Abstract
Objective(s): Erythropoiesis is regulated by some extrinsic and intrinsic factors as microRNAs (miRNAs). miRNAs are endogenously small non-coding regulatory RNAs which play vital roles in the variety of cellular fate, critical processes; growth, apoptosis, metabolism, survival of the cells and specially differentiation. Several miRNAs such as miR-16 and miR-451 have been shown to be correlated with erythroid differentiation. Taking into account the importance of miRNAs in cellular differentiation, the goal of the present study was to examine the role of miRNAs in hematopoietic stem cells (HSC) differentiation into the erythroid cells in the absence of growth factors and stimulatory cytokines.
Materials and Methods: CD133+ stem cells were infected with lentiviruses containing miR-451/miR-16 precursor sequence, erythroid differentiation was evaluated using RT-PCR for hemoglobin chains and surface antigens, also by banzidine staining.
Results: MiR-451up-regulation, but not miR-16, could induce α, β and γ-globin expression in CD133+ cells and have strong correlation with appearance of CD71 and CD235a markers in these cells. Moreover, miR-451 up-regulation increases the banzidine positive cells to ~ %40.
Conclusion: Our results provide strong evidence that miR-451 up-regulation strongly induces erythroid differentiation and maturation of CD133+ stem cells. Hence, this method may provide a useful technique for the production of artificial blood RBC and be used as a new strategy for gene therapy of hemoglobinopathies, such as β-thalassemias and sickle cell anemia.
Key Words: Erythropoiesis, CD133+, microRNA, miR-451, miR-16
Materials and Methods
Recombinant lentiviruses production
The pCDH-451 and pCDH-16 plasmids were generated by ligation of 250 and 458 bp fragments encompassing pri-miR-451 and pri-miR-16 sequences, respectively, into the XbaI /BamHI restriction sites of pCDH-CMV-MCS-EF1-copGFP vector (System Biosciences, USA). These fragments were amplified by PCR reaction using following primers: pri-miR-451 FW: 5′- GGAAGATCTTGACAAGGAGGACAGGAGAG -3′, pri-miR-451 RW: 5′- CCCAAGCTTGCCTTGTTTGAGCTGGAGTC-3′; pri-miR-16 FW: 5′- ACTCTAGAGCAGCACATAATGGTTTG-3′ and pri-miR-16 RW: 5′-TGGATCCCTCTAATGCTGCATAAGC- 3′ on genomic DNA extracted from human whole blood (Cinnagen, Iran). lentivirus production; human embryonic kidney (HEK) 293 cells (3 ×106) were seeded into 10-cm plates containing DMEM medium supplemented with 10% FBS(purchased from Gibco). The next day, pPAX2 plasmid (containing gag and pol genes) and pMD2 plasmid (containing vsv gene) were co-transfected with pCDH-451/ pCDH-16 plasmids also empty vector (pCDH empty backbone) as negative control into seeded 293T cells using lipofectamin 2000 reagent (Invitrogen, USA) according to manufacturer’s protocol. The supernatants containing produced lentiviruses were collected every 12 hours for 2 days after transfection and concentrated by ultracentrifuge at 40.000g for 2 hr. Then for virus titration, 293T cells were transduced with a different concentration of recombinant lentiviruses and the number of viruses in the functional copy was determined using GFP protein and fluorescent microscope forty-eight hours later.
CD133+ cells isolation, culture and infection
Human umbilical cord blood (UCB) was obtained from 3 healthy full-term placentas. Written permission was obtained from healthy pregnant women. Next, to purify CD133+ stem cells, MNCs (mononuclear cells) were isolated from UCB samples by density gradient centrifugation using Ficoll-paque reagent (GE Healthcare, Sweden). CD133+ cells were enriched through positive selection using CD133 magnetic micro-beads and MidiMACS separation systems (Miltenyi Biotec, Germany) as per manufacturer's recommendations. Enriched CD133+ stem cells were expanded into StemSpan media (Stem Cell Technologies, Canada) in the presence of 100ng/mL stem cell factor (SCF), 100 ng/mL thrombopoietin (TPO) and 100ng/mL Flt3-ligand (Stem Cell Technologies) for 5 days. For gain of function studies, expanded CD133+ cells were transferred at density of 1×105 cells per well in 24-well plates containing IMDM media (purchased from Gibco) supplemented with only 5% FBS in the absence of any erythroid stimulatory growth factors and cytokines. The next day, CD133+ cells were infected with a multiplicity of infections (MOI of 30, It means that 30 lentivirus particles were added per one CD133+ stem cell).
The study was performed in four groups: one group was transduced with pCDH-451 lentiviruses (pCDH-451 group), another was infected with pCDH-16 lentiviruses (pCDH-16 group), the third did not receive any treatment (blank control group) and the fourth group was transduced with pCDH-empty lentiviruses (negative control group). After 14 days, the role of miR-16 and miR-451 up-regulation in erythroid differentiation was monitored by hemoglobin chains (α, β, γ and ζ) RT-PCR, erythroid cell surface markers (CD71 and CD235a) RT-PCR and banzidine staining.
RNA extraction and RT-PCR
Total RNA was extracted from test and control groups using Biozol reagent (Bioer,China ) on day 14 post infection. The first strand cDNA was synthesized with AMV reverse transcriptase enzyme from 100 ng of total RNA using cDNA synthesis kit (Bioer) according to manufacturer’s instructions. Next, RT-PCR reactions were performed for hemoglobin genes (α, β, γ and ζ) and erythroid specific cell surface markers (transferrin receptor, CD71:120 bp and glycophorinA, CD235a:167 bp) as erythroid differentiation indexes using following primers: α-globin (annealing temperature: 59ْC, Product size:407 bp): FW, 5′- CCGACAAGACCAACGTCAAGG-3′ and RW, 5′-GGTATTTGGAGGTCAGCACG-3′; β-globin (annealing temperature: 60ْC, Product size:122 bp): FW, FW, 5′-CTCACCTGGACAACCTCAAG -3′; and RW, 5′-TACCCGGTCGTGTGTCTGGT-3′; γ-globin (annealing temperature: 54ْC, Product size: 194 bp): FW, GTCCTCTGC-CTCTGCCATC -3′ and RW, 5′-CGGTCACCAGCACATTTCC-3′; ζ-globin (annealing temperature: 55ْC, Product size: 208 bp): FW, 5′- GGTGAAGAGCATCGACGACA-3′ and RW, 5′-TCTCGGTCAGGACAGAGGA-3′; CD71 (annealing temperature: 60ْC, Product size: 120 bp): FW, 5′-TGAGGTGGCAATGCACACTTC-3′ and RW, 5′- AGGAGTGGCTGCATATGTGTCC-3′; CD235a (annealing temperature: 60ْC , Product size: 167 bp): FW, 5′- GGCTAAGGTCAGACACTGAC-3′ and RW, 5′- TGTGCATTGCCACCTCAGTG-3′; GAPDH (annealing temperature: 58ْC, Product size: 166 bp): FW, 5′- GACAAGCTTCCCGTTCTCAG -3′ and RW, 5′- GAGTCAACGGATTTGGTCGT.
miRNA-qRT- PCR
Total RNA was extracted from all samples at day 4. Then, qRT-PCR for miR-16 and miR-451 were performed using Stratagene kit (USA) according to manufacturer’s protocol. Briefly, first cDNA strand was synthesized through miRNA 1st-strand cDNA synthesis kit (Stratagene) and reverse transcribed into qPCR-ready cDNA. After that, miRNA qRT-PCR analysis was carried out in triplicate on ABI PRISM 7500 real time PCR System (Applied Biosystems, USA) with the high-specifity miRNA qPCR core reagent kit (Stratagene) using following miRNA-specific forward primers: miR-451 FW, 5-′ AAACCGTTACCATTACTGAGATT-3′ and miR-16 FW: 5′- AGCAGCACGTAAATATTGGC-3′ according to manufacturer’s instructions and normalized to U6 small nuclear RNA (snRNA) as endogenous control. U6 primers were: FW: 5′: CTCGCTTCGGCAGCACACATATAC-3′, RW: 5′- ACGCTTCACGAATTTGCGTGTC-3′. The qRT-PCR cycling conditions were 10 minutes at 95°C followed by 10 sec at 95°C, 15 sec at 60°C, and 20 sec at 72°C, repeated for 30 step cycles. Data analyses were performed using the relative quantification of CT method.
Banzidine staining
Banzidine staining was used for calculation of percentage of cells which express hemoglobin chains. For producing benzidine solution, 5 mL of 30% hydrogen peroxide was added to 1 mL of a stock solution of 0.2% benzidine dissolved in 0.5% acetic acid. A number of 105 cells from each test and control groups were resuspended in phosphate buffered saline (PBS) and mixed with the same volume of the benzidine solution for 7 min in room temperature. Then, each sample was monitored by light microscopy and the blue cells (benzidine-positive cells) were counted and expressed as a percentage. 100 cells were counted in each sample and all experiments were performed triplicate.
Statistical analysis
All experiments were repeated three times and data were presented as mean±SD. The comparison between groups was performed by Student's t- test. P- value less than 0.05 designated statistically significant differences.
Results
Erythroid differentiation of CD133+ cells
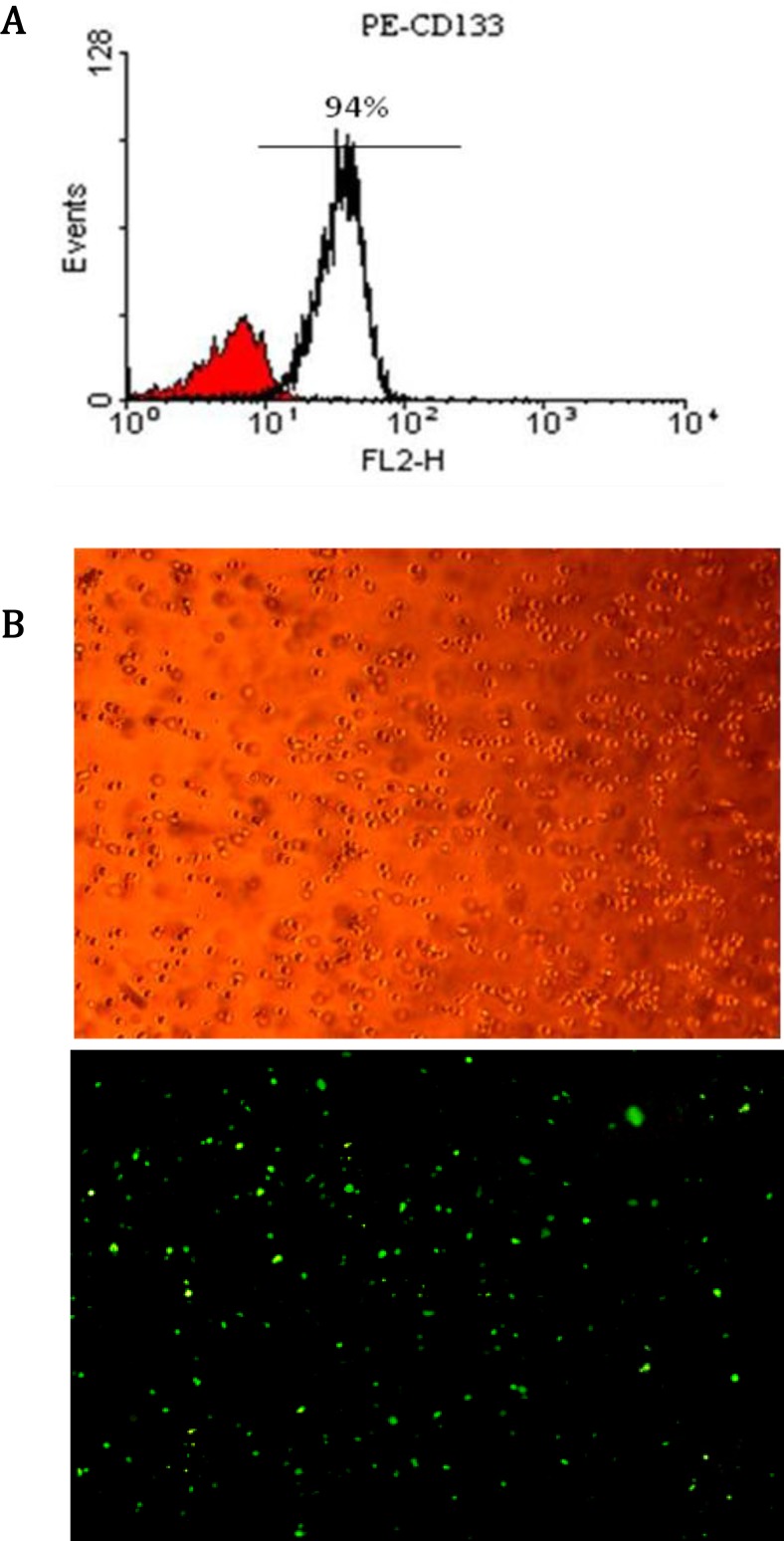
CD133+ stem cells were isolated successfully from each cord blood sample. The purity rate of isolated cells was measured by flow cytometry (Figure 1, A). Quantities over than 90% were selected for further studies. Then, CD133+ stem cells were successfully transduced by lentiviral vectors. Transduction efficiency was checked each time by fluorescent microscopy and determined about 70% (Figure 1, B).
Figure 1.
CD133+ stem cell purification and infection. A: Result of CD133+ cells purification from HCB by flow cytometry. In each case, purification efficiency was above 90%. B: (B, above) Transduced CD133+ cells examined by light microscopy. (B, below Transduced CD133+ cells examined by fluorescent microscopy. Transduction efficiency of CD133+ cells with pCDH-16 and pCDH-451 was obtained approximately more than 70% as determined by fluorescent microscopy
Recombinant lentiviruses increased mature miRNAs level in treated CD133+ cells
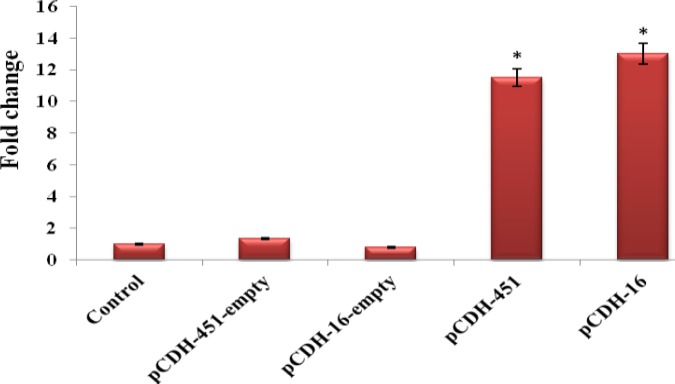
To determine the efficacy of recombinant lentiviruses, we measured the expression level of miR-16 and miR-451 at day 4 after transduction in test and control groups by qRT-PCR. Treatment of CD133+ cells with pCDH-16 lentiviruses, but not with pCDH-empty lentiviruses, led to rise of mature miR-16 by 13-fold relative to the blank control cells 4 days upon transfection. Similar results were obtained when CD133+ cells were infected with pCDH-451 lentiviruses. Over expression of miR-451 in pCDH-451 group enhanced mature miR-451 by 11.5-fold compared with blank control group. As expected, when CD133+ cells were treated with pCDH-empty lentiviruses, miR-451 expression level showed no significant alteration compared to blank control group (P> 0.05, Figure 2). These results indicate that both recombinant lentiviruses are functional and increase mature miRNAs level appreciably.
BFigure 2.
Expression of miR-16 and miR-451 in test and control groups was measured by QRT-PCR at day 4. The expression level of both miRNAs was assessed in the group that infected with pCDH-empty group. All tests were performed in triplicate and data were presented as mean ± SD. Error bars designate SD. * P<0.05
pCDH-451 lentiviruses, but not pCDH-16 lentiviruses, could induce erythroid differention of CD133+ cells
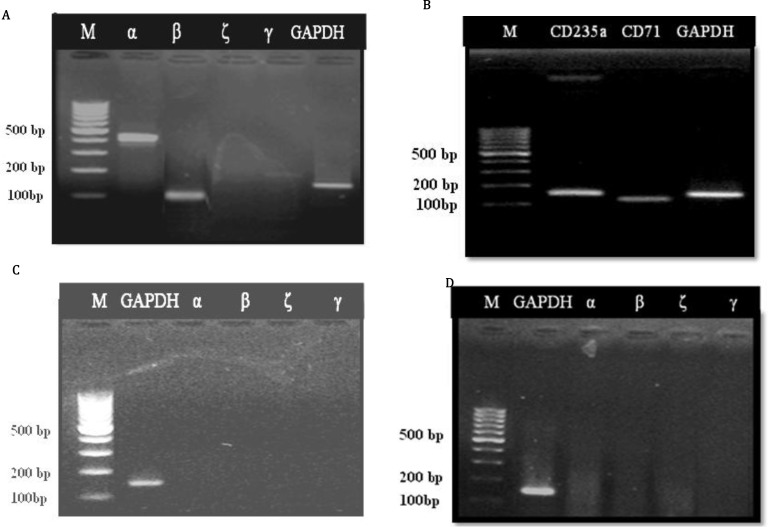
Because miR-16 and miR-451were up-regulated drastically during erythroid differentiation, we sought to determine whether these miRNAs could induce erythropoiesis individually and in the absence of any erythroid stimulatory growth factors and cytokines. So, we evaluated the effect of mentioned miRNAs upregulation on expression of erythroid specific markers. CD133+ cells were infected with pCDH-451, pCDH-16 or pCDH-empty lentiviruses and erythroid differentiation was assessed by RT-PCR for hemoglobin chains and cell surface markers (Figure 2A and B). Over expression of the miR-451 induced expression of α, β and γ- globin, but not the expression of the ζ- globin.
Noteworthy, γ- globin was expressed at lesser extent than α and β chains. However, miR-16 up-regulation in pCDH-16 group had no effect on the hemoglobin chains expression. Similarly, miR-451 up-regulation led to stimulation of CD71 and CD235a expression. Nevertheless, miR-16 up-regulation did not positively effect the expression of either cell surface marker. Since, β-globin and CD235a genes are the important markers of maturing human erythroid cells, these findings suggest that miR-451 up-regulation, but not miR-16 up-regulation, is able to promote erythroid differentiation of the HSCs and even its maturation without adding any stimulatory factors.
miR-451 up-regulation correlated with banzidine positive cells
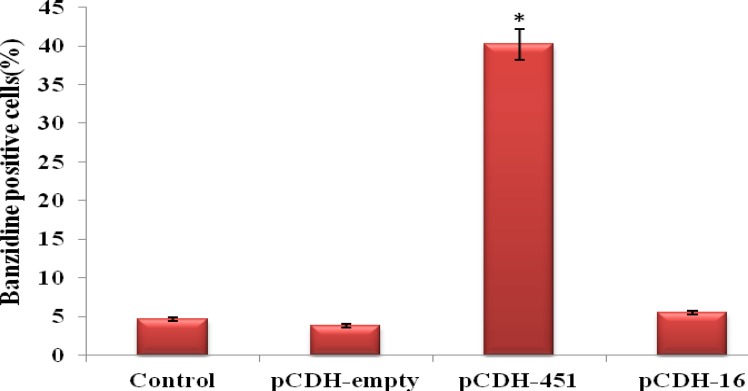
Erythroid differentiation and hemoglobinization were also examined by banzidine staining (Figure 3). MiR-451 over expression significantly increased the proportion of the cells which were benzidine-positive in pCDH-451 group versus control group at day 14 (40.2% vs. 4.7%, respectively), whereas miR-16 up-regulation had no obvious effect on the hemoglobinization (only 5.5% cells was benzidine-positive). Unsurprisingly, no significant alteration was seen between the percentage of the cells that were positive for banzidine staining in pCDH-empty group and blank control group (3.8% vs. 4.7%, respectively, P> 0.05). Taken together, these results further confirmed that miR-451 up-regulation contribute to erythroid differentiation of CD133+ cells and identify this miRNA as one of the main factors that induces hemoglobinization.
Figure 3.
RT-PCR results of hemoglobin chains and surface CD markers expression in pCDH-451 group (A and B), control group (C) and pCDH-16 group (D). α-, β-, γ-globin, CD71 and CD235a were expressed in pCDH-451 group, but not in pCDH-16 group and control group. Product size of α: 407 bp, β: 122 bp, γ: 194 bp, ζ: 208 bp, GAPDH: 166 bp, CD71: 120 bp, CD235a: 167 bp, Marker (M): 100 bp
Discussion
MicroRNAs (miRNAs) are small, non–protein-coding RNAs that recognize target sites, in the 3′ untranslated regions (UTRs) of cognate mRNAs, through imperfect base-pairing, and either destabilize them or inhibit protein translation. miRNAs is involved in different processes in multi-cellular plant and animal species but not in unicellular organisms including cell development, fat storage, apoptosis and differentiation (29-34). Essential roles of miRNAs also were identified in hematopoietic lineage differentiation (23-25). For instance, Bruchova et al (2007) performed gene-expressing profiling using microarray method and indicated that miR-16 and miR-451 up-regulated by 35-fold and 3-fold, respectively, throughout the erythroid differentiation (35-37).
Figure 4.
Evaluation of hemoglobinization using banzidine staining method. Infection of CD133+ cells with miR-451 increased banzidine positive cells to ~ 40%. In other group no obvious changes were observed. Results indicate the mean ± standard deviation of three independent experiments. Error bars represent SD. * P<0.05
In the present study, to determine whether miR-16 and miR-451 could play central roles in erythroid differentiation in the absence of cytokines and growth factors combination, we analyzed the effect of enforced expression of these miRNAs on erythroid differentiation of CD133+ cells. RT-PCR results indicated that, when cells were treated with pCDH-451 lentiviruses, miR-451 up-regulation persuaded the erythroid surface markers CD71 and CD235a. In addition miR-451 overexpression induced hemoglobin α and β expression at high level, whereas miR-451 stimulated γ-globin expression at low level. Findings demonstrate that in this model, adult β-globin (Hb A), with negligible fetal globin (Hb F) was produced that mimics in vivo erythropoiesis. Our results are consistent with a report that states up-regulation of miR-451, increased β-globin and hemoglobinization in MEL cells during DMSO-induced erythroid differentiation (19). Moreover, our data showed that the up-regulation of miR-451caused a significant rise in CD133+ hemoglobinization (~ %40). These findings suggest that miR-451 up-regulation alone and without adding any auxiliary factors could induce erythroid differentiation of CD133+ cells. However, miR-451 target(s) are not yet identified in humans (25).
On the other hand, transduction of the cells with pCDH-16 lentiviruses could not induce hemoglobin and surface antigen genes expression in CD133+ cells. Our observations seem to be in disagreement with a previous study by Choong et al who demonstrated that miR-16 have strong positive correlation to the appearance of erythroid specific cell surface markers such as CD36, CD71 and CD235a and hemoglobin synthesis upon erythroid differentiation of CD34+ cells using cytokines combinations (27). These dissimilarities may be explained by differences in the method of erythropoietic induction of the stem cells and the source of progenitor cells. Thus, up-regulation of miR-16 on their own and without stimulatory cytokines assistance did not promote erythroid differentiation of the CD133+ cells.
Conclusion
Taken together, our evidence suggests that miR-451 up-regulation maybe a critical factor for in vitro erythropoiesis of HSCs and production of artificial blood. On the other hand, in the hemoglobinopathies such as sickle cell anemia and thalassemia, the major problem is failure in the production of adult globin (HbA) (38). Thus, erythropoiesis using miR-451 and other miRNAs may be useful in studying the pathophysiology of hemoglobinopathies and test effective therapeutic strategies for the possibility of reversing these abnormalities by gene therapy. However, it is clear that for clinical applications, complete process of in vitro erythropoiesis using miRNAs expression modulation should be proved functionally and morphologically. Hence, present study is the primary step in this field and further studies would be necessary.
Acknowledgment
Acknowledgment
This research is financially supported by a grant from Tarbiat Modares University, Tehran, Iran.
References
- 1.Chang TM. Red blood cell substitutes. Baillieres Best Pract Res Clin Haematol. 2000;13:651–667. doi: 10.1053/beha.2000.0105. [DOI] [PubMed] [Google Scholar]
- 2.Kim HW, Greenburg AG. Artificial oxygen carriers as red blood cell substitutes: A selected review and current status. Artif Organs. 2004;28:813–828. doi: 10.1111/j.1525-1594.2004.07345.x. [DOI] [PubMed] [Google Scholar]
- 3.Jahr JS, Walker V, Manoochehri K. Blood substitutes as pharmacotherapies in clinical practice. Curr Opin Anaesthesiol. 2007;20:325–330. doi: 10.1097/ACO.0b013e328172225a. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman DS. Toward clinical therapies using hematopoietic cells derived from human pluripotent stem cells. Blood. 2009;114:3513–3523. doi: 10.1182/blood-2009-03-191304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mountford J, Olivier E, Turner M. Prospects for the manufacture of red cells for transfusion. Br J Haematol . 2010;149:22–34. doi: 10.1111/j.1365-2141.2010.08079.x. [DOI] [PubMed] [Google Scholar]
- 6.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: Model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61:800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- 7.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 8.Scicchitano MS, McFarland DC, Tierney LA, Narayanan PK, Schwartz LW. In vitro expansion of human cord blood cd36+ erythroid progenitors: Temporal changes in gene and protein expression. Exp Hematol. 2003;31:760–769. doi: 10.1016/s0301-472x(03)00185-1. [DOI] [PubMed] [Google Scholar]
- 9.Goh SH, Josleyn M, Lee YT, Danner RL, Gherman RB, Cam MC, et al. The human reticulocyte transcriptome. Physiol Genomics. 2007;30:172–178. doi: 10.1152/physiolgenomics.00247.2006. [DOI] [PubMed] [Google Scholar]
- 10.Olivier EN, Qiu C, Velho M, Hirsch RE, Bouhassira EE. Large-scale production of embryonic red blood cells from human embryonic stem cells. Exp Hematol. 2006;34:1635–1642. doi: 10.1016/j.exphem.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Fujimi A, Matsunaga T, Kobune M, Kawano Y, Nagaya T, Tanaka I, et al. In vivo large-scale generation of human red blood cells from cord blood cd34+ cells by co-culturing with macrophages. Int J Hematol. 2008;87:339–350. doi: 10.1007/s12185-008-0062-y. [DOI] [PubMed] [Google Scholar]
- 12.Ma F, Ebihara Y, Umeda K, Sakai H, Hanada S, Zhang H, et al. Generation of functional erythrocytes from human embryonic stem cell-derived definitive hematopoiesis. Proc Natl Acad Sci U S A . 2008;105:13087–13092. doi: 10.1073/pnas.0802220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anstee DJ. Production of erythroid cells from human embryonic stem cells (hesc) and human induced pluripotent stem cells (hips) Transfus Clin Biol. 2010;17:104–109. doi: 10.1016/j.tracli.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Giarratana MC, Kobari L, Lapillonne H, Chalmers D, Kiger L, Cynober T, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 15.Chen CZ, Lodish HF. Micrornas as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Shi B, Prisco M, Calin G, Liu CG, Russo G, Giordano A, Baserga R. Expression profiles of micro rna in proliferating and differentiating 32d murine myeloid cells. J Cell Physiol. 2006;207:706–710. doi: 10.1002/jcp.20613. [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. Microrna fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgantas RW 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, and Civin CI. Cd34+ hematopoietic stem-progenitor cell microrna expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song CZ. Microrna expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garzon R, Croce CM. Micrornas in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 21.Merkerova M, Belickova M, Bruchova H. Differential expression of micrornas in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen CZ, Li L, Lodish HF, and Bartel DP. Micrornas modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 23.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. Micrornas 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruchova H, Yoon D, Agarwal AM, Mendell J, Prchal JT. Regulated expression of micrornas in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35:1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruchova-Votavova H, Yoon D, Prchal JT. Mir-451 enhances erythroid differentiation in k562 cells. Leuk Lymphoma. 2010;51:686–693. doi: 10.3109/10428191003629362. [DOI] [PubMed] [Google Scholar]
- 26.Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, Jun-Shen Huang L, Olson EN. Defective erythroid differentiation in mir-451 mutant ice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614–9. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choong ML, Yang HH, and McNiece I. Micrornaexpression profiling during human cord blood-derived cd34 cell erythropoiesis. Exp Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Masaki S, Ohtsuka R, Abe Y, Muta K, and Umemura T. Expression patterns of micrornas 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509–514. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 29.Cullen BR. Derivation and function of small interfering rnas and micrornas. Virus Res. 2004;102:3–9. doi: 10.1016/j.virusres.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Bushati N, and Cohen SM. Microrna functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, and Zhang GZ. Biological functions of micrornas: A review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 32.He L, and Hannon GJ. Micrornas: Small rnas with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 33.Filipowicz W, Bhattacharyya SN, and Sonenberg N. Mechanisms of post-transcriptional regulation by micrornas: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 34.Nilsen TW. Mechanisms of microrna-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Yang GH, Wang F, Yu J, Wang XS, Yuan JY, and Zhang JW. Micrornas are involved in erythroid differentiation control. J Cell Biochem. 2009;107:548–556. doi: 10.1002/jcb.22156. [DOI] [PubMed] [Google Scholar]
- 36.Havelange V, and Garzon R. Micrornas: Emerging key regulators of hematopoiesis. Am J Hematol. 2010;85:935. doi: 10.1002/ajh.21863. [DOI] [PubMed] [Google Scholar]
- 37.Lawrie CH. Microrna expression in erythropoiesis and erythroid disorders. Br J Haematol. 2010;150:144–151. doi: 10.1111/j.1365-2141.2009.07978.x. [DOI] [PubMed] [Google Scholar]
- 38.Persons DA. Gene therapy: Targeting beta-thalassaemia. Nature. 2010;467:277. doi: 10.1038/467277a. [DOI] [PubMed] [Google Scholar]