The crystal structure of NAD+-bound FDH from P. aeruginosa was determined to 2.7 Å resolution. The overall structure is very similar to that of P. putida FDH. Preliminary kinetics analysis of the enzyme indicated a glutathione-independent bi-substrate ‘ping-pong’ catalytic mechanism.
Keywords: formaldehyde dehydrogenase, Pseudomonas aeruginosa, NAD+
Abstract
Formaldehyde dehydrogenase (FDH) is a member of the zinc-containing medium-chain alcohol dehydrogenase family which oxidizes toxic formaldehyde to formate using NAD+ as an electron carrier. Three-dimensional structures have been reported for FDHs from several different species. Most FDHs are dependent on glutathione for catalysis, but the enzyme from Pseudomonas putida is an exception. In this structural communication, the recombinant production, crystallization and X-ray structure determination at 2.7 Å resolution of FDH from P. aeruginosa are described. Both the tetrameric assembly and the NAD+-binding mode of P. aeruginosa FDH are similar to those of P. putida FDH, which is in good agreement with the high sequence identity (87.97%) between these two proteins. Preliminary enzymatic kinetics studies of P. aeruginosa FDH also revealed a conserved glutathione-independent ‘ping-pong’ mechanism of formaldehyde oxidization.
1. Introduction
Formaldehyde is a toxic compound and most organisms have developed oxidation systems to counteract it. An example is formaldehyde dehydrogenase (FDH), which catalyzes the oxidation of formaldehyde using NAD+ as an electron acceptor and is found in both prokaryotes and eukaryotes (Uotila & Koivusalo, 1989 ▶). FDH belongs to the zinc-containing medium-chain alcohol dehydrogenase (ADH) family, which has long attracted interest in structural and functional studies (Persson et al., 1991 ▶, 1994 ▶; Rossmann et al., 1975 ▶).
Crystal structures of ADHs from a number of species have been reported, revealing the presence of either a dimer (Eklund et al., 1981 ▶; Sanghani et al., 2002 ▶) or a tetramer (Jenkins & Tanner, 2006 ▶; Cowan-Jacob et al., 2003 ▶) as the active form, with each monomer (350–400 amino-acid residues) consisting of catalytic and coenzyme-binding domains together with two bound zinc ions. Functionally, most FDHs are known to be dependent on glutathione during catalysis of formaldehyde oxidation and the reaction product is actually S-formylglutathione rather than free formate (Rose & Racker, 1962 ▶; Johnson & Quayle, 1964 ▶; Strittmatter & Ball, 1955 ▶; Sanghani et al., 2000 ▶). The FDH from Pseudomonas putida is the only member of the ADH family identified to date that can catalyze irreversible oxidation of formaldehyde without glutathione (Ando et al., 1979 ▶; Ito et al., 1994 ▶). In addition, the enzyme can catalyze aldehyde dismutation without releasing NAD(H) (Tanaka et al., 2002 ▶). Its catalytic mechanism of aldehyde dismutation is distinct from most typical ADHs, which use NAD(H) as an exchangeable coenzyme. The crystal structure of this homotetrameric enzyme (Tanaka et al., 2002 ▶) revealed a similar structural arrangement and similar binding modes of NAD+ and zinc ions to these typical ADHs, except that P. putida FDH contains a number of different loop structures. In particular, a long insertion loop (residues 265–279) was found in the cofactor-binding domain and might contribute to the tight binding of the enzyme to NAD+ during catalysis of aldehyde dismutation.
Here, we report the crystal structure of FDH from P. aeruginosa bound to the cofactor NAD+ at 2.7 Å resolution. P. aeruginosa is a ubiquitous environmental Gram-negative bacterium and is a major pathogen that causes opportunistic human infections owing to its intrinsic resistance to antibiotics and disinfectants (Hardalo & Edberg, 1997 ▶; Bodey et al., 1983 ▶). The tetrameric form is observed in the crystal structure as well as in solution. Structural comparison with P. putida FDH indicated high similarity between these two enzymes. The enzymatic kinetics of P. aeruginosa FDH towards the substrate formaldehyde are also reported.
2. Experimental
2.1. Cloning, expression and purification
The FDH gene of P. aeruginosa (strain LESB58) was synthesized based on the protein sequence (residues 1–399; UniProt accession No. B7V5W2) and the DNA sequence was optimized to adapt codon usage to the expression host using JCat (Grote et al., 2005 ▶). An N-terminal His tag (MNHKVHHHHHH) was also introduced into the protein. The synthesized DNA was inserted between the NdeI and HindIII sites of the pCold II vector (Takara) and the resulting plasmid was transformed into Escherichia coli BL21(DE3)/pG-TF2 cells (Novoprotein), which contain a coexpression system for the chaperones GroEL, GroES and Tig. Cultures were grown in LB medium containing ampicillin (100 µg ml−1) and chloramphenicol (34 µg ml−1) at 310 K to an OD600 of 0.5–0.6; 10 ng ml−1 tetracycline was then introduced to produce the chaperones for 30 min. The His-tagged FDH protein was then produced at 289 K for 12 h after adding 0.2 mM IPTG to the culture.
Freeze–thawed cell pellets were resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 250 mM NaCl, 20 mM imidazole) and disrupted by sonication on ice. The crude lysate was centrifuged at 12 000g for 30 min and the supernatant was filtered through a 0.45 µm filter and loaded onto a pre-equilibrated 5 ml Ni–NTA column (GE Healthcare). The column was washed with 20 column volumes of lysis buffer and eluted with an imidazole gradient (50–250 mM). The eluted samples were then dialyzed against buffer A (20 mM Tris–HCl pH 8.0, 1 mM DTT) and further loaded onto a 10 ml Mono Q FF column (GE Healthcare) pre-equilibrated with buffer A. The column was washed with buffer A and was then eluted with a NaCl gradient (0.05–1.0 M) in buffer A. Peak fractions containing FDH were pooled and concentrated to 20 mg ml−1. Finally, the purified and concentrated FDH was dialyzed against 10 mM Tris–HCl pH 8.0, 1 mM DTT, which was used as crystallization buffer, and stored at 253 K.
The multimeric state of P. aeruginosa FDH was analyzed by size-exclusion chromatography (SEC) on a HiLoad 26/60 Superdex 200 column (GE Healthcare). Protein samples at a concentration of 2 mg ml−1 were loaded onto the column and were eluted with buffer A (20 mM Tris–HCl pH 8.0, 1 mM DTT), with detection of the absorbance at 280 nm.
2.2. Enzymatic activity assay
As described by the equation
the enzymatic activity of the FDH was assayed by measuring the formation of NADH in terms of the increase in absorbance at 340 nm (Ando et al., 1979 ▶). A 0.1 ml aliquot of the enzyme (20 µg ml−1) was incubated with 1 ml formaldehyde at various concentrations and NAD+ (20–500 µM) in 50 mM potassium phosphate buffer pH 7.5 at 298 K in a quartz cuvette with 1 cm path length. The change in absorbance at 340 nm was monitored against a blank test at 2 min intervals using a UV-7504 spectrophotometer (Shanghai Xinmao Instrument Co. Ltd). The formation of NADH as a function of time was expressed as ΔE 340 nm min−1. The initial reaction velocity (v 0) was determined from the linear part of the curve and was averaged over three independent assays. The Michaelis constants (K m) of the enzyme for formaldehyde and NAD+ were calculated from a Lineweaver–Burk plot based on a bi-substrate kinetics model.
2.3. Crystallization
To generate the FDH–NAD+ complex, the concentrated FDH was incubated with a solution of NAD+ (dissolved in 10 mM Tris–HCl pH 8.0, 1 mM DTT) at 277 K for 2 h. The protein–NAD+ solution consisting of 15 mg ml−1 FDH and 4 mg ml−1 NAD+ in 10 mM Tris–HCl pH 8.0, 1 mM DTT was then used for crystallization trials. All crystallization screenings were performed at 291 K using the sitting-drop vapour-diffusion method and each drop (1 µl) was prepared by mixing equal volumes of protein–NAD+ solution and reservoir solution. Initial hits were obtained using commercially available crystallization screening kits (Hampton Research Crystal Screen and Crystal Screen 2). Subsequent optimization was performed by setting up 2 µl drops consisting of 1 µl protein–NAD+ solution and 1 µl reservoir solution, and crystals of the FDH–NAD+ complex grew in 0.1 M bis-tris pH 5.5–6.5, 1.5–2.5 M ammonium sulfate at 291 K. Optimum plate-shaped crystals (100 × 50 × 20 µm) were obtained from 0.1 M bis-tris pH 5.5, 1.8 M ammonium sulfate after several rounds of streak-seeding using a cat whisker.
2.4. Data collection and processing
The crystals were cryoprotected in 0.1 M bis-tris pH 5.5, 2.0 M ammonium sulfate with 20%(v/v) glycerol and were harvested into nylon loops prior to flash-cooling in liquid nitrogen. Diffraction data were collected at 100 K with an ADSC Q315 CCD detector using synchrotron radiation (λ = 0.9791 Å) on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF; Shanghai, People’s Republic of China). The crystal diffracted to 2.7 Å resolution and all data processing was performed with HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.5. Structure solution and refinement
The structure of P. putida FDH (PDB entry 1kol, chain A; Tanaka et al., 2002 ▶) was selected as the search model for molecular replacement. The bound NAD+ and all solvent molecules were removed from the model and molecular replacement was carried out with Phaser (McCoy et al., 2007 ▶). Model refinement was performed with REFMAC5 (Murshudov et al., 2011 ▶) followed by employing Coot (Emsley & Cowtan, 2004 ▶) for iterative cycles of rebuilding based on σA-weighted 2F o − F c and F o − F c maps. Noncrystallographic symmetry (NCS) restraints were applied to chains A and D and to chains B and C in the early stages of refinement. Solvent molecules were identified based on the F o − F c difference map. The final steps of refinement also incorporated TLS restraints (Painter & Merritt, 2006 ▶); a total of 19 TLS groups were selected, which were automatically determined by PHENIX (Adams et al., 2010 ▶). The stereochemical quality of the refined structure was validated using MolProbity (Chen et al., 2010 ▶). Data-collection and refinement statistics are presented in Table 1 ▶. The interfaces of the FDH tetramer were analyzed using PDBePISA (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; Krissinel & Henrick, 2007 ▶). Global alignment of the FDH structures was performed using PyMOL (super_align; Schrödinger) and all structural model figures were also created with PyMOL. Structure factors and coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) under accession code 4jlw.
Table 1. Data-collection and refinement statistics for FDHNAD+ .
Values in parentheses are for the highest resolution shell.
| Data-collection statistics | |
| Wavelength () | 0.9791 |
| Resolution () | 50.002.70 (2.802.70) |
| Space group | P21 |
| Molecules per asymmetric unit | 4 |
| Unit-cell parameters (, ) | a = 89.23, b = 98.31, c = 99.62, = = 90.00, = 91.29 |
| No. of observed reflections | 168470 |
| No. of unique reflections | 47323 (4719) |
| Data multiplicity | 3.6 (3.6) |
| Completeness (%) | 99.8 (99.9) |
| Mean I/(I) | 6.43 (2.91) |
| R merge † (%) | 20.3 (63.4) |
| Refinement statistics | |
| Resolution () | 30.002.70 (2.772.70) |
| No. of reflections/No. in test set | 44912/2394 |
| Completeness (%) | 99.30 (92.98) |
| R work/R free ‡ (%) | 21.18/25.94 |
| No. of atoms | |
| Protein | 11704 |
| Ligand/ion | 214 |
| Water | 454 |
| Modelled residues§ | |
| Chains A D | 3397 |
| Overall B factor (2) | |
| Protein | 27.05 |
| Ligand/ion | 35.13 |
| Water | 26.13 |
| R.m.s.d., bond lengths¶ () | 0.012 |
| R.m.s.d., bond angles¶ () | 1.641 |
| Ramachandran plot, residues in†† (%) | |
| Favoured region | 97.1 |
| Allowed region | 2.9 |
| Outlier region | 0 |
R
merge = 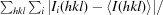
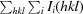 , where I
i(hkl) and I(hkl) are theith and the mean measurement of the intensity of the unique reflection hkl, respectively.
, where I
i(hkl) and I(hkl) are theith and the mean measurement of the intensity of the unique reflection hkl, respectively.
R
work = 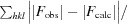
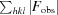 , where F
obs and F
calc are the observed and calculated structure-factor amplitudes for reflection hkl and the summation is over 95% of the reflections in the specified resolution range. The remaining 5% of the reflections were randomly selected before structure refinement and were not included inthe structure refinement. R
free was calculated over these reflections using the same equation as for R
work.
, where F
obs and F
calc are the observed and calculated structure-factor amplitudes for reflection hkl and the summation is over 95% of the reflections in the specified resolution range. The remaining 5% of the reflections were randomly selected before structure refinement and were not included inthe structure refinement. R
free was calculated over these reflections using the same equation as for R
work.
The N-terminal His tag and residues 12 and 398399 of FDH were omitted from the final model owing to poorly resolved electron density.
Root-mean-square deviations from the parameter set for ideal stereochemistry.
As defined by MolProbity.
3. Results and discussion
3.1. Crystallization, data collection and structure determination
Crystals of FDH from P. aeruginosa bound to the cofactor NAD+ were obtained by the sitting-drop vapour-diffusion method. The crystals belonged to the monoclinic space group P21, with unit-cell parameters a = 89.23, b = 98.31, c = 99.62 Å, β = 91.29°. A total of four molecules were observed per asymmetric unit, giving a V M value of 2.62 Å3 Da−1 and a solvent content of 52% (Matthews, 1968 ▶). The structure was solved by molecular replacement using the P. putida FDH structure as a search model. Iterative refinement and manual building of the model resulted in a final R work of 21.18% and an R free of 25.94%. Electron density corresponding to residues 3–397, one NAD+ molecule and two zinc ions was well defined for each of the four crystallographically independent protein subunits. In addition, a total of 454 water molecules and six sulfate ions were included per asymmetric unit. A Ramachandran plot of the final model indicated that 97.1% of the residues were in mostly favoured regions and 2.9% were in additionally allowed regions.
3.2. The overall structure of P. aeruginosa FDH
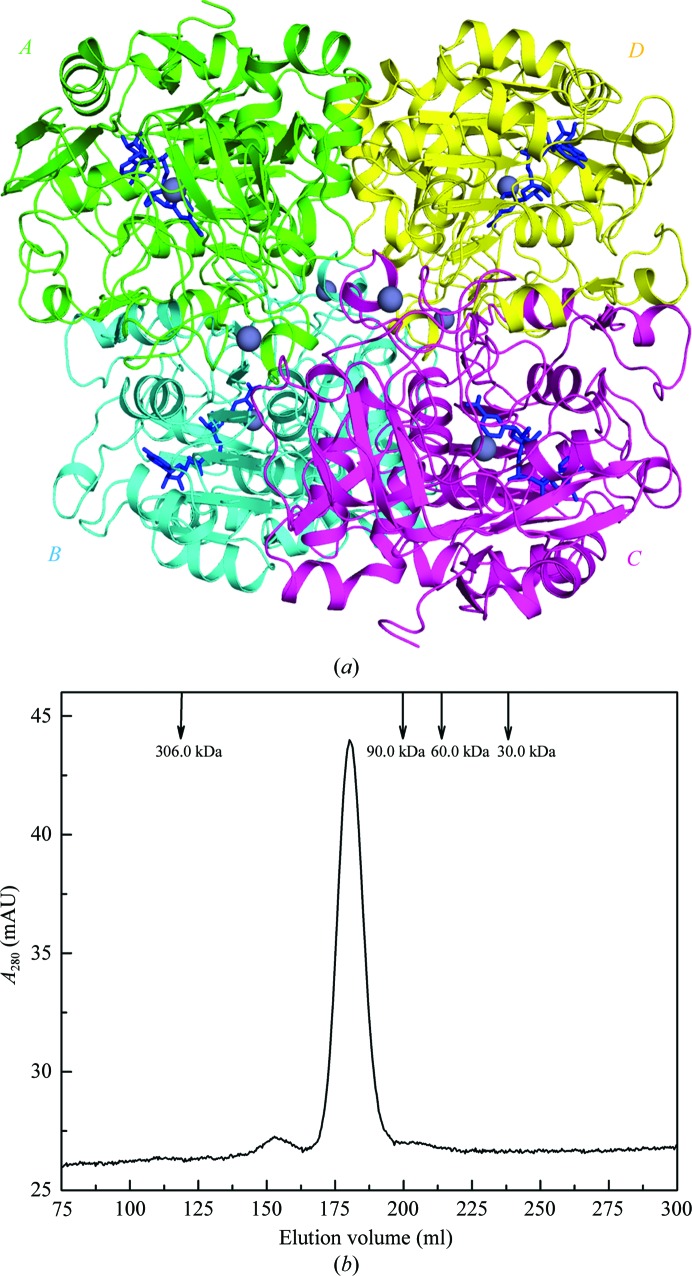
As depicted in Fig. 1 ▶(a), the final model of P. aeruginosa FDH consists of four protein subunits forming a homotetramer with a total buried surface area of ∼12 920 Å2. The tetramer could be considered as a dimer of dimers formed by subunits A (green) and B (cyan) and by subunits C (magenta) and D (yellow). Analogous dimeric interactions have been reported in the structures of mammalian ADHs (Eklund et al., 1981 ▶; Sanghani et al., 2002 ▶). The interface of the A–B (or C–D) dimer is mainly composed of a six-stranded parallel β-sheet from each subunit and has a total buried surface area of ∼3300 Å2. In forming the tetramer, the A–B dimer contacts the C–D dimer by two types of subunit–subunit interfaces. One interface is between subunit A (green) and subunit C (magenta), with a total buried surface area of ∼1600 Å2. The other interface is formed by subunit A (green) and subunit D (yellow) and possesses a total buried surface area of ∼1500 Å2 (Fig. 1 ▶ a). The tetrameric assembly observed here also appears in the structure of P. putida FDH through the [210] crystallographic symmetry operation (Tanaka et al., 2002 ▶). In order to determine whether the tetramer corresponds to a natural state in solution, SEC analysis was performed (Fig. 1 ▶ b). The protein eluted as a single peak with a retention volume of 180.3 ml, corresponding to an estimated molecular mass of ∼150 kDa. This further implies that the enzyme exists in a tetrameric form since the theoretical molecular mass of monomeric P. aeruginosa FDH is 43.4 kDa.
Figure 1.
Tetrameric NAD+-bound formaldehyde dehydrogenase (FDH) from P. aeruginosa. (a) The overall structure of the FDH tetramer. Four subunits (A–D) are shown in ribbon mode in different colours. The zinc ions are depicted as grey spheres and the bound NAD+ ions are shown as blue sticks for each of the four subunits. (b) SEC analysis of the FDH at a concentration of 2 mg ml−1. The protein sample was loaded onto a HiLoad 26/60 Superdex 200 column and was eluted at a flow rate of 3 ml min−1 with detection of the absorbance at 280 nm. The elution profiles of four molecular-mass protein standards are also shown as labelled arrows.
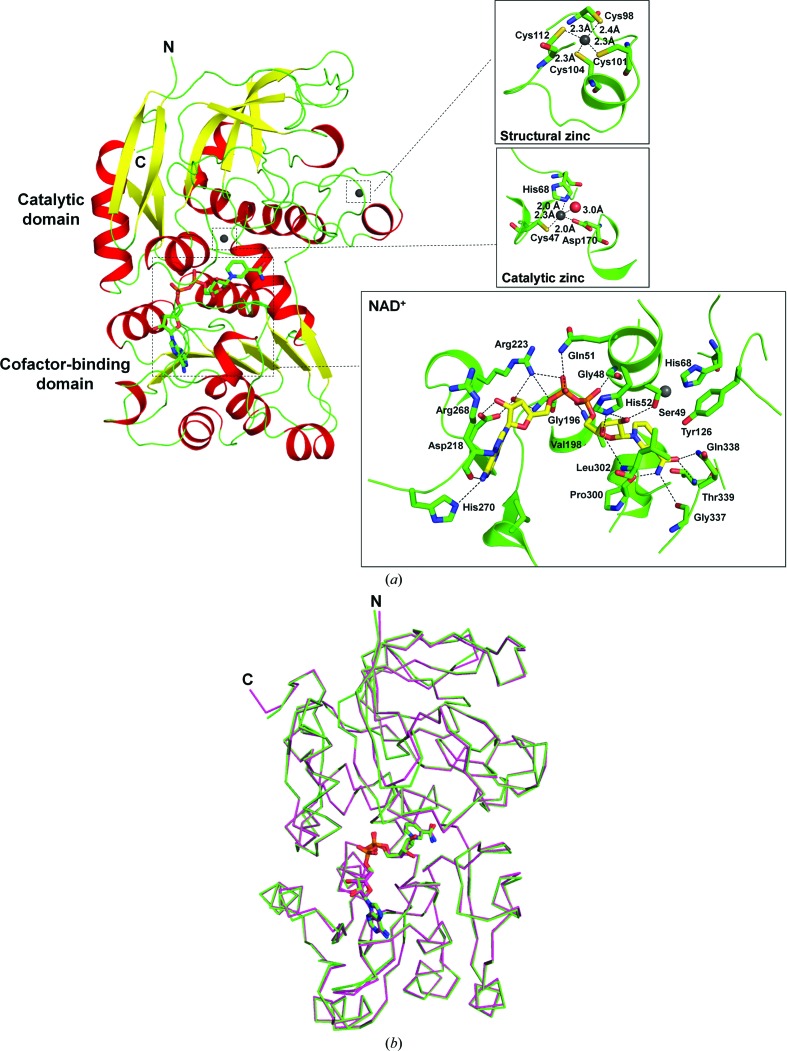
Similar to other ADHs, each subunit of P. aeruginosa FDH is composed of two domains separated by a cleft containing a deep pocket (Fig. 2 ▶ a). Both the N-terminus (residues 1–2) and C-terminus (residues 398–399) of the FDH subunit along with its N-terminal His tag (MNHKVHHHHHH) are not visible in the electron density and were not included in the final model. One of the domains (residues 3–170 and 339–397) is responsible for the binding and catalysis of formaldehyde and is named the catalytic domain; the other domain (residues 171–338) that provides the structural moiety necessary for NAD+ binding is designated the cofactor-binding domain. The catalytic domain mainly consists of eight β-strands and seven α-helices, and all of the α-helices are aligned on the surface of the domain to surround the β-structural core. For the cofactor-binding domain, the overall arrangement of its secondary structures is a six-stranded parallel β-sheet sandwiched by three α-helices on either side, which comprises a characteristic ‘Rossmann fold’ (Rossmann et al., 1974 ▶). The cofactor NAD+ is bound to the central region of the carboxyl end of the parallel β-sheet. A structural comparison of the FDH subunit (chain A) with the structure of P. putida FDH (PDB entry 1kol, chain A) yielded a root-mean-square (r.m.s.) distance of 0.23 Å for all 395 equivalent Cα atoms (Fig. 2 ▶ b), indicating high structural similarity between these two FDHs from different Pseudomonas bacterial strains.
Figure 2.
The structure of the P. aeruginosa FDH subunit. (a) Ribbon drawing of FDH subunit A. The N- and C-termini are labelled by the letters N and C, respectively. α-Helices are coloured red, β-strands yellow and loops green. The NAD+ and zinc ions are highlighted as sticks and spheres, respectively. The insets show the binding modes of two zinc ions (grey spheres) and the cofactor NAD+ (sticks) to the FDH subunit. Hydrogen bonds are indicated by dotted lines. The residues of FDH involved in binding are shown as sticks and the residue numbers are indicated. (b) Superimposition of FDH subunit A (green) with a subunit of P. putida FDH (magenta). Proteins are indicated by Cα traces with N- and C-termini marked and the bound NAD+ ions are shown in stick representation.
3.3. Zinc- and NAD+-binding sites
In the current FDH structure, two firmly bound zinc ions were observed per subunit (Fig. 2 ▶ a, inset). One zinc ion, the so-called structural zinc, is coordinated to the S atoms of Cys98, Cys101, Cys104 and Cys112 from the catalytic domain. Its tetrahedral coordination geometry is well ordered owing to similar zinc–ligand distances among the four Cys ligands (∼2.3 Å). The other zinc ion, called the catalytic zinc, is located at the bottom of the cleft between the two domains of the FDH subunit and has a tetrahedral coordination environment with Cys47, His68 and Asp170 as the three protein ligands together with a water molecule. Compared with the structural zinc, the tetrahedral geometry of the catalytic zinc is more distorted, which might be caused by unequal zinc–ligand distances among the different ligand elements (sulfur, nitrogen and oxygen).
In addition to the two zinc ions, the overall conformation of the cofactor NAD+ and its binding mode to the FDH subunit in the present structure are also similar to those observed for P. putida FDH (Fig. 2 ▶ b). The bound NAD+ adopts an anti conformation, with the nicotinamide and adenine rings orientated roughly perpendicular to the planes of their respective ribose rings (Fig. 2 ▶ a, inset). The nicotinamide nucleoside moiety of the NAD+ interacts with the FDH through eight hydrogen bonds. Four hydrogen bonds are formed from the carboxamide group of the nicotinamide ring to the protein main chain (NAD+ N7N⋯Pro300 O, NAD+ N7N⋯Gly337 O and NAD+ O7N⋯Thr339 N) and side chain (NAD+ O7N⋯Gln338 NE2). The other four were observed between the hydroxyl groups of the ribose ring and the protein: NAD+ O2D⋯Ser49 OG, NAD+ O2D⋯His52 NE2, NAD+ O3D⋯His52 NE2 and NAD+ O3D⋯Leu302 N. In addition, the pyrophosphate moiety of the NAD+ is hydrogen-bonded to the FDH between the O atoms of the nicotinamide or adenine phosphate groups and several protein residues, e.g. NAD+ O1N⋯Gly48 N, NAD+ O2N⋯Val198 N, NAD+ O1A⋯Arg223 NH1, NAD+ O2A⋯Gln51 NE2 and NAD+ O5B⋯Arg223 NH1. Meanwhile, the adenine ribose moiety of the NAD+ also forms extensive interactions with the FDH subunit. In detail, the 2′- and 3′-hydroxyl groups of the ribose ring are recognized by the side chain of Asp218 to form a bifurcated hydrogen bond (NAD+ O2B⋯Asp218 OD1 and NAD+ O3B⋯Asp218 OD2). Two other hydrogen bonds are observed between the ribose ring and residues Gly196 and Arg223 (NAD+ O3B⋯Gly196 N and NAD+ O3B⋯Arg223 NH1). Furthermore, the adenine ring is accommodated in a network of hydrogen bonds which is constituted by the N atoms of the adenine ring and the main chain of Arg268 (NAD+ N6A⋯Arg268 O and NAD+ N7A⋯Arg268 N), as well as the amino group of the adenine ring and the side chain of His270 (NAD+ N6A⋯His270 NE2).
3.4. The enzymatic kinetics of P. aeruginosa FDH
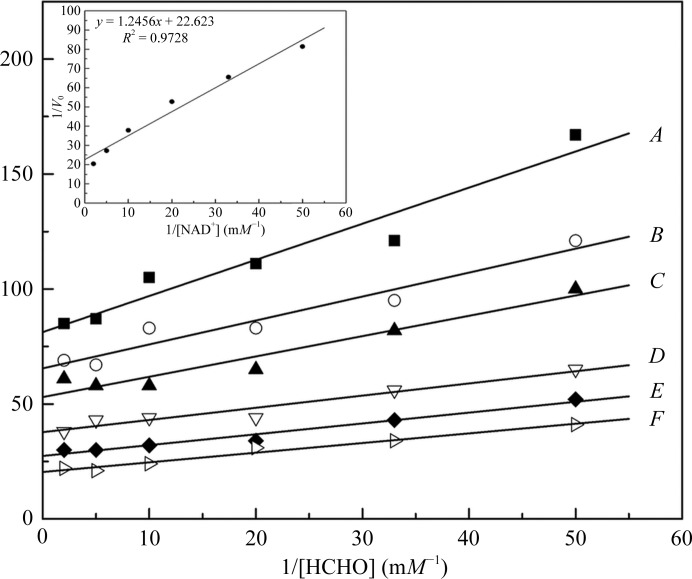
As mentioned above, P. putida FDH is the only identified ADH that is independent of glutathione for catalysis (Ando et al., 1979 ▶). Considering the similarity of our structure to that of P. putida FDH, the enzymatic activity of P. aeruginosa FDH towards the substrate formaldehyde was therefore measured. Firstly, the enzyme required only NAD+ as an electron carrier and addition of glutathione had no effect on the reaction rate (data not shown), indicating glutathione-independent catalysis. The reactions were then kinetically assayed and the results are shown in Fig. 3 ▶. Initial double-reciprocal plots of the reaction velocity versus formaldehyde produced nearly parallel lines for various concentrations of NAD+, implying that the reaction proceeds by a ‘ping-pong’ mechanism as designated by the Michaelis–Menten model of bi-substrate enzymatic kinetics (Cleland, 1967 ▶). The K m values for formaldehyde and NAD+ are 15.3 and 55.0 µM, respectively, which were calculated from the initial plots together with a secondary plot of the intercepts of the parallel lines against the reciprocal of the concentration of NAD+. These results are also comparable with the published data for P. putida FDH (Ando et al., 1979 ▶).
Figure 3.
Enzymatic activity of P. aeruginosa FDH towards the substrate formaldehyde. Double-reciprocal plots of the reaction catalyzed by FDH as a function of the concentration of formaldehyde are shown. The enzyme was incubated with various concentrations of formaldehyde (20–500 µM) and the change in absorbance at 340 nm was monitored. The reaction velocity was expressed as ΔE 340 nm min−1. The concentrations of NAD+ were A, 20 µM; B, 30 µM; C, 50 µM; D, 100 µM; E, 200 µM; F, 500 µM. The inset shows a secondary plot of the intercepts of the parallel lines versus the reciprocal of the concentration of NAD+.
In summary, in this communication we have presented the binary-complex structure of P. aeruginosa FDH bound to the cofactor NAD+ as well as the preliminary characterization of its enzymatic kinetics. Consistent with the published results for the homologous structure of P. putida FDH, the enzyme shows well conserved structural and catalytic features. It will be of interest to determine the structure of a ternary complex of the enzyme with NAD+ and formaldehyde in the future, which could further elucidate the detailed mechanism of glutathione-independent catalysis.
Supplementary Material
PDB reference: NAD+-bound formaldehyde dehydrogenase, 4jlw
Acknowledgments
We are grateful to Li Wang and members of our company for critically reading the manuscript.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Ando, M., Yoshimoto, T., Ogushi, S., Rikitake, K., Shibata, S. & Tsuru, D. (1979). J. Biochem. 85, 1165–1172. [PubMed]
- Bodey, G. P., Bolivar, R., Fainstein, V. & Jadeja, L. (1983). Rev. Infect. Dis. 5, 279–313. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cleland, W. W. (1967). Annu. Rev. Biochem. 36, 77–112. [DOI] [PubMed]
- Cowan-Jacob, S. W., Kaufmann, M., Anselmo, A. N., Stark, W. & Grütter, M. G. (2003). Acta Cryst. D59, 2218–2227. [DOI] [PubMed]
- Eklund, H., Samama, J.-P., Wallén, L., Brändén, C. I., Åkeson, Å. & Jones, T. A. (1981). J. Mol. Biol. 146, 561–587. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Grote, A., Hiller, K., Scheer, M., Münch, R., Nörtemann, B., Hempel, D. C. & Jahn, D. (2005). Nucleic Acids Res. 33, W526–W531. [DOI] [PMC free article] [PubMed]
- Hardalo, C. & Edberg, S. C. (1997). Crit. Rev. Microbiol. 23, 47–75. [DOI] [PubMed]
- Ito, K., Takahashi, M., Yoshimoto, T. & Tsuru, D. (1994). J. Bacteriol. 176, 2483–2491. [DOI] [PMC free article] [PubMed]
- Jenkins, J. L. & Tanner, J. J. (2006). Acta Cryst. D62, 290–301. [DOI] [PubMed]
- Johnson, P. A. & Quayle, J. R. (1964). Biochem. J. 93, 281–290. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Persson, B., Krook, M. & Jörnvall, H. (1991). Eur. J. Biochem. 200, 537–543. [DOI] [PubMed]
- Persson, B., Zigler, J. S. & Jörnvall, H. (1994). Eur. J. Biochem. 226, 15–22. [DOI] [PubMed]
- Rose, Z. B. & Racker, E. (1962). J. Biol. Chem. 237, 3279–3281. [PubMed]
- Rossmann, M. G., Lijas, A., Brändén, C.-I. & Banaszak, L. J. (1975). The Enzymes, 3rd ed., edited by P. D. Boyer, Vol. XI, pp. 61–102. New York: Academic Press.
- Rossmann, M. G., Moras, D. & Olsen, K. W. (1974). Nature (London), 250, 194–199. [DOI] [PubMed]
- Sanghani, P. C., Robinson, H., Bosron, W. F. & Hurley, T. D. (2002). Biochemistry, 41, 10778–10786. [DOI] [PubMed]
- Sanghani, P. C., Stone, C. L., Ray, B. D., Pindel, E. V., Hurley, T. D. & Bosron, W. F. (2000). Biochemistry, 39, 10720–10729. [DOI] [PubMed]
- Strittmatter, P. & Ball, E. G. (1955). J. Biol. Chem. 213, 445–461. [PubMed]
- Tanaka, N., Kusakabe, Y., Ito, K., Yoshimoto, T. & Nakamura, K. T. (2002). J. Mol. Biol. 324, 519–533. [DOI] [PubMed]
- Uotila, L. & Koivusalo, M. (1989). In Glutathione: Chemical, Biochemical, and Medical Aspects, Part A, edited by D. Dolphin, R. Poulson & O. Avramovic. New York: Wiley.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: NAD+-bound formaldehyde dehydrogenase, 4jlw