Abstract
Increased atmospheric pCO2 is expected to render future oceans warmer and more acidic than they are at present. Calcifying organisms such as coccolithophores that fix and export carbon into the deep sea provide feedbacks to increasing atmospheric pCO2. Acclimation experiments suggest negative effects of warming and acidification on coccolithophore calcification, but the ability of these organisms to adapt to future environmental conditions is not well understood. Here, we tested the combined effect of pCO2 and temperature on the coccolithophore Emiliania huxleyi over more than 700 generations. Cells increased inorganic carbon content and calcification rate under warm and acidified conditions compared with ambient conditions, whereas organic carbon content and primary production did not show any change. In contrast to findings from short-term experiments, our results suggest that long-term acclimation or adaptation could change, or even reverse, negative calcification responses in E. huxleyi and its feedback to the global carbon cycle. Genome-wide profiles of gene expression using RNA-seq revealed that genes thought to be essential for calcification are not those that are most strongly differentially expressed under long-term exposure to future ocean conditions. Rather, differentially expressed genes observed here represent new targets to study responses to ocean acidification and warming.
Keywords: coccolithophore, acidification, climate change, RNA-seq, transcriptome
1. Introduction
The oceans have the ability to absorb CO2 from the atmosphere, and together with terrestrial ecosystems are essential components controlling atmospheric pCO2 levels and global climate [1]. When CO2 dissolves in seawater, carbonic acid forms, lowering the pH and altering the absolute and relative abundances of dissolved carbonate species; this process is known as ‘ocean acidification’ (OA) [2]. OA affects photosynthesis and calcification of marine organisms such as coccolithophores [3–5], unicellular microalgae that surround themselves with scales of calcium carbonate (coccoliths). In turn, photosynthesis and calcification affect air–sea CO2 exchange and carbon export to the deep sea [6].
To predict future changes in the marine and global carbon cycles, it is important to understand calcium carbonate and organic matter production by coccolithopores under future ocean conditions. Response of coccolithophores, and especially the cosmopolitan species Emiliania huxleyi ((Lohm.) Hay and Mohler), to OA has been intensively studied, but a unified trend in how these cells perform under future conditions has proved elusive so far [7,8]. One of the first studies on E. huxleyi revealed decreasing calcification rates under elevated pCO2 [5]. While this response was later observed in a number of other studies [9–15], contrasting results have also been reported where elevated pCO2 resulted in either no change in calcification (where in this case the term ‘calcification’ refers to calcium carbon quota and/or calcification rate) [15] or increased calcification [4]. Regardless of outcome, these earlier studies tested the response of E. huxleyi to elevated pCO2 in short-term experiments (less than 20 generations). This is potentially problematic, because organisms with short generation times and large population size, such as E. huxleyi, have high potential for acclimation and adaptation to natural environmental change [16,17], and adaptation to future ocean conditions might change calcification responses compared with short-term acclimation. A few studies have tested the evolutionary responses of E. huxleyi to elevated pCO2 in long-term experiments (several hundreds of generations). Müller et al. [11] observed decreases in growth rate and in the ratio of particulate inorganic carbon (PIC) to particulate organic carbon (POC) after 152 generations. Lohbeck et al. [18] also observed decreased growth and calcification rates after approximately 500 generations. However, cells selected under high pCO2 performed better in short-term assays at high pCO2 than those selected under present-day pCO2, suggesting that adaptation had occurred [18]. Similar responses have been observed in marine diatoms [17].
The response of coccolithophores to elevated pCO2 must be considered in the context of other oceanic variables such as temperature [17,19,20], light availability and nutrient concentrations, all of which are expected to change in the future and hence could have interactive effects on E. huxleyi. While there have been tests of E. huxleyi responses to multiple variables [14,21–23], only one study has tested the effect of multiple environmental factors in long-term experiments [24] and found a synergistic effect of pCO2 and nitrogen source on carbon partitioning in E. huxleyi.
The investigation of genes involved in the process of calcification might also provide information about changes in calcification under future ocean conditions. Genes putatively related to calcification (e.g. calcium and inorganic carbon transport, H + transport and carbonic anhydrases) have been identified via gene expression studies comparing calcifying and non-calcifying E. huxleyi cells [25–29], or in short-term experiments where calcification was regulated by limitation of ions needed for calcification (i.e. Ca2+, HCO3−/CO32− [26,30,31]).
To explore the long-term effect of future ocean conditions on E. huxleyi, we grew strain CCMP 371 in continuous culture under simultaneously elevated pCO2 and temperature: ‘present’ ocean conditions (383 ± 43 µatm pCO2 and 20.0 ± 0.1°C average across all generation points) and ‘future’ ocean conditions (833 ± 68 µatm pCO2 and 24.0 ± 0.2°C average across all generation points; see table 1 for details of conditions and carbonate system parameters). The future ocean scenario of approximately 850 µatm pCO2 and +4.0°C was based on the RCP8.5 model [32]. Samples for physiological parameters were taken after 215, 414 and 703 generations to examine responses to future ocean conditions. Genome-wide transcriptomic changes between present and future ocean conditions are discussed at the 215 generation point, and qPCR for some genes that have been previously identified as involved with calcification [26] was measured across all generation points.
Table 1.
Carbonate system for each generation point in the present ocean condition (383 + 43 µatm pCO2, 20.0 + 0.1°C) and future ocean condition (833 + 68 µatm pCO2, 24.0 + 0.2°C). DIC, dissolved inorganic carbon; TA, total alkalinity. Each value is the mean ± s.d. Salinity was 33.5 ± 0.3.
| generations | 215 |
414 |
703 |
|||
|---|---|---|---|---|---|---|
| treatment | present | future | present | future | present | future |
| temperature (°C) | 20.0 ± 0.1 | 24.0 ± 0.2 | 20.0 ± 0.1 | 24.0 ± 0.2 | 20.0 ± 0.1 | 24.0 ± 0.2 |
| DIC± (µmol kg−1) | 1831a ± 86 | 1763a ± 81 | 1968a ± 33 | 2030b ± 25 | 1842a ± 33 | 1910b ± 53 |
| TA± (µmol kg−1) | 2048a ± 88 | 1876b ± 83 | 2232a ± 50 | 2199a ± 29 | 2077a ± 44 | 2041a ± 52 |
| pCO2± (µatm) | 411a ± 39 | 894b ± 22 | 376a ± 46 | 678b ± 81 | 384a ± 46 | 828b ± 51 |
| pH(total scale) | 8.00a ± 0.02 | 7.69b ± 0.04 | 8.06a ± 0.05 | 7.80b ± 0.09 | 8.03a ± 0.05 | 7.73b ± 0.02 |
| (CO2)± (µmol kg−1) | 13.3a ± 1.2 | 25.6b ± 1.5 | 12.3a ± 1.5 | 21.9b ± 5.1 | 12.5a ± 1.5 | 24.2b ± 1.5 |
| (HCO−3)± (µmol kg−1) | 1674a ± 82 | 1657a ± 77 | 1782a ± 33 | 1886b ± 38 | 1672a ± 33 | 1788b ± 51 |
| (CO32−)± (µmol kg−1) | 144.5a ± 6.8 | 82.0b ± 8.0 | 179.7a ± 19.5 | 123.7b ± 20.8 | 157.3a ± 15.9 | 97.2b ± 2.7 |
| Ωcalcite | 3.5a ± 0.2 | 2.0b ± 0.2 | 4.3a ± 0.5 | 3.0b ± 0.5 | 3.8a ± 0.4 | 2.4b ± 0.1 |
| NO2 + NO3± (µM) | 0.09 ± 0.13 | 0.32 ± 0.47 | 0.38 ± 0.93 | 0 ± 0 | 0.34 ± 0.13 | 0.32 ± 0.06 |
Different superscript letters indicate statistical test results between present and future ocean conditions at each generation time.
2. Material and methods
(a). Culture conditions
Monospecific cultures of E. huxleyi (CCMP 371) from the Provasoli-Guillard National Center for Marine Algae and Microbiota were grown in nitrate-limited chemostats (1 l volume) in duplicate under two different treatments. The first treatment represented ‘present ocean conditions’ with pCO2 and temperature levels of 383±43 µatm and 20.0±0.1°C, respectively (table 1). The second treatment represented ‘future ocean conditions’ with elevated pCO2 and temperature (average 833 ± 68 µatm and 24.0 ± 0.2°C, respectively; table 1). Chemostats were run with a dilution rate (equivalent to growth rate) of 1.1 d−1 using 0.2 µm filtered nutrient-poor, aged seawater with a salinity of 33.5 ± 0.3 (collected offshore of Half Moon Bay, CA, USA; 37° 29′ 31′ N, 122° 30′ 02′ W), enriched with phosphate (14 µM), and with a metal and vitamin mix according to f/2 concentrations [33]. To archive a cell concentration of approximately 500 000 cells ml−1, nitrate concentrations were raised to 30 µM in the ‘present ocean condition’ media and 35 µM in the ‘future ocean condition’. These nitrate concentrations were found in prior tests to yield the target cell concentration. Media were augmented with sodium bicarbonate to compensate for dissolved inorganic carbon (DIC) uptake during calcification (by 245 and 250 µM for the ‘present ocean condition’ and ‘future ocean condition’ media, respectively). This was done after approximately 88 generations to adjust to the DIC uptake as calculated from the carbonate system (see below for carbonate chemistry determination). The 5 µM difference in DIC concentrations in the two media after augmentation is less than 1% of total DIC, and therefore three to eight times lower than the DIC change that is commonly accepted for diluted batch experiments with coccolithophores [12,34].
pCO2 levels in the cultures were controlled by continuously bubbling the culture and the media reservoir with CO2–air mixtures to achieve pCO2 levels of approximately 400 and 900 µatm. Media reservoirs were bubbled for at least 1 day prior to use for pre-equilibration. The pCO2 concentration in gas mixtures was frequently monitored using an infrared gas analyser (LI-820, LI-COR). Cultures were stirred gently throughout the experiment to ensure homogeneous cell suspension and avoid possible light and nutrient gradient effects. Cultures were illuminated on a 16 L : 8 D cycle with cool-white fluorescent lamps (Vita-Lite 5500 K, DUROTEST) at 240–320 µmol photons m−2 s−1. Photon flux density was measured in the centre of each vessel containing E. huxleyi culture using a 4 pi Biospherical instruments probe, model QSP170B. Cell division was synchronized to the photoperiod and a diurnal fluctuation of cell density was found that might have caused a diurnal fluctuation in the carbonate system as well as nutrient concentration.
(b). Sampling and analyses of cultures
Cultures were sampled at three generation points, from 199 to 232 generations (‘215 generations’), from 393 to 436 generations (‘414 generations’) and from 683 to 724 generations (‘703 generations’). Each of these samplings occurred over the course of three to four weeks where various samples were collected on a daily basis (see the electronic supplementary material, table S1). The reason for the extended sampling period was to avoid removing less than 10% of the chemostat volume at any given time, thereby minimizing perturbations to the steady state [35]. The chemostat outflow was not sampled, because it was not maintained under the same temperature, pCO2, and light conditions as the culture. At each generation point, samples for cell density and dilution rate were collected daily (n = 22, 27 and 28 for generation 215, 414 and 703, respectively; electronic supplementary material, table S1). Samples for primary production, calcification rate, DIC, total alkalinity (TA), chlorophyll a (Chl a), and nutrients (n = three per generation point) were collected approximately weekly, and samples for total particulate carbon (TPC) and POC (n = six per generation point) were collected every third or fourth day (see the electronic supplementary material, table S1). Samples for gene expression (n = six per generation point) were collected every 3–6 days (electronic supplementary material, table S1). All samples were collected 14 h after the light onset. Allowing for multiple days to pass between sampling for physiological parameters ensured that the population of cells had been replaced completely between collection points, given the high growth rate in our chemostats of 1.1 d−1 (more than one division per day). This sampling strategy is routinely used for continuous culture experiments, as cells several generations apart represent independent samples of the same population [13,24,31,36].
TA samples (approx. 80 ml) were filtered (combusted GF/F filters, Whatman) and stored in borosilicate glass bottles in the dark at 4°C until measurement. TA was measured on 50 ml of the sample by potentiometric titration [37] and the endpoint determined from Gran plots [38]. DIC samples (24 ml) were sterile filtered (0.2 µm syringe filter, Nalgene) into borosilicate glass vials and stored without headspace in the dark at 4°C until measurement. DIC was measured following Friederich et al. [39], but with pure O2 carrier gas instead of N2. The carbonate system parameters were determined from DIC TA, salinity, temperature, silicate and phosphate concentrations using CO2SYS [40] with constants from Mehrbach et al. [41] refit by Dickson & Millero [42].
Samples for determination of TPC and POC were filtered on pre-combusted GF/F filters (Whatman) (75 and 125 ml, respectively) and dried at 55°C overnight. Prior to analysis, the POC filters were acidified with 230 µl of 1 N HCl to remove inorganic carbon and dried again. TPC and POC were measured on a Costech ECS 4010 CHNSO analyser. PIC was calculated as the difference between TPC and POC. POC and PIC values were normalized either by the volume of water filtered or by the number of cells filtered (determined from sample volume and cell density).
Primary production and calcification rates were determined using 14C incorporation modified after Balch et al. [43]. A 20 ml subsample of the culture was spiked with 6.84 µCi of H14CO3 and incubated for 2 h. Incubations were fixed with 5% HgCl2 after 2 h and 5 ml were filtered four times onto GF/F filters (Whatman) for repeated measurements. Half of the filters were acidified, and radioactivity was measured in Optiphase ‘Hisafe’ 3 scintillation cocktail (Perkin-Elmer Inc., USA) on a Wallac Guardian 1414 liquid scintillation counter (Perkin-Elmer Life and Analytical Sciences Inc.). Calcification rate was calculated with the radioactivity of the acidified (primary production) and non-acidified (total carbon production) filters.
The effect of treatment, chemostat and generation points and their interactions on all response variables was investigated by ANOVA. TukeyHSD post-hoc test was used to characterize pairwise differences across the three generation points in each treatment. Planned comparisons for differences between the treatments at each generation point were performed using unpaired t-tests. Data were analysed by pooling all replicates from one treatment (two chemostats) for each generation point. T-tests were used to test that the two replicate chemostats for each treatment were not significantly different from each other before they were pooled together by treatment. Outliers in each dataset were identified with the Grubbs test for outliers [44]. No more than one outlier was removed from any dataset.
(c). Transcriptomics
One hundred millilitres of culture was filtered onto 1.0 µm polycarbonate filters, which were immediately frozen in liquid nitrogen, then stored at −80°C. Total RNA was extracted from samples using Tri Reagent (Molecular Research Center, Inc.) according to the manufacturer's protocol. RNA quantity was measured using a Qubit v. 2.0 Fluorometer (Life Technologies) and integrity was confirmed using a Bioanalyzer (Agilent 2100) RNA assay.
RNA-seq libraries were constructed using Illumina TruSeq reagents according to the manufacturer's protocol (Illumina, TruSeq RNA Sample Preparation Kit v. 2). Clean-up steps were performed using AMPure XP beads (Agencourt), and first-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Prior to sequencing, cDNA library concentrations were measured via Qubit Fluorometer and fragment size distribution was confirmed by Bioanalyzer high-sensitivity DNA assay. Libraries were sequenced at the Vincent J. Coates Genomics Sequencing Laboratory, University of California Berkeley on the Illumina HighSeq 2000 platform using 100 base pair paired-end sequencing. Multiplexing was at n = 8 libraries per lane.
Raw sequence reads were trimmed for quality using the program DynamicTrim (SolexaQA_v. 2.0, [45]), eliminating sequence with a probability below 0.05 and a quality score below 13 from the analysis. RNA-seq reads were aligned and mapped to the E. huxleyi CCMP1516 draft genome sequence (v. 1.0, Emihu1_best_transcripts.fasta) from the Joint Genome Institute (http://genome.jgi-psf.org/Emihu1/Emihu1.home.html), using Tuxedo Suite software for read alignment and quantification with TopHat (v. 2.0.3, [46]), short read assembly with Bowtie (v. 0.12.2, [47]) and SamTools (v. 0.1.18, [48]). Mapped reads were annotated using the E. huxleyi draft genome sequence and supporting files (including KOG, KEGG and GO identifications). Putative transcripts with fewer than 100 mapped reads were removed from the dataset, and the remaining transcripts were tested for differential expression using edgeR (v. 2.6.12, [49]). Statistical analyses were conducted using pair-wise comparisons of individual chemostats within and between treatments, with a false discovery rate adjusted p-value of 0.05. Genes that differed in at least three of four pairwise comparisons between treatments, but that did not differ in the within-treatment comparison across replicate chemostats were considered differentially expressed in response to present versus future ocean conditions.
Quantitative real time PCR (qPCR) verification of expression in all chemostats across all three generation points was conducted for calcification-related transcripts, a gamma carbonic anhydrase, a cation/H+ exchanger, a putative bicarbonate transporter and proton channel, and normalized to the housekeeping gene actin using published primer sequences (table 2) [26]. RNA was extracted from two samples per chemostat per generation point (see electronic supplementary material, table S1) using Tri Reagent (Molecular Research Center, Inc.) according to the manufacturer's protocol and reverse transcribed into cDNA using Improm-II reverse transcriptase (Promega). Samples were DNAse treated and purified using the RNeasy mini kit (Qiagen). RNA quantity and purity were assessed by NanoDrop spectrophotometer (A260 and A260/A280, respectively) and integrity was assessed by agarose gel electrophoresis. qPCR assays were conducted using SYBR green mastermix (Fermentas Maxima, Thermo Scientific) with the passive reference dye ROX on an ABI7300 real time PCR instrument (Applied Biosystems).
Table 2.
qPCR Primers and amplicon length.
| putative transcript | short name | JGI ID | forward primera | reverse primer | amplicon length |
|---|---|---|---|---|---|
| actin | actin | n.a. | GAC CGA CTG GAT GGT CAA G | GCC AGC TTC TCC TTG ATG TC | 96 |
| gamma carbonic anhydrase | γ-CA | 432493 | TCT CCG CCT CAG TCA ACC | AAG TTG TCG ACT GTG CAA CC | 106 |
| cation/H+ exchanger 3 | CAX3 | 416800 | CTC CTC TGC GTC TTT GCA T | GAG GGC GGT GAT GAG GTA | 90 |
| subunit c of the V0 sector of a vacuolar H+-ATPase | ATPVc/c | 359783 | TAC GGC ACT GCA AAG TCT G | ACG GGG ATG ATG GAC TTC | 83 |
| anion exchanger-like 1 | AEL1 | 198643 | TTC ACG CTC TTC CAG TTC TC | GAG GAA GGC GAT GAA GAA TG | 102 |
aAll primer sequences are given in 5′–3′ direction.
3. Results
(a). Physiological responses to warm and acidified conditions
Physiological responses were not affected by the interaction between treatment and generation point (table 3). Cells had carbon to nitrogen (C : N) ratios (figure 1) of approximately 11 in both treatments at the 215 and 703 generation points, though the ratio was lower in future ocean conditions at the 703 generation point (t-test t8 = 0.62, p = 0.55 at 215 generations; t10 = 2.7, p = 0.026 at 703 generations). The C : N ratio (figure 1) at the 414 generation point was lower at approximately 8.3, but was not significantly different between the treatments (t-test t9 = 0.68, p = 0.52). The sum of nitrate and nitrite concentrations in the chemostats was at all generation points below 1 µM (table 1). PIC content (figure 2a) increased by 26% under future ocean conditions at all three generation points (215, 414 and 703 generations), though this was statistically significant only at the latter two generation points (t-test t14 = 2.0, p = 0.07 at 215 generations; t21 = 4.0, p < 0.001 at 414 generations; t22 = 2.9, p < 0.01 at 703 generations). POC content (figure 2b) did not dramatically differ between present and future ocean conditions at 215 and 414 generations (t-test t20 = 2.7, p = 0.02 at 215 generations, t-test with Welch correction, t16 = 1.5, p = 0.15 at 414 generations), though values were between 10 and 28% higher in the future ocean condition (table 4). At the 703 generation point, POC content (figure 2b) was significantly higher (t-test t22 = 2.0, p = 0.002) by 14% in the future ocean condition. Calcification rate (figure 3a) increased by 26% under future ocean conditions, though this was statistically significant only at the 215 generation point (t-test t9 = 4.5, p < 0.002). Primary production (figure 3b) did not differ between present and future ocean conditions (t-test t9 = 0.29, p = 0.78 at 215 generations; t10 = 0.44, p = 0.67 at 703 generations). PIC : POC cell−1 (table 4) did not differ between treatments at any generation point (t-test t14 = 0.35, p = 0.74 at 215 generations; t22 = 1.8, p = 0.09 at 414 generations; t22 = 0.98, p = 0.34 at 703 generations).
Table 3.
Significance test results for treatment, generation point and their interaction based on a two-way ANOVA. TA, total alkalinity; DIC, dissolved inorganic carbon; pCO2, carbon dioxide concentration; HCO3−, bicarbonate concentration; CO32−, carbonate concentration; POC, particulate organic carbon content per cell; PP, primary production per cell; PIC, particulate inorganic carbon content per cell; CR, calcification rate per cell; Chl a, chlorophyll a content per cell. For units, see table 1.
| parameter | treatment | generation point | treatment × generation point |
|---|---|---|---|
| TA | F1,28 = 15.1, p < 0.001*** | F2,28 = 45.1, p < 0.0001*** | F2,28 = 4.70, p = 0.017* |
| DIC | F1,30 = 1.17, p = 0.29 | F2,30 = 37.9, p < 0.0001*** | F2,30 = 5.42, p < 0.010* |
| pCO2 | F1,27 = 154, p < 0.0001*** | F2,27 = 1.85, p = 0.177 | F2,27 = 0.472, p = 0.629 |
| HCO3− | F1,28 = 11.2, p = 0.002** | F2,28 = 23.9, p < 0.0001*** | F2,28 = 4.86, p = 0.016* |
| CO32− | F1,28 = 166, p < 0.0001*** | F2,28 = 22.4, p < 0.0001*** | F2,28 = 0.161, p = 0.852 |
| pH | F1,28 = 323, p < 0.0001*** | F2,28 = 8.93, p = 0.001*** | F2,28 = 0.847, p = 0.440 |
| Ω | F1,28 = 164, p < 0.0001*** | F2,28 = 22.0, p < 0.0001*** | F2,28 = 0.157, p = 0.855 |
| POC | F1,63 = 16.5, p = 0.0001*** | F2,63 = 19.7, p < 0.0001*** | F2,63 = 0.266, p = 0.767 |
| PP | F1,19 = 0.257, p = 0.618 | F1,19 = 7.25, p = 0.014* | F1,19 = 0.090, p = 0.768 |
| PIC | F1,57 = 23.9, p < 0.0001*** | F2,57 = 12.9, p < 0.0001*** | F2,57 = 0.048, p = 0.954 |
| CR | F1,17 = 5.770, p = 0.028* | F1,17 = 0.018, p = 0.894 | F1,17 = 0.014, p = 0.908 |
| Chl a | F1,28 = 29.7, p < 0.0001*** | F2,28 = 55.6, p < 0.0001*** | F2,28 = 2.35, p = 0.114 |
| POC l–1 | F1,63 = 15.7, p = 0.0002*** | F2,63 = 29.4, p < 0.0001*** | F2,63 = 12.3, p < 0.0001*** |
| PIC l–1 | F1,56 = 15.70, p = 0.002** | F2,56 = 11.0, p = 0.0001*** | F2,56 = 2.10, p = 0.132 |
| Chl a l–1 | F1,30 = 9.59, p = 0.004** | F2,30 = 19.2, p < 0.0001*** | F2,30 = 0.238, p = 0.790 |
Asterisks denote degree of statistical significance (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
Figure 1.
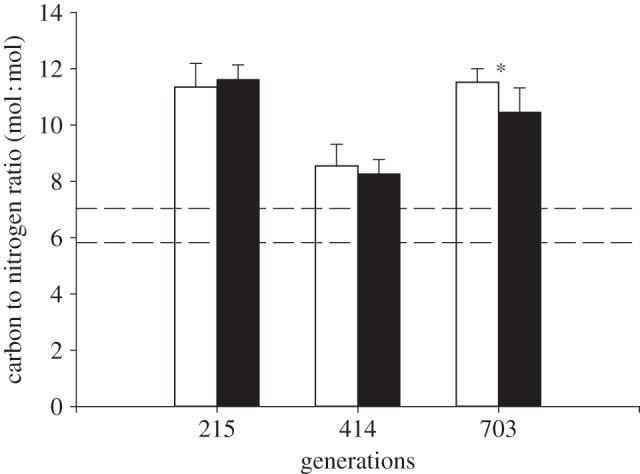
Carbon to nitrogen (C : N) ratio of the E. huxleyi ((Lohm.) Hay and Mohler) cells at 215, 414 and 703 generation points under present ocean condition (383 ± 43 µatm pCO2, 20.0 ± 0.1°C; white bars) and future ocean condition (833 ± 68 µatm pCO2, 24.0 ± 0.2°C; black bars). Values are averages of two replicates with three measurements each; error bars show standard deviation. Asterisks indicate statistically significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) between treatments at each generation point. Dashed lines are the range of the C : N ratio in Feng et al. [23] with the same strain of Emiliania huxleyi.
Figure 2.
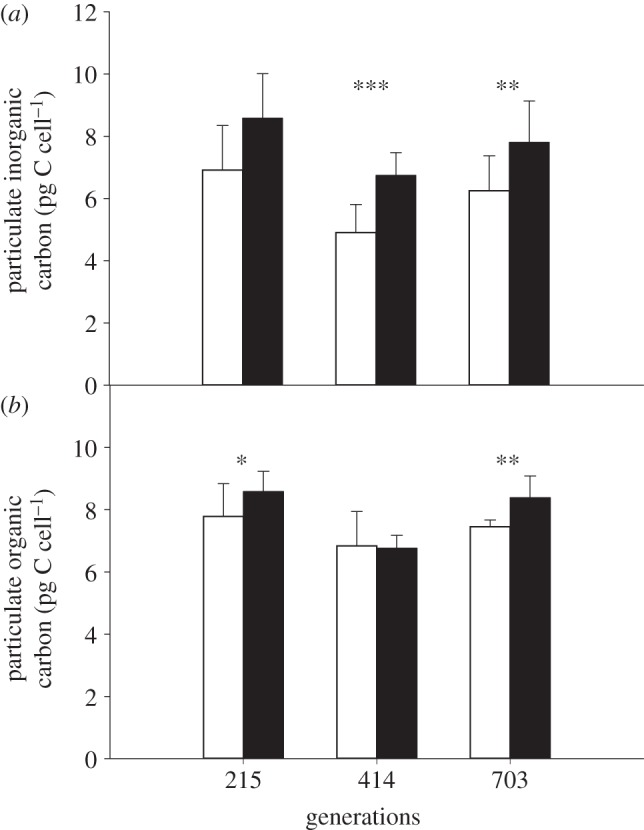
(a) PIC and (b) POC content of Emiliania huxleyi at 215, 414 and 703 generation points under present ocean condition (383 ± 43 µatm pCO2, 20.0 ± 0.1°C; white bars) and future ocean condition (833 ± 68 µatm pCO2, 24.0 ± 0.2°C; black bars). Values are averages of two replicates with three measurements each; error bars show standard deviation. Asterisks indicate statistically significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) between treatments at each generation point.
Table 4.
Chlorophyll a (chl a) content per cell and particulate organic and inorganic carbon (POC and PIC) content per litre of culture at 215, 414 and 703 generations under present ocean condition (383 ± 43 µatm pCO2, 20.0 ± 0.1°C) and future ocean condition (833 ± 68 µatm pCO2, 24.0 ± 0.2°C). Values are averages ± s.d. of two replicates with three to six measurements each.
| generations | 215 |
414 |
703 |
|||
|---|---|---|---|---|---|---|
| treatment | present | future | present | future | present | future |
| Chl a (fg cell−1) | 52 ± 5 | 60 ± 7 | 42 ± 2 | 54 ± 10 | 68 ± 5 | 88 ± 10 |
| Chl a (mg l−1) | 31 ± 12 | 38 ± 9 | 23 ± 2 | 28 ± 2 | 38 ± 3 | 47 ± 5 |
| POC (mg l−1) | 3.86 ± 0.48 | 5.02 ± 0.42 | 3.41 ± 0.45 | 3.60 ± 0.32 | 4.07 ± 0.33 | 4.04 ± 0.51 |
| PIC (mg l−1) | 3.60 ± 0.83 | 4.73 ± 0.92 | 2.91 ± 0.51 | 3.39 ± 0.48 | 3.63 ± 0.57 | 3.89 ± 0.63 |
| PIC : POC (mol : mol) (l−1) | 0.92 ± 0.25 | 0.92 ± 0.19 | 0.87 ± 0.18 | 0.95 ± 0.13 | 0.90 ± 0.18 | 0.98 ± 0.18 |
| PIC : POC (mol : mol) (cell−1) | 0.93 ± 0.25 | 0.96 ± 0.24 | 0.81 ± 0.22 | 0.95 ± 0.16 | 0.90 ± 0.18 | 0.98 ± 0.18 |
Figure 3.
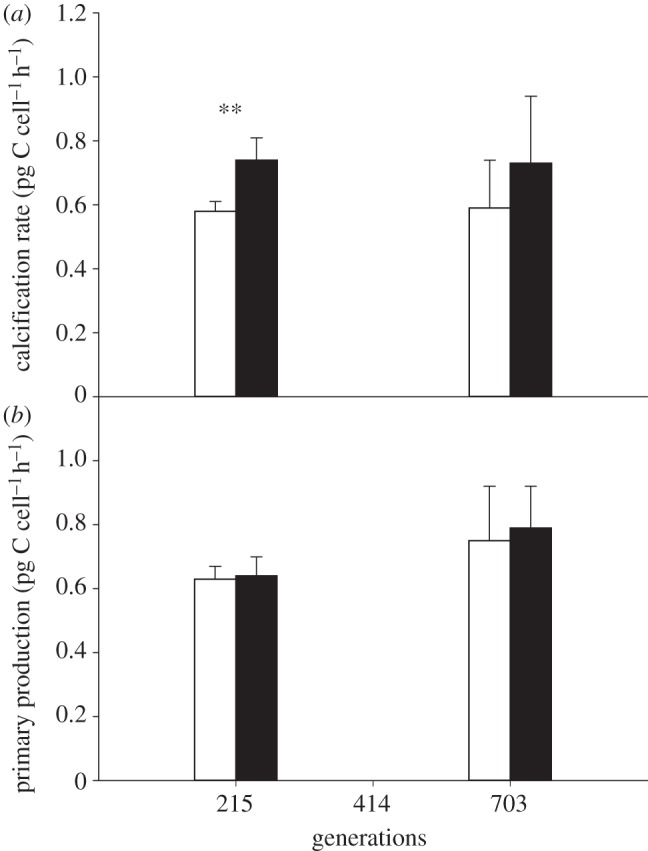
(a) Calcification rate and primary production (b) of Emiliania huxleyi at 215 and 703 generation points under present ocean condition (383 ± 43 µatm pCO2, 20.0 ± 0.1°C; white bars) and future ocean condition (833 ± 68 µatm pCO2, 24.0 ± 0.2°C; black bars). No data for 414 generation point. Values are averages of two replicates with three measurements each; error bars show standard deviation. Asterisks indicate statistically significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) between treatments at each generation point.
Biomass, quantified as POC content per unit volume of culture (table 4), differed between the treatments at the 215 generation point (t-test t19 = 5.9, p < 0.0001), but not at the other two generation points (t-test t22 = 1.1, p = 0.27 at 414 generations; t22 = 0.17, p = 0.87 at 703 generations) and an interactive effect of treatment and generation point was observed (table 3). Chl a (table 4) was not interactively affected by treatment and generation point (table 3) and showed no differences between the treatments at the 215 generations point (t-test t10 = 1.2, p = 0.25), but was different at the other two generation points (t-test t10 = 3.5, p < 0.01 at 414 generations; t10 = 3.7, p < 0.01 at 703 generations).
The total PIC concentration per unit volume of culture (table 4) was not affected by the interaction between treatment and generation point (table 3). Small differences in PIC l−1 were observed between treatments at the first two generation points (t-test t12 = 2.4, p = 0.03 at 215 generations; t22 = 2.4, p = 0.03 at 414 generations; t22 = 1.1, p = 0.30 at 703 generations). PIC : POC l−1 (table 4) did not differ between treatments at any generation point (t-test t12 = 0.06, p = 0.96 at 215 generations; t22 = 1.2, p = 0.23 at 414 generations; t22 = 0.98, p = 0.34 at 703 generations).
(b). Transcriptomic responses to warm and acidified conditions
Sequencing generated 1 B short sequence reads of 100 base pair length, with an average of 43 M reads per sample and 259 M reads per chemostat (see electronic supplementary material, table S2). Sequence reads were mapped to 39 K putative transcripts in the E. huxleyi 1516 reference transcriptome [50], and 17 K putative transcripts had greater than 100 average reads and were considered in the statistical analysis to identify differentially expressed transcripts.
Comparison of replicate chemostats within the future treatment revealed 229 differentially expressed putative transcripts (see electronic supplementary material, table S3). One hundred and three putative transcripts were differentially expressed between replicate chemostats of the present treatment (see electronic supplementary material, table S4), one-third of which were identical to differentially expressed transcripts within the future treatment. For a given condition (present or future), the average fold-changes between replicate chemostats were fourfold or lower, whereas the largest fold-changes between biological replicate chemostats were over 16-fold (see electronic supplementary material, tables S3 and S4).
Of the 121 transcripts differentially expressed between present and future ocean condition treatments, 116 putative transcripts were upregulated in the future ocean condition treatment, whereas five were downregulated (see electronic supplementary material, table S5). Fold expression change between treatments ranged from 1.6-fold to nearly sevenfold.
The five putative transcripts downregulated in the future treatment (figure 4) were related to metabolism, transcription and cellular stress. Downregulated metabolism-related transcripts encode a mitochondrial oxoglutarate/malate carrier protein, a flavin-containing monooxygenase, and an FAD-dependent oxidoreductase. Additionally, a DNA-dependent transcription regulation factor and a molecular chaperone from the HSP70 superfamily were downregulated (figure 4).
Figure 4.

Downregulated putative transcripts in Emiliania huxleyi grown under future ocean conditions at the 215 generation point.
Of the 116 putative transcripts upregulated in the future ocean condition treatment, 44 are putatively involved in cellular processes and signalling, 13 are related to information storage and processing and nine are related to metabolic processes (figure 5). Additionally, three putative transcripts encode viral-related proteins. Putative transcripts without functional annotation (n = 47) represent hypothetical proteins of unknown function or putative transcripts with no annotation (see electronic supplementary material, table S5; figure 5). Genes represented in each of the main functional categories are presented by category below.
Figure 5.
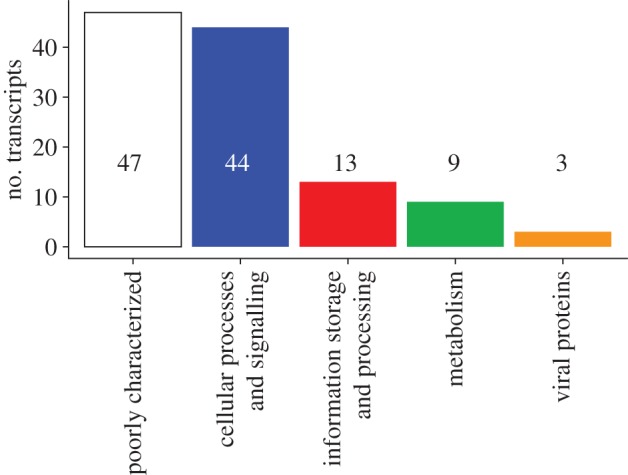
Functional categorization of the upregulated putative transcripts in Emiliania huxleyi grown under future ocean conditions at the 215 generation point. (Online version in colour.)
(i). Cellular processes and signalling
Upregulated putative transcripts from the cellular processes and signalling categorization represent pathways including cell structure (cell wall/membrane/envelope biogenesis, extracellular structures), defence mechanisms, signal transduction mechanisms and protein homeostasis (post-translational modification/protein turnover/chaperones) (figure 6). The 16 transcripts associated with cell wall/membrane/envelope biogenesis include two putative membrane proteins, two junctional membrane protein complexes, and one transcript with homology to a cell wall surface anchored protein (see electronic supplementary material, table S5). Several putative transcripts with homology to chitinase and chitinodextrinase were upregulated in future ocean conditions, as well as two putative transcripts with homology to ankyrins, and one transcript with homology to an ABC transporter solute-binding protein (see electronic supplementary material, table S5).
Figure 6.
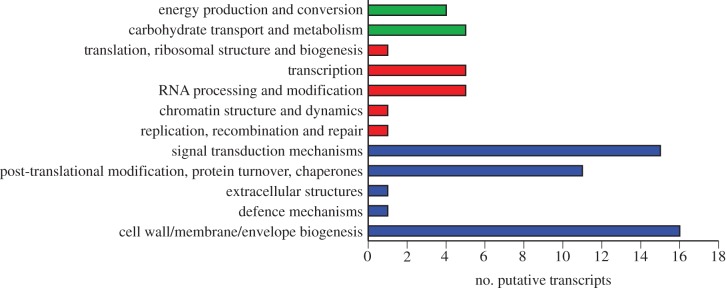
Major pathways in each functional categorization from figure 5 that were upregulated in future ocean conditions at the 215 generation point. For colours follow figure 5. (Online version in colour.)
Transcripts (n = 11) encoding proteins involved in post-translational modification were upregulated in the future ocean condition treatment, including three putative transcripts from the peptidase families S8 and S53 that were upregulated fourfold to sevenfold (see electronic supplementary material, table S5). Additional transcripts encoding proteins involved in protein modification that were upregulated in future ocean conditions include protein tyrosine phosphatase, phospholipase C, serine carboxypeptidase, homocysteine S-methyltransferase, insulysin, thioredoxin/protein disulfide isomerase and two poorly characterized proteins involved in proteolysis and peptidolysis, one with putative subtilase activity and another with cysteine-type endopeptidase activity (see electronic supplementary material, table S5).
Putative transcripts (n = 15) displaying the greatest expression differential between present and future ocean conditions coded for genes involved with signal transduction mechanisms (see electronic supplementary material, table S5). Among these were several putative transcripts with homology to serine/threonine protein kinase, serine/threonine-specific protein phosphatase and phosphoinositide phospholipase C. One of the serine/threonine protein kinases (JGI ID: 432478) was upregulated nearly sevenfold in the future ocean condition treatment, whereas a phosphoinositide phospholipase C (JGI ID: 95985) increased expression sixfold (see electronic supplementary material, table S5). Additional upregulated signal transduction related genes encoded proteins polycystin-1L1, MLP and LIM regulatory proteins, fibrillins or related proteins containing Ca2+-binding EGF-like domains, Ca2+/Mg2+-permeable cation channel, and other differentially expressed putative transcripts that may play a role in cellular signaling, including known extracellular proteins c-type lectin and collagen (see electronic supplementary material, table S5).
(ii). Information storage and processing
Most of the 13 differentially expressed putative transcripts related to information storage and processing encode proteins involved with translation or RNA processing and modification, including a TATA box-binding protein (TBP)-associated factor, a translational repressor of the Puf superfamily (more specifically related to Pumilio/PUF3 and related RNA-binding proteins), a DNA-dependent transcription regulator and three putative transcripts coding for proteins with a Myb-like DNA-binding domain (see electronic supplementary material, table S5). Putative transcripts related to RNA processing and modification included a translation initiation factor (IF-2), two putative DEAD-box RNA helicases (DDX1), a splicing co-activator SRm160/300, subunit SRm160 (contains PWI domain) and a hypothetical protein that possibly binds to RNA (see electronic supplementary material, table S5). A transcript encoding a putative DNA mismatch repair protein was upregulated 4.5-fold, and a transcript with a ubiquitin interaction motif was also upregulated (see electronic supplementary material, table S5).
(iii). Metabolism
There was no strong over-representation of any particular metabolic pathway among the nine upregulated putative transcripts related to metabolism. Those transcripts encoded glycosyl hydrolases, cellulose 1,4-beta-cellobiosidase, myo-inositol-2-dehydrogenase, two putative transcripts with homology to sulfite oxidase, a putative lipoprotein, an endoglucanase 2 precursor, a glycoprotein precursor and a hypothetical protein with homology to a multiple sugar transport system (see electronic supplementary material, table S5).
(iv). Calcification-related transcripts
We screened the RNA-seq data for mapped reads to genes in coccolithophores that have been previously investigated for their potential role in calcification and pH homeostasis, including transcripts associated with calcium and inorganic carbon transport, H+ transport and carbonic anhydrases (table 5). No gene previously shown to differ as a function of coccolithophore calcification was differentially expressed between present and future ocean condition treatments in this study (table 5).
Table 5.
Potential calcification-related genes examined in our study, separated by functional groupings of calcification processes of calcium binding and transport, carbonic anhydrases, inorganic carbon transport and pH homeostasis. Genes with statistically significant differential expression are indicated in italics. Citations indicate genes previously described to be involved with calcification.
| JGI ID | gene description | expression in future conditionsa | expression in present conditionsa | referencec |
|---|---|---|---|---|
| calcium transport | ||||
| 522053 | ER-type CA2+ ATPase 2 | n.d.b | n.d. | [26] |
| 416800 | Ca2+/H+ exchanger (CAX3) (CAX family) | 14 027 ± 4744 | 16 097 ± 2705 | [26,27] |
| 415715 | Ca2+/H+ exchanger (CAX4) (CAX family) | 1100 ± 527 | 836 ± 278 | [26,27] |
| 448526 | four domain voltage-gated Ca2+ channels | 2163 ± 729 | 2287 ± 772 | |
| 448907 | four domain voltage-gated Ca2+channels | 2490 ± 960 | 2175 ± 557 | |
| 66584 | cation/H+ exchanger (CAX family) | 0 ± 1 | 0 ± 1 | [27] |
| 72273 | cation/H+ exchanger (CAX family) | 378 ± 60 | 433 ± 87 | [27] |
| 103021 | cation/H+ exchanger (CAX family) | 128 ± 39 | 137 ± 33 | [27] |
| 223499 | cation/H+ exchanger (CAX family) | 382 ± 86 | 384 ± 93 | [27] |
| 315097 | cation/H+ exchanger (CAX family) | 0 ± 1 | 0 ± 1 | [27] |
| 203920 | cation/Ca2+exchanger | 457 ± 120 | 345 ± 110 | |
| 449053 | cation/Ca2+exchanger | 710 ± 277 | 605 ± 215 | |
| 62350 | calcium pump | 1540 ± 617 | 1611 ± 455 | |
| 101130 | calcium pump | 1 ± 2 | 2 ± 3 | |
| 426283 | calcium pump | 441 ± 319 | 363 ± 488 | |
| 466567 | calcium pump | 1087 ± 274 | 1185 ± 245 | |
| 205210 | K+-dependent Na+/Ca2+ exchanger (NCKX1; NCKX family) | 0 ± 0 | 0 ± 1 | |
| 447939 | K+-dependent Na+/Ca2+ exchanger (NCKX2; NCKX family) | 42 857 ± 15 007 | 45 416 ± 9684 | |
| 454623 | potassium-independent sodium/calcium exchanger | 590 ± 148 | 618 ± 174 | |
| 107737 | Ca2+/Mg2+-permeable cation channels transient receptor potential | 491 ± 354 | 301 ± 132 | |
| 449985 | Ca2+/Mg2+-permeable cation channels transient receptor potential | 2016 ± 670 | 2899 ± 713 | |
| 455760 | Ca2+/Mg2+-permeable cation channels transient receptor potential | 2546 ± 771 | 3610 ± 805 | |
| 468587 | Ca2+/Mg2+-permeable cation channels transient receptor potential | 1964 ± 685 | 2013 ± 459 | |
| 460292 | Ca2+/Mg2+-permeable cation channels (LTRPC family) | 806 ± 590 | 450 ± 135 | |
| 118025 | fibrillins and related proteins containing Ca2+-binding EGF-like domains | 418+282 | 135+113 | |
| 463266 | fibrillins and related proteins containing Ca2+-binding EGF-like domains | 462+302 | 149+123 | |
| 431830 | GPA (glutamic acid, proline and alanine) unknown, calcium binding | 198 ± 113 | 266 ± 186 | [26,51,52] |
| carbonic anhydrases | ||||
| 356859 | carbonic anhydrase, carbon utilization | 77 ± 18 | 95 ± 33 | |
| 56989 | carbonic anhydrase, putative | 72 ± 52 | 77 ± 43 | |
| 233460 | carbonic anhydrase, alpha | 304 ± 108 | 286 ± 117 | |
| 62679 | carbonic anhydrase, alpha | 36 ± 15 | 42 ± 22 | |
| 437452 | carbonic anhydrase, alpha | 567 ± 219 | 623 ± 186 | [30] |
| 456048 | carbonic anhydrase, alpha | 632 ± 239 | 582 ± 206 | [26,51] |
| 558406 | carbonic anhydrase, alpha | n.d. | n.d. | |
| 115240 | carbonic anhydrase, beta | 127 ± 55 | 132 ± 45 | |
| 233823 | carbonic anhydrase, beta | 0 ± 0 | 0 ± 0 | |
| 239690 | carbonic anhydrase, beta | 2 ± 2 | 2 ± 2 | |
| 469462 | carbonic anhydrase, beta | 699 ± 192 | 833 ± 203 | |
| 469783 | carbonic anhydrase, delta | 3459 ± 760 | 4081 ± 838 | |
| 195575 | carbonic anhydrase, delta | 0 ± 1 | 0 ± 1 | |
| 222602 | carbonic anhydrase, delta | 158 ± 67 | 201 ± 75 | |
| 227360 | carbonic anhydrase, delta | 0 ± 1 | 0 ± 1 | |
| 229394 | carbonic anhydrase, delta | 64 ± 36 | 72 ± 32 | |
| 241514 | carbonic anhydrase, delta | 0 ± 1 | 0 ± 1 | |
| 436031 | carbonic anhydrase, delta | 11 553 ± 3592 | 12 767 ± 2783 | |
| 373149 | carbonic anhydrase, gamma | 1778 ± 553 | 1893 ± 510 | |
| 63173 | carbonic anhydrase, gamma | 95 ± 46 | 139 ± 72 | |
| 74995 | carbonic anhydrase, gamma | 195 ± 72 | 193 ± 71 | |
| 432493 | carbonic anhydrase, gamma | 6804 ± 2017 | 7285 ± 1295 | [26,51] |
| 214705 | carbonic anhydrase, gamma | 144 ± 50 | 146 ± 57 | |
| inorganic carbon transport | ||||
| 120259 | bicarbonate transporter, Na+-independent Cl−/HCO3− exchanger AE1 | 14 203 ± 3864 | 15 318 ± 3586 | |
| 200137 | bicarbonate transporter | 23 544 ± 6664 | 23 653 ± 4896 | |
| 469783 | bicarbonate transporter | 3459 ± 760 | 4081 ± 838 | |
| 198643 | anion exchanger-like (AEL1), SLC4 family of solute carrier proteins | 17 579 ± 4885 | 17 422 ± 4136 | [26] |
| 450694 | bicarbonate transporter | 917 ± 191 | 1031 ± 244 | [27,51] |
| 196760 | bicarbonate transporter, anion exchanger-like Cl−/HCO3− exchangers | 48 ± 15 | 61 ± 17 | [51] |
| 99943 | anion exchanger-like, SLC4 Na+ independent Cl−/HCO3− exchangers | 17 072 ± 4609 | 17 922 ± 3914 | [27] |
| 436956 | anion exchanger-like, SLC4 Na+ independent Cl−/HCO3− exchangers | 1219 ± 395 | 1402 ± 299 | [27] |
| 466232 | anion exchanger-like, SLC4 Na+ independent Cl−/HCO3− exchangers | 815 ± 318 | 757 ± 316 | [27] |
| pH homeostasis | ||||
| 359783 | subunit of Vo sector of a vacuolar H+-ATPase | 20 079 ± 6040 | 21 289 ± 4364 | [26] |
| 447659 | bacterial Na+/H+ exchanger | 1992 ± 591 | 2416 ± 520 | [26] |
| 67081 | plasma membrane type H pump/ATPase (PM H+ ATPase) | 985 ± 306 | 1331 ± 390 | [26] |
| 439538 | V-ATPase, A | 69 940 ± 17 871 | 80 305 ± 16 954 | |
| 464767 | V-ATPase, A | 3570 ± 748 | 4143 ± 719 | |
| 435128 | V-ATPase, B | 50 862 ± 15 236 | 58 256 ± 11 228 | |
| 313800 | V-ATPase, C | 2045 ± 738 | 2122 ± 742 | |
| 558234 | V-ATPase, C | n.d. | n.d. | |
| 413949 | V-ATPase, D | 40 516 ± 17 168 | 41 431 ± 12 528 | |
| 420005 | V-ATPase, D | 6759 ± 2391 | 7502 ± 2175 | |
| 433060 | V-ATPase, E | 5739 ± 2573 | 7372 ± 3353 | |
| 352209 | V-ATPase, F | 279 ± 162 | 360 ± 161 | |
| 369392 | V-ATPase, G | 16 496 ± 5372 | 15 882 ± 4869 | |
| 196090 | eukaryotic Na+/H+ exchanger | 591 ± 137 | 708 ± 174 | |
| 434034 | eukaryotic Na+/H+ exchanger | 337 ± 66 | 378 ± 90 | |
| 464223 | eukaryotic Na+/H+ exchanger | 482 ± 178 | 476 ± 123 | |
| 465194 | eukaryotic Na+/H+ exchanger | 566 ± 136 | 591 ± 138 | |
| 467182 | eukaryotic Na+/H+ exchanger | 9889 ± 3800 | 10 049 ± 2595 | |
| 105293 | bacterial Na+/H+ exchanger | 985 ± 248 | 1463 ± 350 | |
| 219535 | bacterial Na+/H+ exchanger | 388 ± 102 | 591 ± 110 | |
| 521936 | bacterial Na+/H+ exchanger | n.d. | n.d. | |
| 76123 | p-type ATPase (PM H+ ATPase) | 0 ± 0 | 0 ± 1 | |
| 415047 | H+ PPase | 11 151 ± 2227 | 12 415 ± 2719 | |
| 439740 | H+ PPase | 17 311 ± 4737 | 20 834 ± 5731 | |
| 631975 | voltage-gated H+ channel | n.d. | n.d. | [53] |
| 75635 | aquaporin | 80 ± 30 | 71 ± 31 | [26] |
| 457738 | low CO2-induced gene | 121 ± 44 | 161 ± 75 | [26] |
aAverage ± s.d. for sequence reads mapped to each reference sequence across all of the n = 12 samples from the two present day or the two future chemostats.
bn.d., not detected: no sequence reads mapped to those genes in our study.
cCitations are from studies that identified these transcripts as important in calcification processes. When exact JGI reference sequence from the Emiliania huxleyi 1516 genome was uncertain, citations have been ascribed for all instances of the gene.
Putative transcripts related to calcium transport that were investigated include four voltage-gated Ca2+ channels, cation/H+ exchangers, putative cation/Ca2+ exchangers, Ca2+ pumps, ER-type Ca2+ ATPases, K+-dependent Na+/Ca2+ exchangers, Ca2+/Mg2+-permeable cation channels and putative transcripts encoding proteins with a Ca2+-binding domain. Of these putative transcripts, the only transcripts with differential expression between the treatments were a Ca2+/Mg2+-permeable cation channel of the LTRPC family (JGI ID: 460292), and two transcripts with homology to fibrillins and related proteins containing Ca2+-binding EGF-like domains (JGI IDs: 118025 and 463266), neither of which had previously been shown to vary in expression with calcification, and all of which were upregulated in the future ocean condition treatment (table 5). Of the non-differentially expressed transcripts, some showed high levels of expression in both treatments, including the Ca2+/H+ exchanger CAX3 (JDI ID: 416800) and the K+-dependent Na+/Ca2+ exchanger NCKX2 (JGI ID: 447939; table 5).
Expression of n = 22 putative carbonic anhydrase transcripts was examined (table 5). These include putative alpha, beta, gamma and delta carbonic anhydrases as well as putative carbonic anhydrases of unknown class. None were differentially expressed, though some were highly expressed in both treatments including two delta carbonic anhydrases (JGI IDs: 469783 and 436031) and two gamma carbonic anhydrases (JGI ID: 373149 and 432493; table 5). Alpha and beta carbonic anhydrases were expressed at relatively low levels (table 5).
Genes putatively involved in pH homeostasis were also not differentially expressed between treatments, but genes with high levels of expression include a subunit of Vo sector of a vacuolar H+-ATPase (ATPVc/c, JGI ID: 359783), multiple V-ATPases (JGI IDs: 439538, 435128, 413949 and 369392) and two H+ PPases (JGI IDs: 415047 and 439740; table 5). The voltage-gated H+ channel investigated in [53] that was shown to be important in calcification either did not map to any of the sequences in the E. huxleyi 1516 reference transcriptome or had zero expression in our treatments (table 5).
Genes putatively related to inorganic carbon transport, namely Cl−/HCO3− exchangers of the SLC4 family of solute carrier proteins, were not differentially expressed between treatments, but the transporter AEL1 examined in [26] (JGI ID: 198643) was highly expressed, as were other transporters of this family (JGI IDs: 99943, 120259 and 200137; table 5).
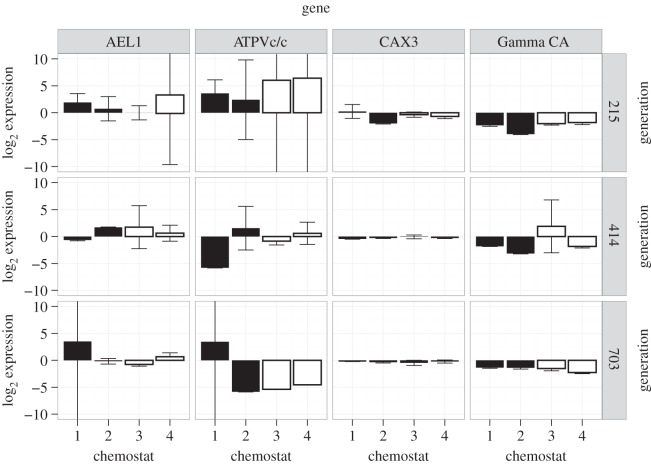
The four transcripts examined by qPCR showed no significant differences in expression between treatments or generation points (figure 7). Expression levels of the examined genes showed a high level of variability in the RNA-seq and qPCR assays.
Figure 7.
Quantitative real-time PCR (qPCR) data for calcification-related genes selected from the literature. Bars represent means±s.d. actin-normalized expression levels for each gene from two filters from each chemostat at each generation point under present ocean condition (383 ± 43 µatm pCO2, 20.0 ± 0.1°C; white bars) and future ocean condition (833 ± 68 µatm pCO2, 24.0 ± 0.2°C; black bars; electronic supplementary material, table S1). Each filter was assayed in three replicate qPCR reactions. If error bar horizontal lines are not visible, the error bars are larger than the y-axis boundary.
4. Discussion
(a). Inorganic carbon physiology
The same trend of increased PIC content and calcification rate was seen at all generation points, even though they were not statistically different at all generations (figures 2a and 3a). These results are in contrast with those of earlier studies examining the combined effects of elevated pCO2 and temperature: one study found decreased PIC content in the same strain (CCMP371) [23], and two studies found no change in PIC content in different strains (RCC1228 and PML B92/11) [18,22] (table 6). Responses of calcification to elevated pCO2 are known to vary across strains of E. huxleyi [7,8,15], and responses to the combination of elevated pCO2 and temperature could also be strain-specific. Therefore, a strain-specific response could explain some of the different results in the experiments under elevated pCO2 and temperature, but not the different response of the same strain (CCMP 371) [7,8,23].
Table 6.
Response of Emiliania huxleyi ((Lohm.) Hay and Mohler) to OA and OA plus elevated temperature in POC content, primary production (PP), PIC content and calcification rate (CR). pH values are in (total scale) and were re-calculated from data in the reference if necessary. present/future represents present and future conditions. Superscript letters and numbers in column 1 indicate the culture method and carbonate system adjustment method used in the experiments. ‘Percent (%) change’ is the change in the average value of the variable under future ocean condition relative to that under present ocean condition. ‘Trend’ is the statistically significant shift in the variable as stated by the author or tested here using data from the source paper. Studies marked with an asterisk in the reference column are long-term experiments (100s+ of generations); all other studies are short-term experiments (10s of generations). Empty cells indicate parameters were not determined.
| condition |
strain | POC |
PP |
PIC |
CR |
references | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | % change | trend | % change | trend | % change | trend | % change | trend | ||
| present/future pH at constant temperature | |||||||||||
| 8.1/7.8 a,(1) | 19 | NZEH | 169 | ↑ | 94 | ↑ | 153 | ↑ | 73 | ↑ | [4] |
| 8.1/7.8 a,(2) | 20 | NZEH | 50 | ↑ | 69 | ↑ | 39 | ↑ | 56 | ↑ | [54] |
| 8.0/7.8 b,(1) | 13 | RCC 1228 | 25 | ↔ | −19 | ↔ | 25 | ↔ | −30 | ↔ | [22] |
| 8.0/7.9 a,(2) | 17 | RCC 1256 | 24 | ↑ | 11 | ↑ | 11 | ↑ | 3 | ↑ | [15] |
| 8.1/7.8 c,(1) | 20 | CCMP 371 | 6 | ↔ | 23 | ↔ | [23] | ||||
| 8.1/7.8 a,(2) | 20 | RCC 1238 | 11 | ↔ | 14 | ↑ | −7 | ↔ | −5 | ↔ | [15] |
| 8.1/7.8 a,(2) | 20 | RCC 1212 | 11 | ↔ | 7 | ↔ | −5 | ↓ | −9 | ↓ | [15] |
| 8.0/7.8 a,(2) | 17 | RCC 1216 | 8 | ↑ | 5 | ↔ | −7 | ↓ | −8 | ↓ | [15] |
| 8.0/7.8 a,(2) | 15 | not given | 22 | ↑ | −15 | ↓ | −9 | ↓ | [9] | ||
| 8.1/7.8 a,(2) | 15 | PML B92/11 | 7 | ↑ | −13 | ↓ | [14] | ||||
| 8.0/7.8 a,(1) | 15 | NZEH | 7 | ↑ | 9 | ↑ | −18 | ↓ | −5 | ↓ | [10] |
| 8.1/7.8 a,(2) | 15 | PML B92/11 | 9 | ↑ | −16 | ↓ | [5] | ||||
| 8.1/7.9 a,(1) | 15 | RCC 1256 | −2 | ↔ | −1 | ↔ | −15 | ↓ | −14 | ↓ | [10] |
| 8.0/7.8 d,(1) | 17 | RCC 1215 | −18 | ↓ | −15 | ↓ | −17 | ↓ | −24 | ↓ | [13] |
| 8.1/7.8 c,(1) | 18 | CS369 | 8 | ↑ | −41 | ↓ | [55] | ||||
| 8.3/7.5 d,(1) | 18 | PML 92A | 18 | ↔ | [31] | ||||||
| 8.2/7.8 c,(1) | 15 | not given | 28 | ↑ | 21 | ↑ | −10 | ↔ | −7 | ↔ | [18]*(4) |
| present/future pH at present and future temperatures | |||||||||||
| 8.0/7.8 b,(1) | 13 and 18 | RCC 1228 | 40 | ↔ | 57 | ↔ | −28 | ↔ | −53 | ↓ | [22] |
| 8.1/7.8 c,(1) | 20 and 24 | CCMP 371 | 26 | ↔ | −27 | ↓ | [23] | ||||
| 8.1/7.5 d,(1) | 14 and 18 | PML B92/11 | −4 | ↔ | −25 | ↔ | [21] | ||||
| 8.0/7.7 d,(3) | 20 and 24 | CCMP 371 | 8 | ↔ | 4 | ↔ | 26 | ↑ | 26 | ↑ | this study*(4) |
adiluted batch
bbatch
csemi-continuous
dchemostat
(1)bubbling
(2)acid/base adjustment
(3)bubbling + bicarbonate adjustment
(4)long-term experiment more than 100s of generations.
Responses to variation in pCO2 and temperature of a given E. huxleyi strain could be modulated by additional experimental variables [7,8]. Light intensity, photoperiod and nutrient levels can influence calcification [12,14,56] and could have interactive effects with pCO2 and/or temperature. Feng et al. [23] used the same temperatures (20°C and 24°C) and similar pCO2 levels (375 and 750 µatm) and light intensity in the high light treatments (400 µmol m−2 s−1) as our study, but the cells were grown under a different photoperiod (12 L : 12 D compared with 16 L : 8 D), under nutrient replete conditions and for only seven generations. Zondervan et al. [14] found no difference in calcification rate of E. huxleyi under a 16 L : 8 D photoperiod or under continuous high light (150 µmol m−2 s−1), suggesting that under high light calcification rate is not influenced by day length. Considering that our experiment used a high-light intensity, observed differences in the calcification result between our experiment and the experiment by Feng et al. [23] are due to factors additional to photoperiod alone.
Hoppe et al. [10] suggest that an increase in PIC per cell can often be explained by the experimental set-up. High cell densities such as those in our experiment (approx. 500 000 cells ml−1) can lead to major changes in water chemistry and nutrient availability and induce the cells to increase the rate of calcium carbonate production due to stress (often resulting in high percentages of detached coccoliths in the culture). We minimized this effect by augmenting the media with sodium bicarbonate and by maintaining Ωcalcite ≥ 2 throughout the experiment (table 1). The chemostats were run under NO3 limitation; the higher cellular C : N ratios in our experiment compared with the C : N ratios reported under N-replete conditions [23] are consistent with N-limitation (figure 1). It has been suggested that calcification increases under N-limitation [57], but this was later attributed to methodological errors in batch experiments [58]. In addition, the C : N ratios did not differ between the treatments in our experiment. Therefore, even if nutrient limitation did affect calcification, such effects were likely the same for both treatments.
Differences in cell size between the treatments are another possible explanation for our results. If the cells were larger in the future ocean treatment, this should result in higher POC content and lower PIC : POC ratio. However, POC content was only different at the 703 generation point (figure 2b), and the PIC : POC ratio was the same at all generation points (table 4). It is therefore unlikely that cell size is responsible for the increased PIC content and calcification rate in the future ocean treatment.
We observed increased PIC content and higher calcification rate under elevated pCO2 and temperature after several hundreds of generations. Calcification rates were not reported by Feng et al. [23], but bulk calcification rates can be calculated using their reported values of PIC per cell and growth rate. The resulting values show no difference between present and future ocean conditions (1.17 ± 0.15 and 1.13 ± 0.22 pg C cell−1 d−1 in ‘ambient’ and ‘greenhouse’ conditions, respectively), and it is possible that the calcification rate per cell in response to elevated pCO2 and temperature had adapted after culturing for more than seven generations [23]. Because these calcification rates of Feng et al. [23] are calculated using a method that differs from that adopted in the present study (isotope incorporation method), comparison of absolute values could be problematic. However, trends in calcification rate within one study should not be altered by possible absolute rate differences stemming from the quantification method.
The adaptive significance of calcification differences is more difficult to ascertain. While growth rate can be used as a proxy for adaptation in single-cell organisms [18], this is not possible in chemostat experiments, since growth rate is set by the dilution rate. Phenotypes that are favoured under different conditions may be selected for, and hence it is possible that calcification rate was a target of selection in our study. Still, if strains are able to tune their calcification responses to elevated pCO2 and temperature after long-term culturing, the predictions drawn using results from short-term acclimation studies [7,8] would not reflect the actual responses of these strains to different ocean conditions expected by the end of the century. The ability of E. huxleyi to adapt to changing environmental conditions was shown by Lohbeck et al. [18]. This study tested the ability of E. huxleyi to adapt to elevated pCO2 on one strain in one experiment and on six strains together in another experiment (all strains isolated from the Raunefjorden in Norway). Approximately after 500 generations, cells adapted to elevated pCO2 exhibited as much as 50% higher calcification rate when compared with those adapted to ambient pCO2 when grown under elevated pCO2. Thus, our results along with those from other long-term culture studies [17] underscore the necessity of long-term studies in order to accurately understand likely responses of coccolithophores to OA.
(b). Organic carbon physiology
There was no difference in the POC content and primary production between the treatments at most generation points (figures 2b and 3b). POC content was significantly higher in the future ocean condition only at the 703 generation point, whereas primary production was the same. The data in this study (figures 2b and 3b) are in agreement with prior studies that examined the combined effect of elevated pCO2 and temperature on POC content [21–23] and primary production [22] (table 6). Only the POC bulk production rate calculated from data reported by Feng et al. [23] increases under elevated pCO2 and temperature (table 6). These results differ from experiments that only examined the effects of elevated pCO2. In these latter experiments, eight out of 15 studies suggest elevated pCO2 increases POC content (table 6, trend in POC), and eight out of 13 studies suggest elevated pCO2 increases primary production (table 6, trend in PP). Thus, although elevated pCO2 seems to increase POC content and production, the combined effect of elevated pCO2 and temperature might weaken this trend. Thermal responses probably depend on the strain- or species-specific temperature optimum [59,60]; if experimental temperatures are shifted towards the optimum, higher growth rates and production are expected. Therefore, one possible explanation for the different response in POC content and primary production in studies that simultaneously exposed E. huxleyi to increased temperature and pCO2 could be offsetting effects of pCO2 and temperature. For example, if increased temperatures cause a shift away from the temperature optimum, thermal decline in growth rate and production could counter the positive effect of elevated pCO2. However, two studies, De Bodt et al. [22] and Borchard et al. [21], observed higher biomass (in POC l−1) and cell density at higher temperatures suggesting that cells grown under elevated temperature were shifted towards the thermal optimum. Shifts away from thermal optima are thus unlikely to be the reason for the absence of a change in POC cell−1 under combined effects of elevated temperature and pCO2, as we observed (figure 3). Whether temperature modulates the effect of pCO2 on primary production, and the cellular mechanisms involved in that modulation, remain to be studied.
(c). Gene expression
Several genes putatively related to calcification (table 3) have been identified via gene expression studies comparing calcifying and non-calcifying E. huxleyi cells [25–29], or in short-term experiments where calcification was regulated by limitation of ions needed for calcification (i.e. Ca2+, HCO3−/CO32− [26,30]). These prior studies examined gene expression associated with large variations in calcification, where other cellular processes were likely to be affected by the treatment conditions and may have accounted for the observed changes in gene expression without being directly involved in the calcification process. Even between calcifying and non-calcifying diploid strains of E. huxleyi [26], differences in gene expression were in most cases not large (typically in the range of 1–4-fold), and the absolute expression levels of these genes are unknown.
None of the genes previously demonstrated to be differentially expressed in cells with differing calcification [25–29] were found to be differentially expressed in this long-term culture study (table 3), despite differences in calcification and calcification rate observed here (figures 2 and 3; table 2). Lack of differential expression in calcification-related genes was demonstrated both in the comprehensive RNA-seq analysis and the more specific examination of selected genes via qPCR. Some of these genes were also investigated in prior studies examining the effects of OA on gene expression. In a short-term study by Richier et al. [51] examining calcification and gene expression (using qPCR), no changes in calcification due to the treatment conditions were observed, and no genes were differentially expressed aside from carbonic anhydrases. Likewise, in short-term cultures with replete DIC and varied carbonate chemistry, expression of these genes was largely unaffected except when DIC or pCO2 was limited, which resulted primarily in upregulated expression [61]. It is possible that the treatment conditions in the present study were not different enough to detect differences in the expression of the previously investigated genes, or that very large differences in calcification must occur in order to observe changes in gene expression of the genes previously hypothesized to be important in calcification. Alternatively, given that the study of cellular mechanisms underlying calcification in coccolithophores is still in its infancy, it is probable that there are additional genes with important roles in calcification. Mechanisms governing fine-scale differences in calcification may not be observable at the transcriptomic level and may be regulated through post-translational modifications and altered cellular signalling. Alternatively, it is possible that small, but carbon pump-significant, variation in the degree of calcification occurs as a result of physicochemical conditions and does not involve changes in cellular physiology.
In this study, we identified differentially expressed genes that have not been previously investigated, but may be involved in calcification: putative transcripts with homology to fibrilins, which have a Ca2+-binding domain, and a Ca2+/Mg2+ channel (table 3). These genes might play a role in calcification or may be involved in cellular signalling pathways that regulate calcification. Several genes involved in post-translational modification and signal transduction that were differentially expressed in this study (see electronic supplementary material, table S5) may likewise play a role either in calcification, pH regulation, or other cellular functions responsive to changing environmental conditions. Putative transcripts that may be involved in the structure and function of cell membranes, such as genes with homology to ankyrin and chitinase (see electronic supplementary material, table S5), may also play a role in calcification, potentially by forming the organic biomineralization surfaces.
We observed the upregulation of several putative transcripts in the future ocean condition treatment that may be involved with cell membrane dynamics and potentially calcification. Several chitinase and chitinodextrinase putative transcripts were upregulated in the future ocean condition treatment. Differential expression of chitinase putative transcripts was previously observed in microarray studies comparing haploid non-calcifying and diploid calcifying E. huxleyi [62], where two putative chitinase transcripts upregulated in the future ocean condition treatment of this study (JGI IDs: 469375 and 417127) were preferentially expressed in the diploid cultures [62]. Chitinase and chitinodextrinase enzymes typically break down the polysaccharide chitin, which is not known to be present in coccolithophores, but has been shown to be associated with the cell wall of some diatoms [63]; these enzymes may regulate other coccolithophore polysaccharides with similar properties to chitin. Upregulation of putative chitinase transcripts during increased calcification suggests that regulation of chitin-like polysaccharides plays a role in coccolith biomineralization, exocytosis of coccoliths to the cell surface or binding of the coccolithosphere to the organic component of the extracellular matrix. Other membrane-related putative transcripts were simultaneously upregulated in the future ocean condition treatment, including two ankyrin putative transcripts, one of which (JGI ID: 116992) was preferentially expressed in diploid when compared with non-calcifying haploid E. huxleyi [62], suggesting a coccolith-related role.
Prior E. huxleyi transcriptome analyses have revealed differential expression of a large number of hypothetical proteins of unknown function, or of putative transcripts without annotation [27–29,62]. The present study likewise revealed many unknown transcripts that may be related to pCO2 level and temperature (see electronic supplementary material, table S5).
Although hypothesized calcification genes were not differentially expressed between treatments, the overall expression levels of these genes may shed light on specific genes that may be involved in calcification and photosynthesis. Genes involved in calcium transport, including a K+-dependent Na+/Ca2+ exchanger NCKX2 (JGI ID: 447939) and Ca2+/H+ exchanger CAX3 (JGI ID: 416800, examined in [26]) were expressed at very high levels in both treatments, whereas the putative calcium-binding protein GPA was expressed at low levels. Several different H+ channels and genes potentially involved in pH homeostasis were expressed at high levels, though their potential role in calcification has not yet been investigated. Potentially, they could be involved in the removal of H+ created during the precipitation of calcite. It is possible that these particular genes are constitutively expressed at high levels in present and predicted future environmental conditions, and that other genes expressed at relatively low levels may be induced upon cellular stress such as low DIC [61].
We observed high expression levels of multiple putative carbonic anhydrases, particularly gamma and delta carbonic anhydrases, and lower expression of alpha and beta carbonic anhydrases. Strong upregulation of a beta carbonic anhydrase has been observed in low DIC conditions, but not when DIC was replete [30,64]. However, those studies [30,64] found no differential expression in a gamma carbonic anhydrase over the tested conditions, in agreement with the study of Sotoj et al. [65]. This suggests that some carbonic anhydrases with low expression levels in our cultures may be induced by conditions such as DIC limitation, whereas other carbonic anhydrases may be constitutively expressed at high levels.
(d). Implications for the future ocean
Export of carbon to the deep sea and sea floor is an important process in the global carbon cycle that can sequester atmospheric CO2 over time scales of centuries or greater. At present, coccolithophores contribute 50% of the pelagic CaCO3 production [66], and up to 80% of the organic carbon export is associated with CaCO3 [67]. In this study, averaged across all generation points, each coccolithophore cell increased its calcification rate (26%) and calcium carbonate quota (26%) in the future ocean treatment (figures 2a and 3a), and the total concentration of calcium carbonate in the culture (PIC l−1) increased 18% in the future ocean condition (table 4). However, the PIC : POC ratio in the future ocean condition, while not statistically different among treatments or generations, increased by only 6% (table 4). Therefore, although our results suggest that coccolithophore calcification will increase in future ocean conditions (table 6), it is unclear whether, or how, such changes might affect carbon export to the deep sea [7,8].
(e). Variability in Emiliania huxleyi responses to future ocean conditions
Studies investigating the effect of elevated pCO2 on the physiology in E. huxleyi have not yielded consistent results. Primary production, calcification rate, POC and PIC content under elevated pCO2 have been shown to increase by as much as 169% or decrease by as much as 43% in future ocean conditions (table 6). Variation in responses could be strain-specific [15], but variable results have been observed for the same strain. Strain NZEH shows the most extreme of such cases in E. huxleyi (table 6), where in one study [4] PIC and POC were 2.5 and 2.7 times higher under elevated pCO2, respectively, while in another study [10] it showed an 18% decrease in PIC and 7% increase in POC. Different pCO2 manipulation methods seem not to influence the results [7,8,10], but less is known about different culturing methods (diluted batch, semi-continuous, chemostat; table 6). Other culturing parameters (e.g. nutrients, light) could have synergistic effect with elevated pCO2 and influence the comparability of experiments. Another strain (PML B92/11) cultured twice under the same conditions and methods (table 6, [5,14]) showed less variability in responses to elevated pCO2 than strain NZEH. Strains could have different natural variability in their response to elevated pCO2, but an insufficient number of experiments have been conducted on the same strain under identical conditions to test this hypothesis [7,8]. Therefore, it remains to be discovered whether different strains show greater variability in their response to pCO2. All except one of the experiments in table 6 have used strains isolated from coastal areas that may have evolved under conditions of highly variable pCO2 compared with what is typical for the open ocean [68,69].
An additional challenge in describing general response patterns of E. huxleyi to future ocean conditions is high variability among experimental replicates. For example, De Bodt et al. [22] found a 57% increase in photosynthetic rate under elevated pCO2 that was non-statistically significant (table 6), whereas Langer et al. [15] found a 3% increase in calcification rate under elevated pCO2 that was statistically significant (table 6). Inconsistency in the variability of experimental replicates among studies may be reduced with increased rigor of experimental design, or may persist if that variability reflects genetic differentiation among E. huxleyi strains.
5. Conclusion
We have shown that long-term multi-parameter culture studies could shift our prediction of E. huxleyi responses to future ocean conditions. Unfortunately, these studies are difficult and time consuming to conduct, but they yield critical information to correctly predict additive or synergistic effects of elevated pCO2 and other changing parameters that will characterize the future oceans. Elevating pCO2 concentration and shifting nitrogen source have a synergistic effect on calcification [24], and here we indicate that the responses of E. huxleyi to elevated pCO2 might be offset by simultaneously increasing temperature. Responses of E. huxleyi to simultaneous shifts in multiple environmental parameters add to the complexity of understanding how coccolithophores will respond to the future ocean and the subsequent feedback to the global carbon cycle. The high variability in studies with E. huxleyi (table 6) shows that additional multivariate and replicated studies are needed to examine the response of E. huxleyi to future ocean conditions. Our study also suggests that cellular responses to future ocean conditions are not likely to be straightforward, as shifts in calcification may have less to do with calcification-related genes and more to do with intracellular regulatory processes in cells grown under continuous culture for many hundreds of generations.
Acknowledgements
We thank Kristine Okimura, Joëlle Tirindelli, Robert Hausman, Michelle Drake, and Oscar Steiner for assistance with chemostat culture, and Scott Fay for assistance with bioinformatic analyses.
Funding statement
This work was funded by the NSF grant BIO-OCE 0723908 to E.J.C., J.H.S. and T.K.
References
- 1.Sabine CL, et al. 2004. The oceanic sink for anthropogenic CO2. Science 305, 367–371 (doi:10.1126/science.1097403) [DOI] [PubMed] [Google Scholar]
- 2.Feely RA, Sabine CL, Lee K, Berelson W, Kleypas J, Fabry VJ, Millero FJ. 2004. Impact of anthropogenic CO2 on the CaCO3 system in the oceans. Science 305, 362–366 (doi:10.1126/science.1097329) [DOI] [PubMed] [Google Scholar]
- 3.Beaufort L, et al. 2011. Sensitivity of coccolithophores to carbonate chemistry and ocean acidification. Nature 476, 80–83 (doi:10.1038/nature10295) [DOI] [PubMed] [Google Scholar]
- 4.Iglesias-Rodriguez MD, et al. 2008. Phytoplankton calcification in a high-CO2 world. Science 320, 336–340 (doi:10.1126/science.1154122) [DOI] [PubMed] [Google Scholar]
- 5.Riebesell U, Zondervan I, Rost B, Tortell PD, Zeebe RE, Morel FMM. 2000. Reduced calcification of marine plankton in response to increased atmospheric CO2. Nature 407, 364–367 (doi:10.1038/35030078) [DOI] [PubMed] [Google Scholar]
- 6.Holligan PM, Robertson E. 1996. Significance of ocean carbonate budgets for the global carbon cycle. Glob. Change Biol. 2, 85–95 (doi:10.1111/j.1365-2486.1996.tb00053.x) [Google Scholar]
- 7.Findlay HS, Calosi P, Crawfurd K. 2011. Determinants of the PIC : POC response in the coccolithophore Emiliania huxleyi under future ocean acidification scenarios. Limnol. Oceanogr. 56, 1168–1178 (doi:10.4319/lo.2011.56.3.1168) [Google Scholar]
- 8.Ridgwell A, Schmidt DN, Turley C, Brownlee C, Maldonado MT, Tortell P, Young JR. 2009. From laboratory manipulations to Earth system models: scaling calcification impacts of ocean acidification. Biogeoscience 6, 2611–2623 (doi:10.5194/bg-6-2611-2009) [Google Scholar]
- 9.Barcelos e Ramos J, Müller MN, Riebesell U. 2010. Short-term response of the coccolithophore Emiliania huxleyi to an abrupt change in seawater carbon dioxide concentrations. Biogeoscience 7, 177–186 (doi:10.5194/bg-7-177-2010) [Google Scholar]
- 10.Hoppe CJM, Langer G, Rost B. 2011. Emiliania huxleyi shows identical responses to elevated pCO2 in TA and DIC manipulations. J. Exp. Mar. Biol. Ecol. 406, 54–62 (doi:10.1016/j.jembe.2011.06.008) [Google Scholar]
- 11.Müller MN, Schulz KG, Riebesell U. 2010. Effects of long-term high CO2 exposure on two species of coccolithophores. Biogeoscience 7, 1109–1116 (doi:10.5194/bg-7-1109-2010) [Google Scholar]
- 12.Rost B, Zondervan I, Riebesell U. 2002. Light-dependent carbon isotope fractionation in the coccolithophorid Emiliania huxleyi. Limnol. Oceanogr. 47, 120–128 (doi:10.4319/lo.2002.47.1.0120) [Google Scholar]
- 13.Sciandra A, Harlay J, Lefèvre D, Lemée R, Rimmelin P, Denis M, Gattuso J-P. 2003. Response of coccolithophorid Emiliania huxleyi to elevated partial pressure of CO2 under nitrogen limitation. Mar. Ecol. Prog. Ser. 261, 111–122 (doi:10.3354/meps261111) [Google Scholar]
- 14.Zondervan I, Rost B, Riebesell U. 2002. Effect of CO2 concentration on the PIC/POC ratio in the coccolithophore Emiliania huxleyi grown under light-limiting conditions and different daylengths. J. Exp. Mar. Biol. Ecol. 272, 55–70 (doi:10.1016/S0022-0981(02)00037-0) [Google Scholar]
- 15.Langer G, Nehrke G, Probert I, Ly J, Ziveri P. 2009. Strain-specific responses of Emiliania huxleyi to changing seawater carbonate chemistry. Biogeoscience 6, 2637–2646 (doi:10.5194/bg-6-2637-2009) [Google Scholar]
- 16.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4, 457–469 (doi:10.1038/nrg1088) [DOI] [PubMed] [Google Scholar]
- 17.Tatters AO, et al. 2013. Short- and long-term conditioning of a temperate marine diatom community to acidification and warming. Phil. Trans. R. Soc. B 368, 20120437 (doi:10.1098/rstb.2012.0437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohbeck KT, Riebesell U, Reusch TBH. 2012. Adaptive evolution of a key phytoplankton species to ocean acidification. Nat. Geosci. 5, 1–6 (doi:10.1038/ngeo1441) [Google Scholar]
- 19.Godbold JA, Solan M. 2013. Long-term effects of warming and ocean acidification are modified by seasonal variation in species responses and environmental conditions. Phil. Trans. R. Soc. B 368, 20130186 (doi:10.1098/rstb.2013.0186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell BD, Connell SD, Findlay HS, Tait K, Widdicombe S, Mieszkowska N. 2013. Ocean acidification and rising temperatures may increase biofilm primary productivity but decrease grazer consumption. Phil. Trans. R. Soc. B 368, 20120438 (doi:10.1098/rstb.2012.0438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borchard C, Borges AV, Händel N, Engel A. 2011. Biogeochemical response of Emiliania huxleyi (PML B92/11) to elevated CO2 and temperature under phosphorous limitation: a chemostat study. J. Exp. Mar. Biol. Ecol. 410, 61–71 (doi:10.1016/j.jembe.2011.10.004) [Google Scholar]
- 22.De Bodt C, Van Oostende N, Harlay J, Sabbe K, Chou L. 2010. Individual and interacting effects of pCO2 and temperature on Emiliania huxleyi calcification: study of the calcite production, the coccolith morphology and the coccosphere size. Biogeoscience 7, 1401–1412 (doi:10.5194/bg-7-1401-2010) [Google Scholar]
- 23.Feng Y, Warner ME, Zhang Y, Sun J, Fu F-X, Rose JM, Hutchins DA. 2008. Interactive effects of increased pCO2, temperature and irradiance on the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae). Eur. J. Phycol. 43, 87–98 (doi:10.1080/09670260701664674) [Google Scholar]
- 24.Lefebvre SC, Benner I, Stillman JH, Parker AE, Drake MK, Rossignol PE, Okimura KM, Komada T, Carpenter EJ. 2011. Nitrogen source and pCO2 synergistically affect carbon allocation, growth and morphology of the coccolithophore Emiliania huxleyi: potential implications of ocean acidification for the carbon cycle. Glob. Change Biol. 18, 493–503 (doi:10.1111/j.1365-2486.2011.02575.x) [Google Scholar]
- 25.Mackinder L, Wheeler G, Schroeder D, Riebesell U, Brownlee C. 2010. Molecular mechanisms underlying calcification in coccolithophores. Geomicrob. J. 27, 585–595 (doi:10.1080/01490451003703014) [Google Scholar]
- 26.Mackinder L, Wheeler G, Schroeder D, von Dassow P, Riebesell U, Brownlee C. 2011. Expression of biomineralization-related ion transport genes in Emiliania huxleyi. Environ. Microb. 13, 3250–3265 (doi:10.1111/j.1462-2920.2011.02561.x) [DOI] [PubMed] [Google Scholar]
- 27.von Dassow P, Ogata H, Probert I, Wincker P, Da Silva C, Audic S, Claverie JM, de Vargas C. 2009. Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol. 10, R114 (doi:10.1186/gb-2009-10-10-r114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahlund TM. 2005. Molecular characterization of genes and proteins involved in inorganic carbon acquisition and metabolism in the marine alga Emiliania huxleyi during calcite biomineralization. Phycologia 44, 108–109 [Google Scholar]
- 29.Wahlund TM, Zhang XY, Read BA. 2004. Expressed sequence tag profiles from calcifying and non-calcifying cultures of Emiliania huxleyi. Micropaleontology 50, 145–155 (doi:10.2113/50.Suppl_1.145) [Google Scholar]
- 30.Bach LT, Mackinder L, Schultz KG, Wheeler G, Schroeder DC, Brownlee C. 2013. Dissecting the impact of CO2 and pH on the mechanisms of photosynthesis and calcification in the coccolithophore Emiliania huxleyi. New Phytol. 199, 121–134 (doi:10.1111/nph.12225) [DOI] [PubMed] [Google Scholar]
- 31.Leonardos N, Geider RJ. 2005. Elevated atmospheric carbon dioxide increases organic carbon fixation by Emiliania huxleyi (Haptophyta), under nutrient-limited high-light conditions. J. Phycol. 41, 1196–1203 (doi:10.1111/j.1529-8817.2005.00152.x) [Google Scholar]
- 32.Meinshausen M, et al. 2011. The RCP greenhouse gas concentrations and their extensions from 1765 to 2300. Climate Change 109, 213–241 (doi:10.1007/s10584-011-0156-z) [Google Scholar]
- 33.Guillard RRL, Ryther JH. 1962. Studies of marine planktonic diatoms I. Cyclotella nanna (Hustedt) and Detonula convervacea (Cleve). Can. J. Microb. 8, 229–239 (doi:10.1139/m62-029) [DOI] [PubMed] [Google Scholar]
- 34.Langer G, Geisen M, Baumann K-H, Kläs J, Riebesell U, Thoms S, Young JR. 2006. Species-specific responses of calcifying algae to changing seawater carbonate chemistry. Geochem. Geophys. Geosyst. 7, Q09006 (doi:10.1029/2005GC001227) [Google Scholar]
- 35.Hutchins DA, Pustizzi F, Hare CE, DiTullio GR. 2003. A shipboard natural community continuous culture system for ecologically relevant low-level nutrient enrichment experiments. Limnol. Oceanogr. Methods 1, 82–91 (doi:10.4319/lom.2011.1.82) [Google Scholar]
- 36.Lefebvre SC, Harris G, Webster R, Leonardos N, Geider RJ, Raines CA, Read BA, Garrido JL. 2010. Characterization and expression analysis of the Lhcf gene family in Emiliania huxleyi (Haptophyta) reveals differential responses to light and CO2. J. Phycol. 46, 123–134 (doi:10.1111/j.1529-8817.2009.00793.x) [Google Scholar]
- 37.Bradshaw AL, Brewer PG, Shaffer DK, Williams RT. 1981. Measurements of total carbon dioxide and alkalinity by potentiometric titration in the GEOSECS program. Earth Planet Sci. Lett. 55, 99–115 (doi:10.1016/0012-821X(81)90090-X) [Google Scholar]
- 38.Gran G. 1952. Determination of the equivalence point in potentiometric titrations of seawater with hydrochloric acid. Oceanol. Acta 5, 209–218 [Google Scholar]
- 39.Friederich GE, Walz PM, Burczynski MG, Chavez FP. 2002. Inorganic carbon in the central California upwelling system during the 1997–1999 El Niño–La Niña event. Prog. Oceanogr. 54, 185–203 (doi:10.1016/S0079-6611(02)00049-6) [Google Scholar]
- 40.Lewis E, Wallace DWR. 1998. Program developed for CO2 system calculations ORNL/CDIAC-105. Oak Ridge, TN: Carbon Dioxide Information and Analysis Center, Oak Ridge Nat. Lab., US Department of Energy [Google Scholar]
- 41.Mehrbach C, Culberson CH, Hawley JE, Pytkovicz RM. 1973. Measurement of the apparent dissociation constants of carbonic acid in seawater at atmospheric pressure. Limnol. Oceanogr. 18, 897–907 (doi:10.4319/lo.1973.18.6.0897) [Google Scholar]
- 42.Dickson AG, Millero FJ. 1987. A comparison of the equilibrium constants for the dissociation of carbon acid in seawater media. Deep-Sea Res. 34, 1733–1743 (doi:10.1016/0198-0149(87)90021-5) [Google Scholar]
- 43.Balch WM, Holligan PM, Kilpatrick KA. 1992. Calcification, photosynthesis and growth of the bloom-forming coccolithophore, Emiliania huxleyi. Cont. Shelf Res. 12, 1353–1374 (doi:10.1016/0278-4343(92)90059-S) [Google Scholar]
- 44.Grubbs FE. 1969. Procedures for detecting outlying observations in samples. Technometrics 11, 1–21 (doi:10.2307/1266761) [Google Scholar]
- 45.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinf. 11, 485 (doi:10.1186/1471-2105-11-485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformation 25, 1105–1111 (doi:10.1093/bioinformatics/btp120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Bio. 10, R25 (doi:10.1186/gb-2009-10-3-r25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformation 25, 2078–2079 (doi:10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformation 26, 139–140 (doi:10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Read BA, et al. 2013. Pan genome of the phytoplankton Emiliania drives its global distribution. Nature 499, 209–213 (doi:10.1038/nature12221) [DOI] [PubMed] [Google Scholar]
- 51.Richier S, Fiorini S, Kerros ME, von Dassow P, Gattuso JP. 2011. Response of the calcifying coccolithophore Emiliania huxleyi to low pH/high pCO2: from physiology to molecular level. Mar. Biol. 158, 551–560 (doi:10.1007/s00227-010-1580-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroeder DC, Biggi GF, Hall M, Davy J, Martinez JM, Richardson AJ, Malin G, Wilson WH. 2005. A genetic marker to separate Emiliania huxleyi (Prymnesiophyceae) morphotypes. J. Phycol. 41, 874–879 (doi:10.1111/j.1529-8817.2005.000100.x) [Google Scholar]
- 53.Taylor AR, Chrachri A, Wheeler G, Goddard H, Brownlee C. 2011. A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores. PLoS Biol. 9, e1001085 (doi:10.1371/journal.pbio.1001085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi D, Xu Y, Morel FMM. 2009. Effects of the pH/pCO2 control method on medium chemistry and phytoplankton growth. Biogeoscience 6, 1199–1207 (doi:10.5194/bg-6-1199-2009) [Google Scholar]
- 55.Gao K, Ruan Z, Villafañe VE, Gattuso J-P, Helbling EW. 2009. Ocean acidification exacerbates the effect of UV radiation on the calcifying phytoplankter Emiliania huxleyi. Limnol. Oceanogr. 54, 1855–1862 (doi:10.4319/lo.2009.54.6.1855) [Google Scholar]
- 56.Paasche E. 1998. Roles of nitrogen and phosphorus in coccolith formation in Emiliania huxleyi (Prymnesiophyceae). Eur. J. Phycol. 33, 33–42 [Google Scholar]
- 57.McConnaughey TA, Whelan JF. 1997. Calcification generates protons for nutrient and bicarbonate uptake. Earth Sci. Rev. 42, 95–117 (doi:10.1016/S0012-8252(96)00036-0) [Google Scholar]
- 58.Langer G, Oetjen K, Brenneis T. 2013. Coccolithophores do not increase particulate carbon production under nutrient limitation: a case study using Emiliania huxleyi (PML B92/11). J. Exp. Mar. Biol. Ecol. 443, 155–161 (doi:10.1038/ngeo1750) [Google Scholar]
- 59.Eppley RW. 1972. Temperature and phytoplankton growth in the sea. Fish. Bull. 70, 1063–1085 [Google Scholar]
- 60.Fisher NS, Honjo S. 1989. Intraspecific differences in temperature and salinity responses in the coccolithophore Emiliania huxleyi. Biol. Oceanogr. 6, 355–361 [Google Scholar]
- 61.Bach LT, Riebesell U, Schulz KG. 2011. Distinguishing between the effects of ocean acidification and ocean carbonation in the coccolithophore Emiliania huxleyi. Limnol. Oceanogr. 56, 2040–2050 (doi:10.4319/lo.2011.56.6.2040) [Google Scholar]
- 62.Rokitta SD, de Nooijer LJ, Trimborn S, de Vargas C, Rost B, John U. 2011. Transcriptome analyses reveal differential gene expression patterns between the life-cycle stages of Emiliania huxleyi (Haptophyta) and reflect specialization to different ecological niches. J. Phycol. 47, 829–838 (doi:10.1111/j.1529-8817.2011.01014.x) [DOI] [PubMed] [Google Scholar]
- 63.Durkin CA, Mock T, Armbrust EV. 2009. Chitin in diatoms and its association with the cell wall. Euk. Cell 8, 1038–1050 (doi:10.1128/ec.00079-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bach LT, Mackinder L, Schultz KG, Wheeler G, Schroeder DC, Brownlee C, Riebesell U. 2012. Calcification and photosynthesis in Emiliania huxleyi: physiological and genetic responses to individual carbonate system parameters. PhD thesis, University of Kiel, Kiel, Germany [Google Scholar]
- 65.Sotoj AR, Zheng H, Shoemaker D, Rodriguez J, Read BA, Wahlund TM. 2006. Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliania huxleyi. Appl. Environ. Microb. 72, 5500–5511 (doi:10.1128/aem.00237-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baumann K-H, Andruleit H, Böckel B, Geisen M, Kinkel H. 2005. The significance of extant coccolithophores as indicator of ocean water masses, surface water temperature, and palaeoproductivity: a review. Paläontol. Zeitschr. 79, 93–112 [Google Scholar]
- 67.Klaas C, Archer DE. 2002. Association of sinking organic matter with various types of mineral ballast in the deep sea: implications for the rain ratio. Glob. Biogeochem. Cycles 16, 1116 (doi:10.1029/2001GB001765) [Google Scholar]
- 68.Bates NR. 2007. Interannual variability of the oceanic CO2 sink in the subtropical gyre of the North Atlantic Ocean over the last 2 decades. J. Geophys. Res. 112, C09013 (doi:10.1029/2006JC003759) [Google Scholar]
- 69.Wootton JT, Pfister CA, Forester JD. 2008. Dynamic patterns and ecological impacts of declining ocean pH in a high-resolution multi-year dataset. Proc. Natl Acad. Sci. USA 105, 18 848–18 853 (doi:10.1073/pnas.0810079105) [DOI] [PMC free article] [PubMed] [Google Scholar]