Abstract
Background
The herpes simplex virus (HSV-1) is a virus that manifests itself in viral infection with painful, watery blisters in the skin or on the genitals as well as mucous membrane such as the mouth or lips. During an outbreak, the disease is contagious particularly and is irredeemable with present technology. Genetic studies of HSV-1 have shown that ICP22 (US1) gene is an immediate early gene and is responsible for genome replication and also has contribution in viral infection.
Method
For disease diagnosis, ICP22 (US1) gene may be suitable target. Viral activity can be controlled through RNA interference technology, a significant method for the post-transcriptional gene silencing. However, in different viral isolates there is a genetic variability; it is very challenging to design possible siRNA molecules which can silence the respective target genes. The work was done by using various computational tools as similarity search, target alignment, secondary structure prediction and RNA interaction evaluation.
Result
In our study two effective siRNA molecules for ICP22 (US1) gene silencing of seven different strains of HSV-1 were rationally designed and authenticated using computational methods, which might lead to knockdown the viral activity.
Conclusion
siRNA molecules were foreseen against ICP22 (US1) gene of different strains of HSV-1 as effective aspirant using computational methods. Thus, the approach may deliver a vision for the chemical synthesis of antiviral RNA molecule for treatment of HSV-1, at genomic level.
Keywords: HSV-1, Antiviral, ICP22 (US1) gene, RNAi, siRNA
1. Introduction
Herpes simplex virus (HSV) can infect both human and animal but only human shows symptom of diseases like cold sore or fever blister, so they are mainly called human herpes simplex.1 HSV mainly divided in two major types; they are HSV-1 and HSV-2. At present there is no mentionable cure but some treatments are available for HSV-1.2 Generally this HSV-1 is transmitted by contact with infected area.3 Their entry into the host cell occurs through surface glycoprotein.4 After infection a cascade of herpes viral protein called immediate early gene, early and late gene are produced. Immediate early genes encode protein that regulates the early and late gene expression.5 ICP22 (US1), an immediately early gene found in different strains of HSV-1, plays significant role in transcription of early and late genes.6 Small interfering RNA (siRNA) is a double stranded RNA with 21 base pairs in length. The main role of siRNA is post transcriptional gene silencing.7 By using nanoparticles like polyethyleneimine and other methods, siRNA can be introduced into the cell for knockdown of a gene of interest.8 siRNA incorporates with RNAi induced silencing complex (RISC) which seeks out the target mRNA. Ultimately antisense strand of siRNA directs degradation of target mRNA using different enzymes. siRNA may also be used for therapeutic purpose like vaccine and chemical drugs.9 Our present study is aimed to design a potential siRNA for inhibiting the translation of this ICP22 (US1) gene and hinder the overall replication process.
2. Materials and methods
2.1. Sequence retrieval and analysis
Seven complete cds of ICP22 (US1) genes of different strains of human herpes simplex virus 1 were collected from the viral gene bank database available at National Centre for Biotechnological Information (http://www.ncbi.nlm.nih.gov/). The accession numbers of these seven complete cds are JF511475.1, JF511471.1, JF511473.1, JF511469.1, JF511470.1, JF511474.1 and JF511472.1 for different strain named TFT401, OD4, KOS, CJ394, CJ311, 994 and 970 of human herpes simplex 1 respectively.
2.2. Multiple sequence alignment
All the seven complete cds were aligned with each other using clustalW (http://www.genome.jp/tools/clustalw/).
2.3. Target identification and potential siRNA designing
For target identification and designing of potential siRNA siDirect 2.0 was used. It is available on website (http://siDirect2.RNAi.jp/). It considers some rules as parameter like Ui-Tei, Amarzguioui, Renold rules and melting temperature (Tm) should be below 21.5 °C for potential siRNA duplex. Dharma siRNA technology (http://www.dharmacon.com/designcentre/) and GeneScript siRNA target finder (http://www.genescript.com/index.html) was used for confirmation of predicted molecules. Besides these other factors were also taken on the concept of algorithms [Table 1].
Table 1.
Algorithms or rules for rational design of siRNA molecules.
| Ui-Tei rules | Amarzguioui rules | Reynolds rules |
|---|---|---|
| A/U at the 5′ terminus of the sense | Duplex end A/U differential >0 | Each rule is assigned a score which is summed up to a total duplex score to improve the efficiency of siRNA. |
| Strand | Strong binding of 5′ sense strand | |
| G/C at the 5′ terminus of the anti- | No U at position 1. Presence of A at position 6. | |
| Sense strand | ||
| At least 4 A/U residues in the 5′ terminal | Weak binding of 3′ sense strand. No | |
| 7 bp of sense strand. | G at position 19 | |
| No GC stretch longer than 9 nt |
2.4. Similarity search and target alignment
For checking any off target sequence resemblance in genome of other non-targeted organisms, blast tool (http://www.ncbi.nlm.nih.gov/blast) was used against whole Genebank database by applying expected thresholds value 10 and BLOSUM 62 matrix as parameter. MSA (http://www.ebi.ac.uk/Tools/msa/clustalw2/) of these selected siRNA targets showed that this sequence is divided into two groups.
2.5. GC calculation and secondary structure prediction
For GC content calculation of predicted siRNA, DNA/RNA GC content calculator (http://www.endmemo.com>Tools>Biology) was used while mfold server (http://www.mfold.rna.albany.edu/) was used for secondary structure prediction aimed to compute the free energy of folding.
2.6. Calculation of RNA-RNA interaction through thermodynamics
To study the thermodynamics of interaction between predicted siRNA and target gene, RNAcofold program (http://rna.tbi.univie.ac.at/cgi-bin/RNAcofold.cgi) was used. It calculates the hybridization energy and base-pairing form of two RNA sequences. It functions as extension of McCaskill's partition function algorithm to compute probabilities of base pairing, realistic interaction energies and equilibrium concentrations of duplex structures. Flow chart shows the complete approach used for screening of effective siRNA molecules in this study [Fig. 1].
Fig. 1.
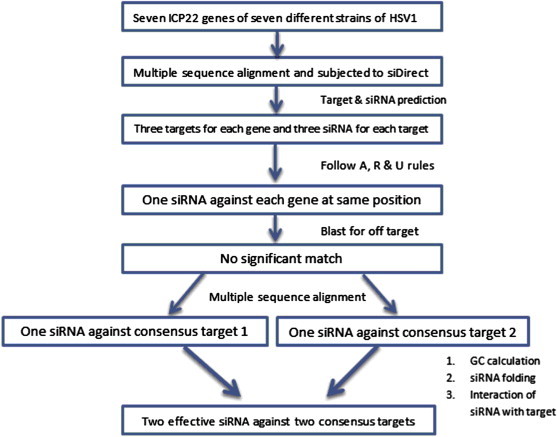
Flow chart showing the complete approach used for screening of effective siRNA molecules in this study.
3. Result and discussion
This study was conducted with ICP22 (US1) gene from seven different strains of human herpes simplex 1 virus. Gene sequences available in the viral gene bank database from NCBI were taken and analyzed their similarity using clustalW which specified a great similarity among seven gene sequences. The similarity among sequences was in a range of 98-100% and a phylogenetic tree was established to observe the evolutionary relationship among these seven strains [Fig. 2]. Then all seven sequences were subjected to siDirect to produce efficient, target specific siRNA with reduced probability of off target silencing. Individual ICP22 (US1) gene from seven different strains showed three target sequences at the same position of each gene. siDirect provided putative siRNA maintaining all rules of Ui-Tei, Amarzguioui and Reynolds. siRNA that follows all the rules and algorithms of Ui-Tei, Amarzguioui and Reynolds are supposed to be most effective.10 According to siDirect result three siRNA against three target sequences of individual gene was found and one siRNA for each gene was found that follows all rules of Ui-Tei, Amarzguioui and Reynolds. MSA was performed to classify these siRNA targets into groups and to design a common siRNA against more than one target. Multiple sequence alignment of all seven siRNA targets produced a result, in which five siRNA targets consist of identical sequences which we identified as consensus siRNA target 1 [Table 2] and remaining two are also identical to each other identified as consensus siRNA target 2 [Table 3]. Sequence alignment of these two consensus groups of siRNA target with clustalW2 depicted that there is only one mismatch between these two siRNA target groups. Alignment of predicted siRNA with clustalW2 also revealed one base pair mismatch almost at the middle [Fig. 3]. For the seed target duplex, Tm was calculated and also thermodynamics parameters for RNA duplex formation were studied because Tm should be less than 21.5 °C.11 The formulation for calculating Tm is:
| (1) |
where ΔH (kcal/mol) is the amount of the nearest neighbor enthalpy change, A is the helix initiation constant (−10.8), ΔS are the quantity of the nearest neighbor entropy change, R is the gas constant (1.987 cal/deg/mol), and CT is the entire molecular concentration of the strand (100 μM). [Na+] was fixed at 100 mM.
Fig. 2.
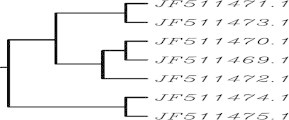
Phylogenetic tree for ICP22 (US1) gene of seven different strains of human herpes simplex virus 1.
Table 2.
Predicted siRNA target for ICP22 (US1) gene-consensus target 1.
| Accession no. | Target | Location of target within the gene | siRNA target sequence within gene |
|---|---|---|---|
| JF511470.1 | Target 1 | 20-42 | GCGCTTTTGCGCCTTGTGTAAAA |
| JF511469.1 | Target 2 | 20-42 | GCGCTTTTGCGCCTTGTGTAAAA |
| JF511472.1 | Target 3 | 20-42 | GCGCTTTTGCGCCTTGTGTAAAA |
| JF511471.1 | Target 4 | 20-42 | GCGCTTTTGCGCCTTGTGTAAAA |
| JF511474.1 | Target 5 | 20-42 | GCGCTTTTGCGCCTTGTGTAAAA |
Table 3.
Predicted siRNA target for ICP22 (US1) gene-consensus target 2.
| Accession no. | Target | Location of target within the gene | siRNA target sequence within gene |
|---|---|---|---|
| JF511473.1 | Target 1 | 20-42 | GCGCTTTTGTGCCTTGTGTAAAA |
| JF511475.1 | Target 2 | 20-42 | GCGCTTTTGTGCCTTGTGTAAAA |
Fig. 3.
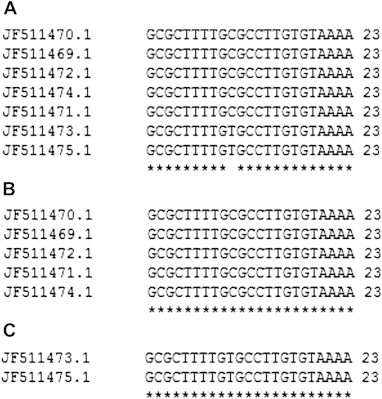
Multiple sequence alignment of all predicted siRNA target sequence. (A) All aligned siRNA sequences; (B) consensus target 1; (C) consensus target 2.
Apart from this, to plaid the precision of results GeneScript target Finder and Dharma siRNA technology was also applied.7 siDirect selects siRNAs with minimum Tm value at the seed region, that contains 7 nucleotides at positions 2-8 from 5′ end of the guide strand. The siRNA targeted region on the mRNA sequence of a gene should not share significant homology with other genes or sequences in the human genome, so off target silencing is a great problem in designing of a potential siRNA.12 Result of siDirect defines no chances for off target silencing, further it was clarified by subjecting the target sequences in Blast similarity search from NCBI. Two siRNA molecules against ICP22 (US1) gene from seven different strains of human herpes simplex virus 1 were used for next study with different parameters to determine their perfection. The GC content of the siRNA is an apparent contender for a parameter that might correlate with siRNA functionality. There is a connection between target site accessibility and GC content. It is generally recommended to pick sequences with low GC content (between 31.6% and 57.9%), because there is a considerable negative correlation between GC-content and RNAi activity. In our study GC content of predicted siRNA was 52% for consensus siRNA target 1 and 47% for consensus siRNA target 2 which indicates the feasibility of two siRNA.13 For measuring the stability of structure of the guide strand, the minimum free energy (MFE) of the optimal folding was computed with mfold. mfold follows the most broadly used algorithms for the prediction of RNA secondary structure, which are based on a quest for the minimal free energy state. Here, both siRNA molecules are having zero free energy of folding at 37 °C [Table 4]. Previous studies have suggested that an RNA molecule must have smallest free energy of folding for their stability.14 Therefore, the molecule with negative energy may have lower accessibility for target site. For prediction of thermodynamics of RNA-RNA interaction which is another parameter for siRNA efficiency both siRNA was subjected to Vienna RNA webservers. The Vienna web server is an ample collection of programs, web services, tools that offer algorithms for RNA folding, assessment and prediction of RNA-RNA interactions and databases, related to our work on RNA secondary structures. The RNAcofold web server from Vienna RNA webservers used to figures the hybridization energy and base-pairing pattern of two RNA sequences. The two sequences are concatenated and the point of concatenation is specified by an ampersand. In addition to ΔGAB, the free energy of the heterodimer of sequence A and sequence B can be calculated using the equation ΔGbinding = ΔGAB − ΔGA − ΔGB (Equation 2).15
Table 4.
Two effective siRNA molecule with GC%, free energy of folding and free energy of binding with target.
| Target | Location of target within mRNA | siRNA target within consensus target | Predicted siRNA duplex siRNA candidate at 37 °C | GC% | Free energy of folding with target | Free energy of binding |
|---|---|---|---|---|---|---|
| Consensus Target 1 |
20-42 | GCGCTTTTGCGCCTTGTGTAAAA | UUACACAAGGCGCAAAAGCGC GCUUUUGCGCCUUGUGUAAAA |
52 | 0.00 | −27.16 |
| Consensus Target 2 |
20-42 | GCGCTTTTGTGCCTTGTGTAAAA | UUACACAAGGCACAAAAGCGC GCUUUUGUGCCUUGUGUAAAA |
47 | 0.00 | −31.78 |
So, for the interaction of consensus target 1 and its predicted siRNA, free energy of binding is
For consensus siRNA target 2 the calculation is
| (2) |
All parameter used; support the efficiency of siRNA against their target. So it can be a potential way for these two siRNA to play role in advance treatment method against seven different strains of human herpes simplex 1 virus. This finding is helpful to meet the demand of same treatment against different strains of same virus or against different viruses.
The advance approach, RNAi technology for post-transcriptional silencing of specific gene is successfully adopted in several cases such as colorectal cancer therapeutics, therapeutics of hepatitis C virus,16 for CNS disorder,17 siRNA therapeutics can now possible in metabolic diseases of liver and are being evaluated in clinical trials for hypercholesterolemia.18 Synthetic siRNAs targeted against the viral structural Env proteins produced by HIV-1 can specifically suppress the expression of HIV-1 genes. Therefore, the use of synthetic siRNAs provides a rapid and cost-effective tool for new anti-HIV-1 gene therapeutics.19 The ability of siRNA to control ischemia reperfusion injury related transcription factors, apoptosis, oxidative stress molecules, and complement factors supports the observation that RNAi-based therapeutics epitomize a novel and promising strategy for the control of IRI.20
4. Conclusion
With RNAi technology it is possible to design a number of siRNA molecules for silencing of substantial genes in numerous biological systems. Their interactions with target can also be calculated computationally. Hence, in our study two siRNA molecules were foreseen against ICP22 (US1) gene of different strains of HSV-1 as effective aspirant using computational methods. These molecules may prime to a novel antiviral therapy against HSV-1. The outcome of this study would also afford a foundation to the researchers and pharmaceutical industry to develop antiviral therapeutics at genomic level.
Conflicts of interest
All authors have none to declare.
References
- 1.Huff J., Barry P. B-virus (Cercopithecine herpesvirus 1) infection in humans and macaques: potential for zoonotic disease. Emerg Infect Dis. 2003;9:246–250. doi: 10.3201/eid0902.020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGeoch D.J., Rixon F.J., Davison A.J. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Koelle D.M., Corey L. Herpes simplex: insights on pathogenesis and possible vaccines. Annu Rev Med. 2008;59:381–395. doi: 10.1146/annurev.med.59.061606.095540. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian R.P., Geraghty R.J. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matis J., Kúdelová M. Early shutoff of host protein synthesis in cells infected with herpes simplex viruses. Acta Virol. 2001;45:269–277. [PubMed] [Google Scholar]
- 6.Rajcáni J., Andrea V., Ingeborg R. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes. 2004;28:293–310. doi: 10.1023/b:viru.0000025777.62826.92. [DOI] [PubMed] [Google Scholar]
- 7.Hoque K.M., Azim M.F., Mia M.R. Design of potential siRNA molecules for T antigen gene silencing of Merkel Cell Polyomavirus. Bioinformation. 2012;8:924–930. doi: 10.6026/97320630008924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saengkrit N., Sanitrum P., Woramongkolchai N. The PEI-introduced CS shell/PMMA core nanoparticle for silencing the expression of E6/E7 oncogenes in human cervical cells. Carbohydr Polym. 2012;90:1323–1329. doi: 10.1016/j.carbpol.2012.06.079. [DOI] [PubMed] [Google Scholar]
- 9.Geall A.J., Verma A., Otten G.R. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA. Sep 4, 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taxman D.J., Livingstone L.R., Zhang J. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ui-Tei K., Naito Y., Nishi K., Juni A., Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008;36:7100–7109. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 13.Chan C.Y., Carmack C.S., Long D.D. A structural interpretation of the effect of GC-content on efficiency of RNA interference. BMC Bioinform. 2009;10:S33. doi: 10.1186/1471-2105-10-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh S., Gupta S.K., Nischal A. Design of potential siRNA molecules for hepatitis delta virus gene silencing. Bioinformation. 2012;8:749–757. doi: 10.6026/97320630008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernhart S.H., Tafer H., Mückstein U., Flamm C., Stadler P.F., Hofacker I.L. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motavaf M., Safari S., Alavian S.M. Therapeutic potential of RNA interference: a new molecular approach to antiviral treatment for hepatitis C. J Viral Hepat. 2012;19:757–765. doi: 10.1111/jvh.12006. [DOI] [PubMed] [Google Scholar]
- 17.Boudreau R.L., Davidson B.L. RNAi therapeutics for CNS disorders. Brain Res. 2010;1338:112–121. doi: 10.1016/j.brainres.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Czech M.P., Aouadi M., Tesz G.J. RNAi-based therapeutic strategies for metabolic disease. Nat Rev Endocrinol. 2011;7:473–484. doi: 10.1038/nrendo.2011.57. [DOI] [PubMed] [Google Scholar]
- 19.Park W.S., Miyano-Kurosaki N., Hayafune M. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 2002;30:4830–4835. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z.X., Min W.P., Jevnikar A.M. Use of RNA interference to minimize ischemia reperfusion injury. Transplant Rev (Orlando) 2012;26:140–155. doi: 10.1016/j.trre.2011.03.001. [DOI] [PubMed] [Google Scholar]


