In this study, the variable domain of the collagen-like protein Scl2 from invasive M3-type S. pyogenes has successfully been crystallized. Single-wavelength anomalous dispersion experiments have been carried out to obtain experimental phases by preparing crystal derivatives with lanthanides. Model building and refinement, which are in progress, will provide the first structural clues for Scls.
Keywords: collagen-like protein, Scl2, group A streptococcus, Streptococcus pyogenes
Abstract
Streptococcal collagen-like proteins (Scls) are widely expressed by the well recognized human pathogen Streptococcus pyogenes. These surface proteins contain a signature central collagen-like region and an amino-terminal globular domain, termed the variable domain, which is protruded away from the cell surface by the collagen-like domain. Despite their recognized importance in bacterial pathogenicity, no structural information is presently available on proteins of the Scl class. The variable domain of Scl2 from invasive M3-type S. pyogenes has successfully been crystallized using vapour-diffusion methods. The crystals diffracted to 1.5 Å resolution and belonged to space group H32, with unit-cell parameters a = 44.23, b = 44.23, c = 227.83 Å. The crystal structure was solved by single-wavelength anomalous dispersion using anomalous signal from a europium chloride derivative.|
1. Introduction
Collagen-like proteins that form stable triple helices have been shown to be present in many bacterial species (Rasmussen et al., 2003 ▶) and to play a role in pathogenicity. The best characterized prokaryotic collagens are the two collagen-like proteins Scl1 and Scl2, which have been demonstrated to be simultaneously expressed on the cell surface of Streptococcus pyogenes and to promote bacterial adhesion to the host (Humtsoe et al., 2005 ▶; Rasmussen & Björck, 2001 ▶). Both Scl1 and Scl2 proteins contain a signal sequence, an N-terminal variable globular domain (V), a highly charged collagen-like triple-helix domain (CL) consisting of (Gly-Xaa-Yaa)n triplet repeats and a C-terminal Gram-positive cell-wall attachment domain. The Scl1 and Scl2 proteins form stable triple-helical structures when expressed as recombinant proteins (Han et al., 2006 ▶; Xu et al., 2002 ▶) and their N-terminal globular V domain adjacent to the triple-helix domain appears to be important for efficient triple-helix assembly (Lukomski et al., 2000 ▶; Mohs et al., 2007 ▶; Xu et al., 2002 ▶).
Bacterial adherence to host tissues, an early critical step in the infection process, often involves surface proteins (Esposito et al., 2008 ▶; Chagnot et al., 2012 ▶). Among these, Scl (streptococcal collagen-like) proteins of S. pyogenes are crucial to host–pathogen recognition (Berisio & Vitagliano, 2012 ▶). It has been demonstrated that Scl1 can bind selected human extracellular matrix components (Caswell et al., 2010 ▶), cellular integrin receptors (Humtsoe et al., 2005 ▶; Caswell, Barczyk et al., 2008 ▶; Caswell et al., 2007 ▶) and plasma components (Gao et al., 2010 ▶; Caswell, Han et al., 2008 ▶; Reuter et al., 2010 ▶; Påhlman et al., 2007 ▶). Importantly, human collagen receptors, such as integrin α2β1, recognize the triple-helix CL domain of Scl1 and this event results in cell signalling, indicating that collagen-like bacterial proteins display not only structural but also functional similarities to human collagens (Humtsoe et al., 2005 ▶; Caswell, Barczyk et al., 2008 ▶; Caswell et al., 2007 ▶). Despite the key role of Scl proteins in bacterial pathogenicity, their three-dimensional structure has not been determined. In addition, whereas the triple-helical structure of the CL domain of Scls can be predicted based on sequence identity to collagen (Xu et al., 2002 ▶), no structural clues regarding the V domain can be obtained from Scl sequences owing to poor sequence identity with known structures. Here, we report the crystallization and preliminary crystallographic investigations of the V domain of Scl2 from invasive M3-type S. pyogenes (Scl2.3-V).
2. Experimental procedures
2.1. Cloning, expression and purification of recombinant rScl2.3-V protein
Recombinant rScl2.3-V protein was produced in the Escherichia coli periplasm using the Strep-tag II expression and purification system (IBA GmbH, Göttingen, Germany) as reported previously (Han et al., 2006 ▶). Briefly, the 5′-portion of the scl2.3 gene from strain MGAS315, encoding the amino-terminal Scl2.3-V region, was PCR-amplified using the forward primer scl2-M3VF (5′-GAGATGGCCGATGGTGAAGATGCCCAAAAAAG) and the reverse primer scl2-M3VR (5′-CAGCGTCTCAGCGCTATCAAGGACATGATCTTGTATGCC) and was cloned into pASK-IBA2 vector, resulting in plasmid pSL155. E. coli strain DH5α was used for cloning and E. coli strain BL21 was used for protein expression. E. coli harbouring plasmid pSL155, which encodes the rScl2.3-V protein, was grown in Luria–Bertani liquid medium (BD Biosciences) supplemented with ampicillin (100 µg ml−1). Plasmid construct pSL155 was confirmed by DNA sequencing and the identity of the purified recombinant protein rScl2.3-V was confirmed by N-terminal Edman degradation.
2.2. Crystallization experiments
Crystallization trials were performed at 293 K using the hanging-drop vapour-diffusion method. Preliminary crystallization conditions were set up using a robotic station for high-throughput crystallization screening (Hamilton STARlet NanoJet 8+1) and commercially available sparse-matrix kits (Crystal Screen, Crystal Screen 2 and Index, Hampton Research). Optimization of the crystallization conditions was performed manually by fine-tuning the protein and precipitant concentrations.
2.3. Data collection and processing
Diffraction data were collected to 1.52 Å resolution in-house from a native crystal at 100 K using a Rigaku MicroMax-007 HF generator producing Cu Kα radiation and equipped with a Saturn 944 CCD detector. Cryoprotection of the crystals was achieved without the addition of further cryoprotectants, given the composition of the crystallization mother liquor, which contained the cryoprotectant pentaerythritol ethoxylate (15/4 EO/OH) at 30%(v/v). For phasing purposes, native crystals were soaked in solutions containing between 2 and 8 mM EuCl3 for different soaking times. Data were collected from several crystals to identify the best single-wavelength anomalous diffraction (SAD) signal. The data sets were scaled and merged using the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). Statistics of data collection are reported in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outermost resolution shell.
| EuCl3 derivative | Native | |
|---|---|---|
| Space group | H32 | H32 |
| Unit-cell parameters (Å, °) | a = b = 44.26, c = 228.01, γ = 120 | a = b = 44.23, c = 227.83, γ = 120 |
| Resolution (Å) | 1.87 (1.90–1.87) | 1.52 (1.55–1.52) |
| Average multiplicity | 9.5 (7.5) | 5.3 (2.6) |
| Unique reflections | 7545 | 13802 |
| Completeness (%) | 100 (99.9) | 99.2 (86.7) |
| R merge † (%) | 0.043 (0.419) | 0.061 (0.345) |
| Average I/σ(I) | 47.7 (5.1) | 49.2 (3.1) |
R
merge = 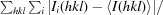
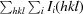 .
.
2.4. Structure determination and refinement
Phasing was achieved using in-house SAD data. A preliminary evaluation of the anomalous signal was performed for all tested crystals using the SCALEPACK software implemented in HKL-2000 (Otwinowski & Minor, 1997 ▶). SHELXD was used to identify europium-ion sites (Sheldrick, 2008 ▶). Phases were then improved by solvent-flattening density modification and phase extension by RESOLVE (Terwilliger, 2004 ▶). The obtained model was further improved using ARP/wARP (Langer et al., 2008 ▶).
3. Results and discussion
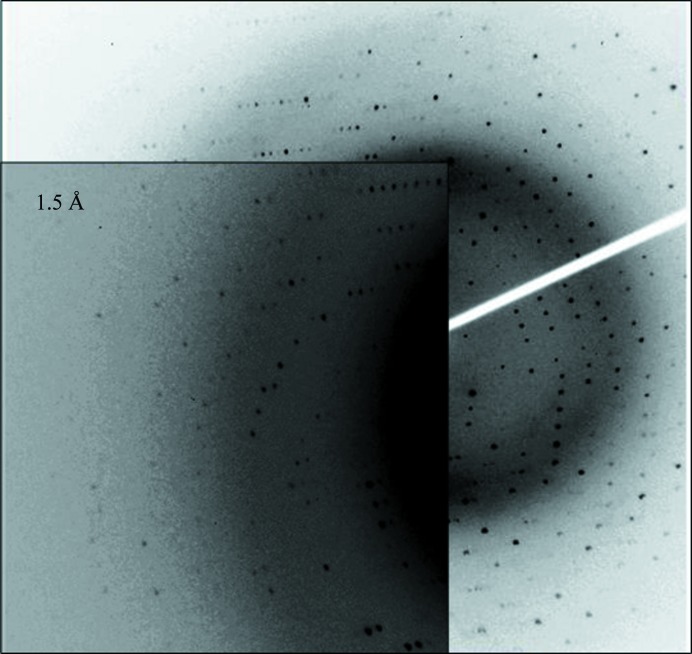
The recombinant rScl2.3-V protein corresponds to the N-terminal part (residues 1–77) of the Scl2.3 variant from the invasive S. pyogenes M3-type strain MGAS315 (Beres et al., 2002 ▶). This protein construct has successfully been purified and crystallized using vapour-diffusion methods. The purified rScl2.3-V showed a single band of approximately 10 kDa on SDS–PAGE, which is in good agreement with the predicted molecular mass of 10 105 Da. The initial automated crystallization screening using commercially available solutions provided the first hints of crystallization conditions. Small crystals that were not appropriate for diffractometric experiments were obtained in the presence of pentaerythritol ethoxylate. The quality of these crystals was improved by fine-tuning the concentration of the protein and of the precipitants. Crystals suitable for X-ray diffraction experiments (Fig. 1 ▶) were obtained in 3 d using a protein concentration of 5 mg ml−1 and 0.05 M ammonium sulfate, 0.05 M bis-tris pH 6.5, 30%(v/v) pentaerythritol ethoxylate (15/4 EO/OH). These crystals, which diffracted to 1.5 Å resolution (Fig. 2 ▶), showed threefold symmetry and belonged to space group H32, with unit-cell parameters a = 44.23, b = 44.23, c = 227.83 Å (Table 1 ▶). Matthews coefficient calculations (Matthews, 1968 ▶) suggested the presence of one molecule per asymmetric unit (V M = 2.14 Å3 Da−1 with 42.6% solvent content).
Figure 1.

Image of typical rScl2.3-V crystals grown using 5 mg ml−1 protein solution and 0.05 M ammonium sulfate, 0.05 M bis-tris pH 6.5, 30%(v/v) pentaerythritol ethoxylate (15/4 EO/OH).
Figure 2.
Diffraction pattern of a native rScl2.3-V crystal. Diffraction data are detectable to 1.5 Å resolution.
Lanthanides can yield high-phasing-power derivatives using in-house copper sources (Ruggiero et al., 2011 ▶; Pérez-Dorado, González et al., 2010 ▶; Pérez-Dorado, Sanles et al., 2010 ▶). Europium chloride derivative crystals were prepared by soaking the native crystals in stabilizing solutions containing between 2 and 8 mM EuCl3 for increasing soaking times. SAD data were collected at 100 K using a Rigaku MicroMax-007 HF generator producing Cu Kα radiation. The best SAD data were obtained upon crystal soaking in a solution consisting of 8 mM EuCl3, 0.05 M ammonium sulfate, 0.05 M bis-tris, 30%(v/v) pentaerythritol ethoxylate (15/4 EO/OH) at pH 6.5 for 3 h. The data sets were scaled and merged using the HKL-2000 program package (Otwinowski & Minor, 1997 ▶; Table 1 ▶). Using SHELXD (Sheldrick, 2008 ▶), we could identify four europium sites in the asymmetric unit of the protein. With this substructure, a correlation coefficient of 31.4% was calculated (CCall, calculated with all data). The obtained phases were improved by phase extension and density modification using RESOLVE (Terwilliger, 2003 ▶, 2004 ▶) and ARP/wARP (Langer et al., 2008 ▶). Using this approach, about 80% of the residues present in the asymmetric unit could be automatically modelled. Manual model-building sessions (Jones, 2004 ▶) aimed at defining the complete Scl2.3-V structure are in progress.
Acknowledgments
The authors thank the Ministero Italiano dell’Istruzione, dell’Università e della Ricerca (PRIN 2009 – prot. 200993WWF9), the COST Action BM1003 (COST-Grants-BM1003-00772) and the Mizutani Foundation for glycoscience for financial support (to RB). This work was supported in part by Public Service grant No. AI50666 from the National Institutes of Health (to SL).
References
- Beres, S. B. et al. (2002). Proc. Natl Acad. Sci. USA, 99, 10078–10083.
- Berisio, R. & Vitagliano, L. (2012). Curr. Protein Pept. Sci. 13, 855–865. [DOI] [PMC free article] [PubMed]
- Caswell, C. C., Barczyk, M., Keene, D. R., Lukomska, E., Gullberg, D. E. & Lukomski, S. (2008). J. Biol. Chem. 283, 36168–36175. [DOI] [PMC free article] [PubMed]
- Caswell, C. C., Han, R., Hovis, K. M., Ciborowski, P., Keene, D. R., Marconi, R. T. & Lukomski, S. (2008). Mol. Microbiol. 67, 584–596. [DOI] [PubMed]
- Caswell, C. C., Lukomska, E., Seo, N.-S., Höök, M. & Lukomski, S. (2007). Mol. Microbiol. 64, 1319–1331. [DOI] [PubMed]
- Caswell, C. C., Oliver-Kozup, H., Han, R., Lukomska, E. & Lukomski, S. (2010). FEMS Microbiol. Lett. 303, 61–68. [DOI] [PMC free article] [PubMed]
- Chagnot, C., Listrat, A., Astruc, T. & Desvaux, M. (2012). Cell. Microbiol. 14, 1687–1696. [DOI] [PubMed]
- Esposito, C., Pethoukov, M. V., Svergun, D. I., Ruggiero, A., Pedone, C., Pedone, E. & Berisio, R. (2008). J. Bacteriol. 190, 4749–4753. [DOI] [PMC free article] [PubMed]
- Gao, Y., Liang, C., Zhao, R., Lukomski, S. & Han, R. (2010). FEMS Microbiol. Lett. 309, 55–61. [DOI] [PMC free article] [PubMed]
- Han, R., Zwiefka, A., Caswell, C. C., Xu, Y., Keene, D. R., Lukomska, E., Zhao, Z., Höök, M. & Lukomski, S. (2006). Appl. Microbiol. Biotechnol. 72, 109–115. [DOI] [PubMed]
- Humtsoe, J. O., Kim, J. K., Xu, Y., Keene, D. R., Höök, M., Lukomski, S. & Wary, K. K. (2005). J. Biol. Chem. 280, 13848–13857. [DOI] [PubMed]
- Jones, T. A. (2004). Acta Cryst. D60, 2115–2125. [DOI] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Lukomski, S., Nakashima, K., Abdi, I., Cipriano, V. J., Ireland, R. M., Reid, S. D., Adams, G. G. & Musser, J. M. (2000). Infect. Immun. 68, 6542–6553. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mohs, A., Silva, T., Yoshida, T., Amin, R., Lukomski, S., Inouye, M. & Brodsky, B. (2007). J. Biol. Chem. 282, 29757–29765. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Påhlman, L. I., Marx, P. F., Mörgelin, M., Lukomski, S., Meijers, J. C. & Herwald, H. (2007). J. Biol. Chem. 282, 24873–24881. [DOI] [PubMed]
- Pérez-Dorado, I., González, A., Morales, M., Sanles, R., Striker, W., Vollmer, W., Mobashery, S., García, J. L., Martínez-Ripoll, M., García, P. & Hermoso, J. A. (2010). Nature Struct. Mol. Biol. 17, 576–581. [DOI] [PMC free article] [PubMed]
- Pérez-Dorado, I., Sanles, R., González, A., García, P., García, J. L., Martínez-Ripoll, M. & Hermoso, J. A. (2010). Acta Cryst. F66, 448–451. [DOI] [PMC free article] [PubMed]
- Rasmussen, M. & Björck, L. (2001). Mol. Microbiol. 40, 1427–1438. [DOI] [PubMed]
- Rasmussen, M., Jacobsson, M. & Björck, L. (2003). J. Biol. Chem. 278, 32313–32316. [DOI] [PubMed]
- Reuter, M., Caswell, C. C., Lukomski, S. & Zipfel, P. F. (2010). J. Biol. Chem. 285, 38473–38485. [DOI] [PMC free article] [PubMed]
- Ruggiero, A., Squeglia, F., Marasco, D., Marchetti, R., Molinaro, A. & Berisio, R. (2011). Biochem. J. 435, 33–41. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Terwilliger, T. C. (2003). Methods Enzymol. 374, 22–37. [DOI] [PubMed]
- Terwilliger, T. (2004). J. Synchrotron Rad. 11, 49–52. [DOI] [PubMed]
- Xu, Y., Keene, D. R., Bujnicki, J. M., Höök, M. & Lukomski, S. (2002). J. Biol. Chem. 277, 27312–27318. [DOI] [PubMed]