Tn3 proteins, a novel class of binding molecules based on a fibronectin type III domain, have been cloned, expressed, purified and crystallized in complex with their cognate target.
Keywords: fibronectin type III domains, CD40L
Abstract
Tn3 proteins are a novel class of binding molecules based on the third fibronectin type III domain of human tenascin C. Target-specific Tn3 proteins are selected from combinatorial libraries in which three surface-exposed loops have been diversified. Here, the cocrystallization of two different Tn3 proteins in complex with CD40L, a therapeutic target for immunological disease, is reported. These crystal structures are the first to be reported of Tn3 proteins and will help to reveal how these engineered molecules achieve specific recognition of a cognate target.
1. Introduction
Alternative scaffolds have recently emerged as a new class of binding proteins for research and therapeutic applications (Skerra, 2007 ▶; Löfblom et al., 2011 ▶). They are based on small, non-antibody protein scaffolds that can be engineered for specific binding to targets of interest. Alternative scaffolds are typically small monomeric protein modules of <200 amino acids that exhibit high structural stability and can be expressed at high levels in bacteria. Combinatorial libraries of scaffold proteins are generated by introducing amino-acid sequence diversity at exposed positions that form a contiguous molecular surface. Powerful display technologies, such as phage display, are then used to select specific binding proteins against targets of interest from these large molecular repertoires. The ease of manufacturing, excellent stability and ability to further engineer scaffold proteins for multispecificity or enhanced in vivo half-life have led to significant interest in their therapeutic potential. Several drug candidates based on this technology have entered clinical trials and many more are in preclinical development (Wurch et al., 2012 ▶).
One of the more commonly used alternative scaffolds is based on the fibronectin type III domain (FnIII), a ubiquitous module found in many proteins across diverse phyla (recently reviewed in Koide et al., 2012 ▶). This alternative scaffold has an Ig-like β-sandwich fold with seven β-strands connected by six loops. Most combinatorial libraries of FnIII scaffolds have been generated by introducing sequence and length diversity into three of the CDR-like loops at one end of the β-sandwich. These libraries can yield FnIII-based binders in which the loops engage cleft-like epitopes in a ‘head-on’ manner, but surprisingly ‘side-on’ recognition of convex surfaces has also been reported via interaction of a single loop together with undiversified β-sheet regions (Gilbreth & Koide, 2012 ▶). Given the potential for these unique modes of target binding, there is considerable interest in solving the structures of additional FnIII scaffold proteins bound to their cognate targets.
We have developed an FnIII alternative scaffold based on the third such module in human tenascin C (Leahy et al., 1992 ▶). This scaffold, which we have called Tn3, is approximately 90 amino acids long and has been engineered for enhanced stability through the introduction of a disulfide bond (Swers et al., 2013 ▶). Large libraries of phage-displayed Tn3 variants have been created by introducing molecular diversity into the BC, DE and FG loops which connect strands at one end of the β-sandwich. These libraries were used to identify Tn3 proteins that bind human CD40L, a member of the TNF superfamily that has been implicated in the pathogenesis of several autoimmune diseases (for a recent review, see Law & Grewal, 2009 ▶). A number of these Tn3 proteins were found to inhibit the biological activity of CD40L by preventing the binding of this ligand to CD40, its naturally occurring receptor.
Here, we report the expression and purification of two Tn3 proteins that bind CD40L and inhibit its biological activity. We also describe the formation, purification and subsequent crystallization of the two Tn3–CD40L complexes. We expect that elucidation of these crystal structures will help in understanding how these Tn3 proteins are able to neutralize CD40L activity and will provide the first insights into modes of molecular recognition by the Tn3 scaffold.
2. Materials and methods
2.1. Expression and purification of Tn3 molecules and human soluble CD40L
Tn3 proteins that specifically bind CD40L were isolated from large phage-display libraries (Swers et al., 2013 ▶). Two independent clones, Tn3A and Tn3B, were selected for crystallization studies. These were expressed in Escherichia coli using an in-house vector designed to secrete proteins into the periplasmic space. This vector has a Ptac promoter, a signal peptide derived from the E. coli oppA gene product and a C-terminal 8×His tag. In addition, a thrombin cleavage site is located immediately upstream of the C-terminal tag, allowing proteolytic removal. The Tn3 sequences were subcloned between the signal peptide and thrombin cleavage site. Expression plasmids were transformed into E. coli and cultures were grown overnight in auto-inducing MagicMedia (Invitrogen, Carlsbad, California, USA). Secreted His-tagged Tn3 proteins were found to accumulate in the bacterial media, presumably via leakage from the periplasmic space, and were purified using Ni–NTA resin according to the manufacturer’s instructions (Qiagen, Valencia, California, USA). Cleavage of the C-terminal His tag was effected by treatment with bovine thrombin (Sigma–Aldrich, St Louis, Missouri, USA) and the tag fragment was removed by Ni–NTA affinity capture. Untagged Tn3 proteins were further purified by ion-exchange chromatography using a HiTrap Q column (GE Healthcare, Piscataway, New Jersey, USA) performed on an ÄKTApurifier system (GE Healthcare). Pooled fractions containing Tn3 were dialyzed against 50 mM Tris–HCl pH 8. The purified Tn3 proteins showed greater than 95% purity based on SDS–PAGE and analytical size-exclusion chromatography (SEC) and had the expected molecular masses as determined by electrospray mass spectrometry.
A synthetic gene encoding a soluble extracellular fragment of human CD40L (amino acids 113–261; UniProt P29965) fused to an N-terminal 6×His tag was synthesized by GeneArt (Invitrogen). This was cloned into an in-house mammalian expression vector and transiently transfected into HEK293F suspension cells as described elsewhere (Dimasi et al., 2009 ▶). The media were harvested after 4 and 8 d, pooled and soluble CD40L was purified using Ni–NTA resin. Further purification was effected by an ion-exchange step using a HiTrap SP FF column (GE Healthcare) performed on an ÄKTApurifier system (GE Healthcare). Pooled fractions containing CD40L were dialyzed against 50 mM Tris–HCl pH 7.5, 50 mM NaCl. The purified CD40L protein showed greater than 95% purity based on SDS–PAGE and analytical SEC.
To prepare complexes, each Tn3 protein was mixed with CD40L homotrimer in a 3.3:1 molar ratio, concentrated to approximately 10 mg ml−1 using Vivaspin concentrators (30 000 Da cutoff; GE Healthcare) and subjected to SEC using a Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.02% NaN3. After the separation step, the complex was concentrated to 18 mg ml−1 and subjected to crystallization.
2.2. Crystallization screening and optimization
Sitting-drop crystallization experiments were set up in 96-well Intelli-Plates (Art Robbins Instruments, Sunnyvale, California, USA) using a Phoenix crystallization robot (Art Robbins Instruments) by mixing 300 nl volumes of well solution and protein complex solution in the drop compartment and letting the drop equilibrate against 50 µl well solution. Commercial crystallization screens from Hampton Research (Aliso Viejo, California, USA), Emerald BioSystems (Bainbridge Island, Washington, USA) and Molecular Dimensions (Apopka, Florida, USA) were used.
Crystallization of Tn3A–CD40L required an optimization step which included additional screening using Additive Screen HT (Hampton Research). In the optimization step, the well solution of the 96-well plate was filled with 90% of the successful solution from the initial screening and 10% of the respective additive. The drop was made of 300 nl protein solution and 300 nl of the new well solution after thorough mixing of the latter. The diffraction-quality crystals were harvested directly from the 96-well plate. For cryo-preservation, the crystal was transferred into three consecutive solutions of mother liquor with increasing glycerol concentrations.
The crystals of the Tn3B–CD40L complex obtained from the initial screening conditions were of sufficient quality to enable collection of diffraction data without optimization.
2.3. X-ray diffraction and data collection
X-ray diffraction data for the Tn3A–CD40L complex were collected on beamline 5.0.3 of the Advanced Light Source at Lawrence Berkeley National Laboratory (University of California, Berkeley, USA) using an ADSC Q315r CCD detector (Area Detector Systems Corporation, Poway, California, USA). A total of 360 images were collected with 0.8 s exposure, 0.5° oscillation and a 300 mm crystal-to-detector distance.
Diffraction data for the Tn3B–CD40L complex were collected from a single crystal on beamline 31-ID-D of the Advanced Photon Source at Argonne National Laboratory (University of Chicago, Chicago, Ilinois, USA) using a Rayonix 225 HE detector (Rayonix LLC, Evanston, Illinois, USA). A total of 180 images were collected with 0.8 s exposure, 1° oscillation and a 300 mm crystal-to-detector distance.
Data reduction and scaling for both data sets were performed using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
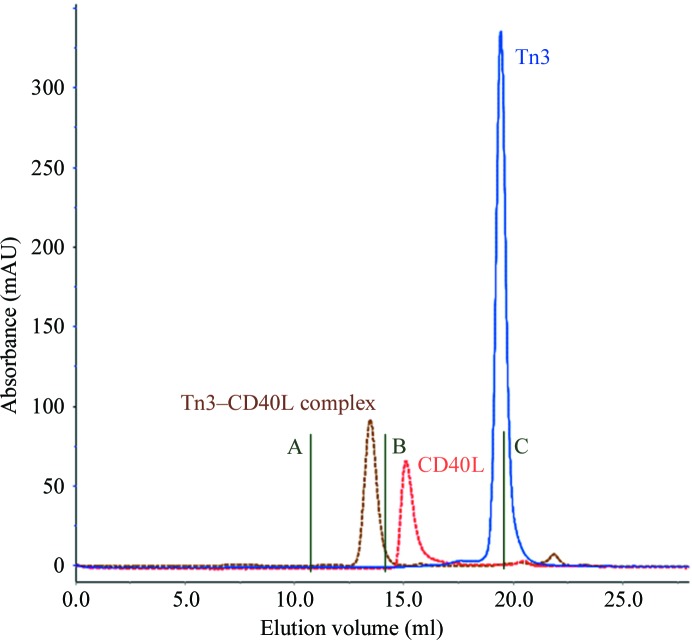
Tn3A and Tn3B, unique Tn3 proteins that specifically bind and inhibit CD40L, were expressed in E. coli and secreted into the periplasmic space (Fig. 1 ▶). Interestingly, bacterial secretion of Tn3 proteins results in their leakage into the culture medium, which eliminates the need to lyse the cells or produce periplasmic extracts prior to downstream purification. Following metal-affinity purification, thrombin digestion to remove the tag sequence and final anion-exchange chromatography, purified Tn3A and Tn3B were shown to be monomeric by SEC and to have the expected molecular masses by electrospray mass spectrometry. A soluble His-tagged fragment of human CD40L protein was produced in HEK293F cells and secreted into the culture medium. Following purification and concentration, this material was used directly in the formation of Tn3 complexes, which were purified by SEC prior to crystallization (Fig. 2 ▶). No effort was made to enzymatically remove the N-linked glycan from Asn240 within the CD40L homotrimer.
Figure 1.

Aligned amino-acid sequences of the Tn3A and Tn3B molecules are presented. Identical amino acids are shown in red.
Figure 2.
Superdex 200 size-exclusion chromatographic profiles of Tn3 (Tn3A), CD40L and the complex between them. Molecular-weight standards A, B and C correspond to ferritin (440 kDa), BSA (67 kDa) and aprotinin (6.5 kDa), respectively.
The most reproducible crystallization condition for the Tn3A–CD40L complex was B5 (0.2 M NaNO3, 20% PEG 3350) in the PEG/Ion screen (Hampton Research). Further optimization with Additive Screen yielded diffraction-quality crystals from condition A1 (0.1 M BaCl2.2H2O). The crystal shown in Fig. 3 ▶(a) was harvested from the 96-well plate and cooled in liquid nitrogen after transfer to mother-liquor solution supplemented with 20% glycerol. The crystal had the symmetry of the orthorhombic space group P212121, with unit-cell parameters a = 85.69, b = 90.64, c = 95.56 Å, and diffracted to 3.1 Å resolution.
Figure 3.
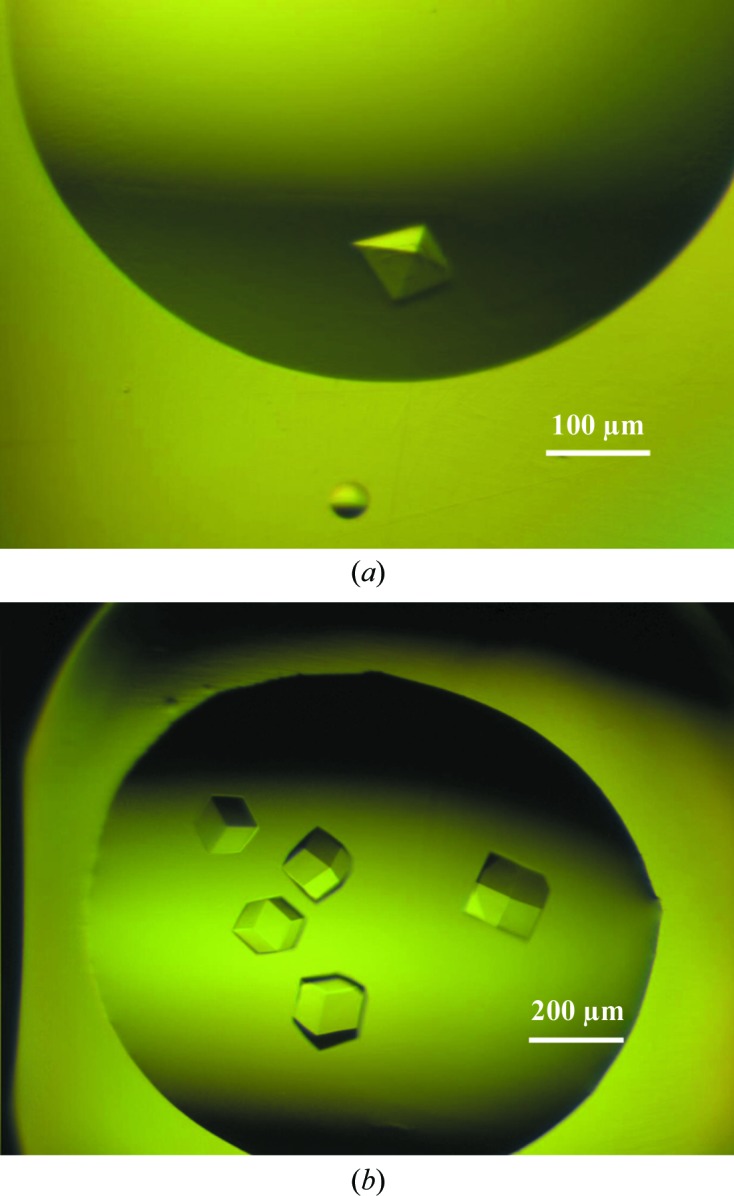
Crystals of the Tn3A–CD40L and Tn3B–CD40L complexes. (a) A crystal of Tn3A–CD40L, which grew to dimensions of 0.15 × 0.15 × 0.1 mm. Conditions were 45 µl reservoir solution (0.2 M NaNO3, 20% PEG 3350) mixed with 5 µl 0.1 M BaCl2.2H2O; 300 nl of this mixture was then combined with 300 nl protein stock solution. (b) Crystals of Tn3B obtained during the initial screening phase using Wizard Cryo 1 and 2 (Emerald BioStructures) under condition F7 (40% PEG 600, 0.1 M sodium acetate, 0.2 M MgCl2).
Crystals of the Tn3B–CD40L complex were obtained under a number of conditions within the Wizard Cryo 1 and 2 screen (Emerald BioStructures) without the need for optimization. A single crystal (Fig. 3 ▶ b) from condition F7 (40% PEG 600, 0.1 M sodium acetate, 0.2 M MgCl2) was used for data collection. The crystal had the symmetry of the cubic space group P213, with unit-cell parameter 97.62 Å, and diffracted to 2.9 Å resolution. Data statistics for crystals of each complex are shown in Table 1 ▶.
Table 1. X-ray data statistics.
Values in parentheses are for the outermost resolution shell.
| Tn3A–CD40L | Tn3B–CD40L | |
|---|---|---|
| Wavelength (Å) | 0.9793 | 0.9793 |
| Resolution (Å) | 50.0–3.05 (3.16–3.05) | 50.0–2.94 (3.05–2.94) |
| Space group | P212121 | P213 |
| Unit-cell parameters (Å) | a = 85.69, b = 90.64, c = 95.56 | a = 97.62 |
| Total reflections | 94024 | 128140 |
| Unique reflections | 14555 | 6720 |
| Average multiplicity | 6.5 (6.4) | 19.2 (19.7) |
| Completeness (%) | 100.0 (100.0) | 99.4 (100.0) |
| R merge † | 0.097 (0.443) | 0.114 (0.785) |
| Mean I/σ(I) | 17.2 (4.6) | 20.1 (2.4) |
R
merge = 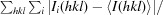
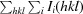 .
.
For molecular replacement, the molecule from PDB entry 2rb8 (Hu et al., 2007 ▶) was used as a template for Tn3 after removing all residues corresponding to the modified loop regions. The model for CD40L used in molecular replacement was taken from PDB entry 1aly (Karpusas et al., 1995 ▶) and was used without modification.
The asymmetric unit of the Tn3A–CD40L crystal is most likely to contain one CD40L homotrimer and three Tn3A molecules, which yields a V M value (Matthews, 1968 ▶) of 2.3 Å3 Da−1, which is within the range expected for protein crystals. The asymmetric unit of the Tn3B–CD40L crystal is most likely to contain one copy of each molecule, which yields a V M of 2.9 Å3 Da−1, which is also within the expected range for protein crystals. These expectations were confirmed by computing molecular-replacement solutions using Phaser (McCoy et al., 2005 ▶). Electron-density maps calculated using the phases generated by Phaser showed a good match to the template models. Refinement and building of the missing parts of these models is under way for both structures.
References
- Dimasi, N., Gao, C., Fleming, R., Woods, R. M., Yao, X.-T., Shirinian, L., Kiener, P. A. & Wu, H. (2009). J. Mol. Biol. 393, 672–692. [DOI] [PubMed]
- Gilbreth, R. N. & Koide, S. (2012). Curr. Opin. Struct. Biol. 22, 413–420. [DOI] [PMC free article] [PubMed]
- Hu, X., Wang, H., Ke, H. & Kuhlman, B. (2007). Proc. Natl Acad. Sci. USA, 104, 17668–17673. [DOI] [PMC free article] [PubMed]
- Karpusas, M., Hsu, Y.-M., Wang, J.-H., Thompson, J., Lederman, S., Chess, L. & Thomas, D. (1995). Structure, 3, 1031–1039. [DOI] [PubMed]
- Koide, S., Koide, A. & Lipovšek, D. (2012). Methods Enzymol. 503, 135–156. [DOI] [PubMed]
- Law, C. L. & Grewal, I. S. (2009). Adv. Exp. Med. Biol. 647, 8–36. [DOI] [PubMed]
- Leahy, D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. (1992). Science, 258, 987–991. [DOI] [PubMed]
- Löfblom, J., Frejd, F. Y. & Ståhl, S. (2011). Curr. Opin. Biotechnol. 22, 843–848. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Skerra, A. (2007). Curr. Opin. Biotechnol. 18, 295–304. [DOI] [PubMed]
- Swers, J. S., Grinberg, L., Wang, L., Feng, H., Lekstrom, K., Carrasco, R., Xiao, Z., Inigo, I., Leow, C. C., Wu, H., Tice, D. A. & Baca, M. (2013). Mol. Cancer Ther. 12, 1235–1244. [DOI] [PubMed]
- Wurch, T., Pierré, A. & Depil, S. (2012). Trends Biotechnol. 30, 575–582. [DOI] [PubMed]