubMed] [Google Scholar]
- 18.Baker SE, et al. 2009. Hydrogen production by a hyperthermophilic membrane-bound hydrogenase in water-soluble nanolipoprotein particles. J. Am. Chem. Soc. 131, 7508–7509. ( 10.1021/ja809251f) [DOI] [PubMed] [Google Scholar]
- 19.Kovačič J, Božič B, Svetina S. 2010. Budding of giant unilamellar vesicles induced by an amphitropic protein β2-glycoprotein I. Biophys. Chem. 152, 46–54. ( 10.1016/j.bpc.2010.07.005) [DOI] [PubMed] [Google Scholar]
- 20.Mavelli F, Ruiz-Mirazo K. 2013. Theoretical conditions for the stationary reproduction of model protocells. Integr. Biol. 5, 324–341. ( 10.1039/C2IB20222K) [DOI] [PubMed] [Google Scholar]
- 21.Tsai F-C, Stuhrmann B, Koenderink GH. 2011. Encapsulation of active cytoskeletal protein networks in cell-sized liposomes. Langmuir 27, 10 061–10 071. ( 10.1021/la201604z) [DOI] [PubMed] [Google Scholar]
- 22.Charalambous K, Booth PJ, Woscholski R, Seddon JM, Templer RH, Law RV, Barter LMC, Ces O. 2012. Engineering de novo membrane-mediated protein–protein communication networks. J. Am. Chem. Soc. 134, 5746–5749. ( 10.1021/ja300523q) [DOI] [PubMed] [Google Scholar]
- 23.Freisleben HJ, Zwicker K, Jezek P, John G, Bettin-Bogutzki A, Ring K, Nawroth T. 1995. Reconstitution of bacteriorhodopsin and ATP synthase from Micrococcus luteus into liposomes of the purified main tetraether lipid from Thermoplasma acidophilum: proton conductance and light-driven ATP synthesis. Chem. Phys. Lipids 78, 137–147. ( 10.1016/0009-3084(95)02491-Z) [DOI] [PubMed] [Google Scholar]
- 24.Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD, Gingeras TR. 1990. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc. Natl Acad. Sci. USA 87, 7797 ( 10.1073/pnas.87.19.7797-a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avnir Y, Barenholz Y. 2005. pH determination by pyranine: medium-related artifacts and their correction. Anal. Biochem. 347, 34–41. ( 10.1016/j.ab.2005.09.026) [DOI] [PubMed] [Google Scholar]
- 26.Clement NR, Gould JM. 1981. Pyranine (8-hydroxy-1,3,6-pyrenetrisulfonate) as a probe of internal aqueous hydrogen ion concentration in phospholipid vesicles. Biochemistry 20, 1534–1538. ( 10.1021/bi00509a019) [DOI] [PubMed] [Google Scholar]
- 27.Báez-Viveros JL, Flores N, Juárez K, Castillo-España P, Bolivar F, Gosset G. 2007. Metabolic transcription analysis of engineered Escherichia coli strains that overproduce L-phenylalanine. Microb. Cell Fact. 6, 30 ( 10.1186/1475-2859-6-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, Van Oudenaarden A. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31, 69–73. ( 10.1038/ng869) [DOI] [PubMed] [Google Scholar]
- 29.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297, 1183–1186. ( 10.1126/science.1070919) [DOI] [PubMed] [Google Scholar]
- 30.Pereira de Souza T, Stano P, Luisi PL. 2009. The minimal size of liposome-based model cells brings about a remarkably enhanced entrapment and protein synthesis. ChemBioChem 10, 1056–1063. ( 10.1002/cbic.200800810) [DOI] [PubMed] [Google Scholar]