Abstract
The development of the emerging field of ‘paleovirology’ allows biologists to reconstruct the evolutionary history of fossil endogenous retroviral sequences integrated within the genome of living organisms and has led to the retrieval of conserved, ancient retroviral genes ‘exapted’ by ancestral hosts to fulfil essential physiological roles, syncytin genes being undoubtedly among the most remarkable examples of such a phenomenon. Indeed, syncytins are ‘new’ genes encoding proteins derived from the envelope protein of endogenous retroviral elements that have been captured and domesticated on multiple occasions and independently in diverse mammalian species, through a process of convergent evolution. Knockout of syncytin genes in mice provided evidence for their absolute requirement for placenta development and embryo survival, via formation by cell–cell fusion of syncytial cell layers at the fetal–maternal interface. These genes of exogenous origin, acquired ‘by chance’ and yet still ‘necessary’ to carry out a basic function in placental mammals, may have been pivotal in the emergence of mammalian ancestors with a placenta from egg-laying animals via the capture of a founding retroviral env gene, subsequently replaced in the diverse mammalian lineages by new env-derived syncytin genes, each providing its host with a positive selective advantage.
Keywords: endogenous retrovirus, envelope protein, syncytin, placenta, cell–cell fusion
1. Introduction
Much to the surprise of biologists, the determination of genomic sequences from eukaryotes revealed that, in the course of evolution, the DNA of almost all eukaryotic organisms had been colonized by countless mobile genetic elements [1,2]. Mobile elements, also known as transposable elements (TEs), can both multiply copies of themselves and integrate these copies into the genome of their host's germline, become ‘fixed’ and be transmitted vertically to the offspring in a Mendelian fashion. They could be identified at the nucleotide sequence level as repeated sequences with specific features and/or by homology with extant, active mobile elements. Similar to paleontological fossil records that are used to reconstitute the evolutionary history of species, more or less well-conserved traces of these elements could be retrieved, which indicated that they had been invading eukaryotic genomes over millions of years and had coevolved with them.
Two main classes of TEs can be distinguished: class I that includes long terminal repeat (LTR) retrotransposons, non-LTR retrotransposons and endogenous retroviruses (ERVs), and class II that comprises DNA transposons [3]. Whereas DNA transposons propagate via an excision and reintegration mechanism using a transposase enzyme they encode, autonomous retrotransposons and ERVs have an RNA genome and code for the enzymes allowing them to make a cDNA copy of their genomic RNA (reverse transcriptase) and integrate it into the genome of their host (endonuclease or integrase). Non-autonomous retrotransposons are trans-mobilized by the replicative machinery of active autonomous retrotransposons. Recently, both DNA viruses and RNA viruses (other than retroviruses) were also found to have left within eukaryotic genomes integrated DNA copies, designated as endogenous viral elements (EVEs; [4,5]), albeit at a frequency much lower than that of TEs. EVEs are thought to result either from direct insertion by host recombination mechanisms such as non-homologous end joining in the case of DNA viruses or from transposition through the use of the enzymatic machinery of retrotransposons in the case of RNA viruses [4]. Altogether, these elements occupy a substantial fraction of many eukaryotic genomes. For instance, they make up at least 45% and 40% of the human and mouse genomes, respectively [6,7]. Most of the TE sequences have degenerated and have been extensively rearranged in evolution compared with the ancestral inserted elements, but the youngest are still intact and active in a number of host species [8].
Transposons are potentially pathogenic elements that can be gene-disruptive depending on their integration sites and all the more so when amplification bursts occurred, as can be traced through evolution by the accumulation of multiple copies that inserted during the same time interval. But host organisms have developed multiple defence mechanisms to counteract and restrict TE invasions. In turn, TEs are subject to high mutation rates that allow them to escape host restriction factors, which consequently require new restriction mechanisms to be selected for by the host. The outcome of this struggle for survival on both sides turns out to be a motor for evolution [9–11].
Although endogenous mobile elements can be considered as mere ‘genetic parasites’, generator of the so-called ‘junk’ DNA, there is accumulating evidence that some of them were repeatedly co-opted by their host genomes to play beneficial functional roles [12–14]. Actually, the fact that such an extensive fraction of many eukaryotic genomes is composed of TE sequences should have led to the inference that their presence could be accommodated and was not irremediably detrimental to the fitness of the organisms harbouring them. In several cases, they even appear to have provided ultimately a selective advantage. The effects of endogenous TEs encompass genome remodelling owing to DNA recombination and unequal crossover, changes in gene expression through transcription regulation and splicing variations, and even contribution to protein-coding sequences [15–18]. In the context of the latter, the present review will focus on the ERV class of TEs and address the process of endogenization of retroviruses, the co-optation of protein-coding genes from ERVs in diverse mammalian species and more specifically the diversion of retroviral gene functions to perform specialized tasks in the placenta.
2. Retrovirus endogenization
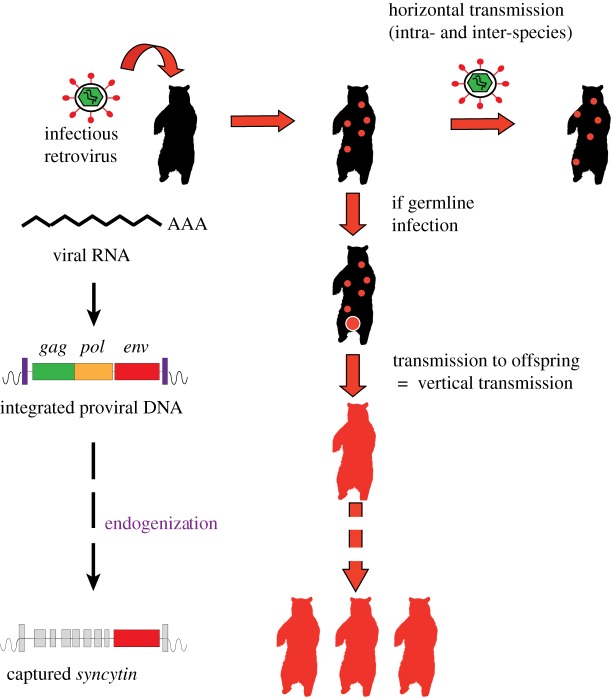
As mentioned above, retroviruses replicate via an RNA intermediate [19]. The viral genomic RNA present in the infectious particles serves as a template for the synthesis of a cDNA copy which is then integrated into the genome of the target cell in the form of a ‘provirus’. Repeated transcription of the provirus allows amplification of viral genomes and production of new infectious virus particles. Infection of the germline, integration and fixation of proviruses at defined positions in the host genome and vertical Mendelian transmission to the offspring constitute the successive stages of the process of endogenization (figure 1). The resulting ERVs that have conserved a recognizable full-length structure display the typical genetic organization of proviruses from known infectious retroviruses. Most of them harbour only the three genes contained in the genome of simple retroviruses, gag, pol and env, flanked by two LTRs. The gag gene encodes a precursor of the subunits that serve as components of the viral capsid, the pol gene a polyprotein that is processed into the constituents of the viral enzymatic machinery required to convert the viral RNA into the double-stranded proviral DNA form, including reverse transcriptase, RNase H and integrase, and the env gene, a glycoprotein which is inserted into the virion membranous envelope and forms spikes at the surface of the retrovirus particles.
Figure 1.
Retrovirus endogenization and syncytin gene capture in mammalian species. Retroviruses are able to reverse transcribe their genomic RNA into a cDNA copy that integrates into the genome of infected cells as a ‘provirus’ which contains the three viral genes gag, pol and env (left). Retroviral infection leads to proviral insertion into a limited number of cells of the infected animal. Production of new infectious virions results in horizontal transmission of the virus (top right). In occasional cases where a retrovirus infects germline cells, the integrated proviral DNA is transmitted vertically to the offspring (centre). The retrovirus has been ‘endogenized’ and is now present in all the somatic and germline cells of the animal. During evolution, most of the endogenous retrovirus-derived genes are disrupted by multiple mutational events but occasionally one of them, such as the env gene exemplified here (bottom left), may be preserved and remains functional over several million years, playing a role in the physiology of its host.
During evolution, successive bursts of amplification within host germlines have led to the formation of ERV repeat families that occupy a significant fraction of mammalian genomes (e.g. 8% and 10% in humans and mice, respectively) [19,20]. Over time, almost all ERVs not subject to any selective pressure have been mutated to various degrees, including large deletions, insertions and extensive sequence rearrangements. Most have been left as scattered and truncated fragments. Very often, solitary LTRs have been generated by homologous recombination between the two LTRs from single proviruses and loss of the internal sequences.
However, in some mammalian species, fully functional proviruses persist. Such ‘young’ ERVs can be reactivated to produce infectious virions that are transmitted horizontally and can even reinfect the germline to form proviruses that can become fixed as new ERVs at novel positions in the host genome. This is notably the case for the mouse leukemia viruses (MLVs) and the mouse mammary tumor viruses [21], the endogenous Jaagsiekte sheep retroviruses (enJSRVs) [22] and the more recently identified koala retrovirus (KoRV) [23]. The last is spreading among koala populations where it may lead to leukaemia and lymphoma, although some preserved, isolated populations are still virus-free. KoRV, present in affected animals as both an infectious exogenous retrovirus and an endogenized ERV within the koala genome, is being considered as an attractive model to study the process of endogenization in the evolution of a species.
In other rare situations, single genes from endogenized ERVs have been conserved over millions of years of evolution, whereas the rest of the proviral sequences have degenerated or been lost (figure 1). Maintenance of a functional gene of retroviral origin in a group of species belonging to the same clade strongly suggests that such a gene provides a selective advantage to its hosts. Remarkably, retroviral env genes were found to have been repeatedly ‘captured’ and ‘domesticated’ to fulfil critical physiological functions in the placenta of several mammalian species belonging to separate clades.
3. The retroviral envelope glycoprotein
The envelope glycoprotein (Env) of enveloped viruses is essential for viral entry by recognizing susceptible cells and inducing fusion of the virion envelope with the cell plasma membrane, allowing the release of the nucleocapsid into the cell cytoplasm [24]. The env gene of retroviruses encodes a precursor protein which is cleaved, during transport through the host cell secretory pathway, by a cellular furin protease to generate two fragments, the surface (SU) and the transmembrane (TM) subunits (figure 2a) [25]. Both subunits remain associated as heterodimers which are further assembled into Env trimers at the surface of the virion envelope membrane. The SU subunit displays a domain which specifically interacts with a receptor on the surface of cells susceptible to the virus, whereas the TM subunit triggers the fusion of the virus and cell membranes by insertion of its N-terminal fusion peptide into the membrane of target cells (figure 2a,b). The TM subunit also contains a transmembrane domain anchoring the Env heterodimer into the virion envelope and a highly conserved immunosuppressive domain (ISD).
Figure 2.
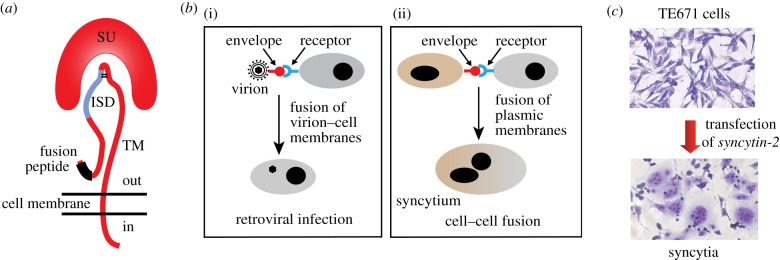
The retroviral envelope glycoprotein. (a) Structure of the retroviral envelope protein with the SU and TM subunits, the fusion peptide and the immunosuppressive domain (ISD). (b) Consequences of the interaction between a retroviral envelope protein and its cognate receptor: virion–cell membrane fusion and virus entry into the target cell (i) or cell–cell membrane fusion and formation of a syncytium (ii). (c) Cell–cell fusion and formation of multinucleated syncytia induced by transfecting human TE671 cells with a syncytin-2 expression vector (May–Grünwald–Giemsa staining).
The immunosuppressive activity of retroviral Env proteins had been first reported in ex vivo studies [26] and was further substantiated in our laboratory using in vivo tumour rejection assays that measured the ability of the proteins, when expressed in allogeneic tumour cells normally rejected by engrafted mice, to allow these cells to escape, at least transiently, immune rejection [27]. This approach led to the delineation of the ISD within a 20 amino acid-long peptide and to the identification of two amino acid positions where introduction of specific mutations could abolish the immunosuppressive activity without affecting the fusogenicity and infectivity of the protein [28]. The ISD was next demonstrated to be critical for the propagation of the retrovirus itself [29]: the presence of the ISD-inactivating mutations within the env gene fully impaired the replication of Friend MLV in normal immunocompetent mice, whereas the mutant virus replicated with the same efficiency as the wild-type, both ex vivo in cultured cells and in vivo in irradiated immunocompromized mice. Finally, specific cell depletion experiments carried out using the above virus–host murine model demonstrated that the identified ISD had a pleiotropic effect, with impairment of both the innate and adaptive immune responses. Thus, retroviral Envs are more complex than initially suspected, being multifunctional proteins that can act both as mechanical effectors for membrane fusion, and as immunological effectors required for repressing the anti-viral host immune response.
Interestingly, the Env glycoproteins encoded by several endogenized ERV env genes have kept some of the properties initially displayed by the Env of their ancestral infectious retrovirus, such as recognition of a specific cell receptor, fusogenicity and/or immunosuppressive activity. Conservation of such features may indicate a significant physiological role in the host cells where the genes are expressed. In the mouse, for instance, the ERV env-derived genes, Fv4 and Rcmf, were shown to be involved in a process of receptor interference, conferring to the cells where they are expressed resistance to superinfection with exogenous retroviruses by downregulating their receptor on the target cell surface [30]. The same phenomenon was found to occur in the case of the enJSRV env genes [22]. In the case of several enveloped viruses, the Env protein synthesized by infected cells and exposed at their surface can confer on them the ability to fuse with neighbouring uninfected cells that express the appropriate receptor, resulting in the formation of multinucleated giant cells called syncytia (figure 2b) [31]. In this context, several placenta-specific genes encoding fusogenic Env proteins co-opted from members of distinct ERV families have been identified within the genome of various mammalian species.
4. Human endogenous retrovirus env genes
Comprehensive screening for identifying human ERV (HERV) env genes with long open reading frames (ORFs) within the human genome disclosed 18 genes encoding a putative envelope protein [32,33]. Among those potentially functional, the first to be described was ERV3, a proviral gene specifically expressed in the placenta and conserved in the Hominoid and Old World monkey families [34]. The ERV3 protein lacks a hydrophobic transmembrane anchoring domain and therefore the ability to operate as a fusogenic protein, and was suggested to be involved in the proliferation and/or differentiation of the trophoblast. However, the absence of the ERV3 gene in the gorilla [34] and the fact that a premature stop codon was found to be present in a homozygous state in 1% of the human population ruled out that this otherwise highly conserved gene might be critical for reproduction or survival [35]. However, one could envision that the human ERV3 gene has played a role in the past, but may now be undergoing a process of progressive loss of function with compensatory replacement by another gene(s).
Next, attention was focused on two envelope glycoproteins, one encoded by a member of the HERV-W family [36,37] and the other by a member of the HERV-FRD family [38]. Both env genes display placenta-specific expression and induce syncytium formation when introduced into cultured cells (figure 2c), which led to their designation as syncytin-1 and syncytin-2, respectively. Each protein binds a distinct receptor: syncytin-1 interacts with the neutral amino acid transporter ASCT2 [36] and syncytin-2 with the multipass transmembrane protein MFSD2 [39]. Comparative analysis of the syntenic regions within the genomes from various species revealed that the syncytin-1 gene entered the genome of primates before the separation of Hominoids from Old World monkeys, being conserved only in Hominoids [40], whereas syncytin-2 was found to be conserved in the genome of all primates, except prosimians [38].
Determination of the presence or absence of each syncytin gene in various extant species provides a paleovirological record of its evolutionary history which, together with the otherwise determined phylogeny of the present-day species, allows the dating of the insertion of its parental ERV into the genome of a common ancestor. Thus, the age of the syncytin-1 gene could be estimated to be around 30 million years (Myr) and that of syncytin-2 to be older, at more than 45 Myr. Compellingly, for both genes, the conservation of an intact ORF in multiple related species, with a low ratio of non-synonymous versus synonymous mutation rates (dN/dS < 1) indicative of a ‘purifying selection’ acting on the genes, and the very low levels of polymorphism within the human population argue for an essential physiological role [41]. Several features of both human syncytin proteins indicated that they are instrumental in the formation of a major component of the placenta, i.e. the syncytiotrophoblast.
5. Syncytins in the human placenta
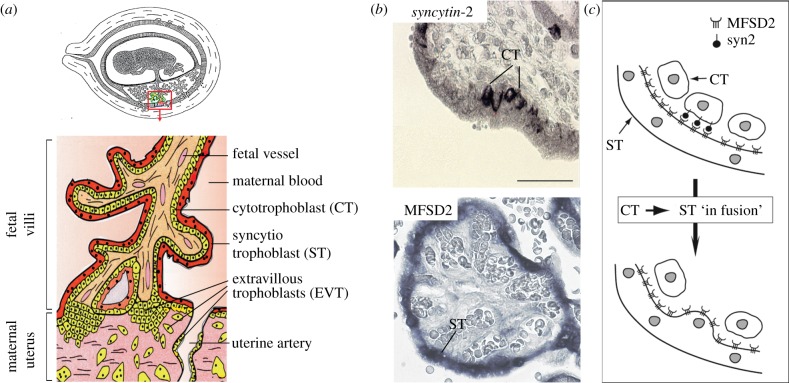
The placenta is a transient autonomous organ of embryonic origin whose main function is to mediate mutual metabolic exchanges between fetus and mother. It also provides an environment of immune tolerance ensuring the protection of the ‘fetal allograft’, which expresses ‘foreign’ paternal antigens, within the mother's body. The placental tissue, or trophoblast, is formed by peripheral cells of the blastocyst, the cytotrophoblasts, which attach to the uterine wall. Cytotrophoblasts are among the rare cells of the human body, together with the myoblasts, the macrophages and the gametes, able to undergo cell–cell fusion. Beginning at embryonic day 6 (E6), mononucleated cytotrophoblasts fuse together into the syncytiotrophoblast, a highly invasive multinucleated syncytial layer that mediates implantation of the embryo into the maternal endometrium [42]. From E15, the basic functional unit of the placenta composed of the chorionic villi, starts to develop. The villi either float freely in maternal blood spaces or are anchored into the uterine wall (figure 3a). At this stage, fetal blood vessels are embedded in the internal mesenchymal stroma, whereas the syncytiotrophoblast covers the outside surface of the villi, in direct contact with maternal blood. The syncytiotrophoblast regenerates and expands by fusion of underlying cytotrophoblasts. This essential tissue plays multiple roles, including exchange functions between mother and fetus, hormone secretions, regulation of the immune response and protection against pathogens [43]. Finally, extravillous cytotrophoblasts originating from an independent differentiation pathway and located at the base of anchoring villi gain an invasive phenotype, migrate into the maternal decidua and later differentiate into giant cells in the myometrium. They also invade the uterine ‘spiral arteries’ where they replace the endothelial cells and remodel them into dilated vessels, thus increasing the maternal blood flow to the fetus (figure 3a) [44].
Figure 3.
Involvement of syncytin-2 and its cognate receptor MFSD2 in human placentation. (a) Schematic of a human feto–placental unit and (enlarged) of a placental villus. (b) In situ analysis of human placental villi sections for syncytin-2 and MFSD2 expression. MFSD2 expression is detected exclusively within the syncytiotrophoblast layer (ST), whereas syncytin-2 expression is restricted to underlying mononucleated cytotrophoblasts (CT). (c) ‘In fusion’ model for syncytiotrophoblast formation, where interaction between syncytin-2 and MFSD2 results in polarized fusion of the cytotrophoblasts into the syncytiotrophoblast. Adapted from Esnault et al. [39].
In situ analysis of syncytin expression suggested that both genes were involved in placental development. Syncytin-1 was found to be expressed throughout the placenta in all the trophoblast cell lineages [45,46]. Interestingly, the placental transcription factor GCM1 was shown to control syncytin-1 expression and to display an expression profile coinciding with that of syncytin-1 in placental tissues [47]. However, no clear results have yet been reported with regard to the localization of the expression sites for ASCT-2, the syncytin-1 receptor, in trophoblast cells, which seems to depend on the antibodies used, so that evidence for syncytin-1 involvement in syncytiotrophoblast formation is not fully conclusive at this time. By contrast, syncytin-2 expression takes place in only a fraction of the villous cytotrophoblasts [48], whereas expression of the gene for its receptor, MFSD2, is detected in the syncytiotrophoblast (figure 3b) [39]. Coordinated expression of syncytin-2 and MFSD2 in distinct cell types may drive the incorporation by fusion of the mononucleated cytotrophoblasts into the adjacent syncytiotrophoblast, while impairing fusion of the cytotrophoblasts between themselves. Such a process of polarized ‘in-fusion’ may promote the growth and renewal of the syncytiotrophoblast during pregnancy (figure 3b) [39]. In primary cultures of spontaneously differentiating cytotrophoblasts, silencing of syncytin-2 and, to a lesser extent of syncytin-1, either by the addition of antisense oligonucleotides or by RNA interference with small inhibitory RNAs, greatly interferes with cell–cell fusion [49,50]. Altogether, these data provide evidence for a major role of syncytin-2 (likely in concert with syncytin-1) in the differentiation of the syncytiotrophoblast. Although additional cellular factors have been previously reported to contribute to the fusion process (e.g. phosphatidylserine externalization, the genes encoding connexin 43, cadherin 11 and CD 98, the zona occludens-1 (ZO-1) gene) [51,52], the real effectors are most probably the syncytin genes, owing to their intrinsic fusogenicity. It has to be stressed that it has not been possible to substantiate a role for syncytins in syncytiotrophoblast formation at early implantation (i.e. at approx. E6), owing to the limited availability of placental material at such an early stage in humans.
6. Mouse syncytins: a genetic clue for a role in placentation
With the purpose of developing an animal model to investigate the role of syncytin proteins in placental development, the mouse genome was screened for syncytin-encoding genes. Remarkably, bioinformatic analysis of the fully sequenced mouse genome led to the discovery of two genes encoding retroviral Env proteins, designated syncytin-A and -B, distinct from the human syncytin-1 and -2 genes, but sharing the same characteristics [53]: they display placenta-specific expression, are fusogenic, and have been highly conserved since their integration into the genome of a muroid ancestor more than 25 Myr ago (Ma).
In the mouse placenta, the fetal capillaries are separated from the maternal blood lacunae by two distinct layers of syncytiotrophoblast (ST-I and ST-II; figure 4). Although the gross architecture of the mouse and human placentae differ, a functional analogy can be made between the single human syncytiotrophoblast layer and the murine ST-I and ST-II layers: the two murine layers could be considered as a unique structure because they are in intimate contact, with gap junctions establishing continuity between their respective cytoplasm (for general reviews comparing the structure and function of the mouse and human placentae, see Georgiades et al. [56] and Watson & Cross [57]). The mouse syncytin-A gene was found to be expressed in the ST-I layer, proximal to maternal blood spaces, whereas syncytin-B expression was detected in the ST-II layer, close to fetal blood vessels [58]. As was the case for human syncytin-1, syncytin-B expression is controlled by the Gcm1 transcription factor, in agreement with the fact that the syncytin-B and Gcm1 genes are both expressed in the ST-II layer. Knocking out the syncytin-A gene in mice had dramatic effects, since homozygous mutant embryos died at mid-gestation [54], displaying disruption of the early placental architecture, accumulation of unfused cytotrophoblasts and defective cell–cell fusion within the ST-I layer (figure 4). By contrast, invalidation of the syncytin-B gene had only mild effects, since syncytin-B null embryos were viable, showing only limited late-onset growth retardation and a slight decrease in the number of neonates [55]. Yet, the syncytin-B null placenta displayed an altered ST-II layer with unfused, apposed cytotrophoblasts and enlarged maternal blood spaces (figure 4).
Figure 4.
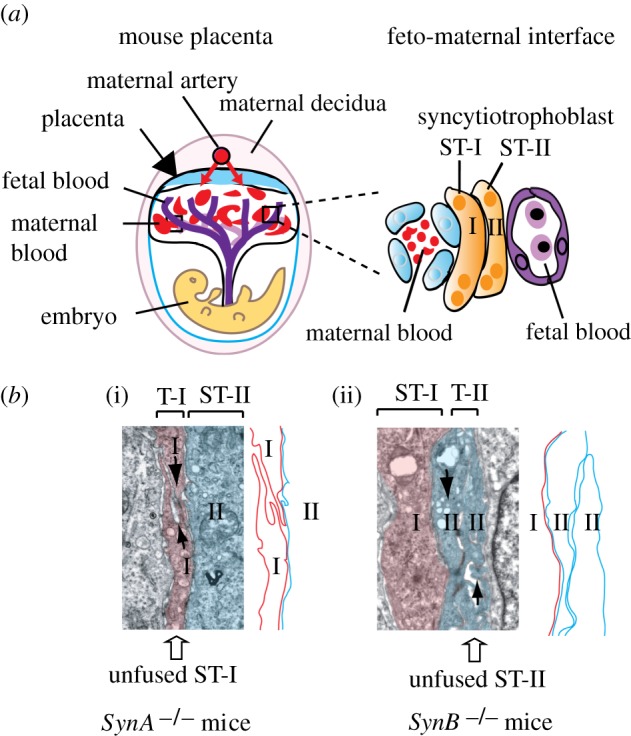
Both murine syncytin genes are required for the formation of the murine syncytiotrophoblast double layer. (a) Scheme of the mouse placenta with an enlarged representation of the feto-maternal interface highlighting the presence of two syncytiotrophoblast layers (ST-I and ST-II). (b) Electron microscopy analysis of the placental feto-maternal interface of mice deficient for syncytin-A (i) or syncytin-B (ii), showing fusion defects of syncytiotrophoblast layers ST-I and ST-II, respectively. Adapted from Dupressoir et al. [54,55].
Analysis of the syncytin-B null mutant transcriptome, compared with that of wild-type placenta, showed induction of the gene encoding connexin-30, a gap junction protein, which was found to accumulate at the maternal–fetal interface within the area of the defective unfused ST-II cell layer. These results suggested that compensatory mechanisms mediated by gap junctions, which promote direct intercellular communications, might counteract the fusion defects and allow embryos lacking the syncytin-B gene to survive. Remarkably, double knockout embryos lacking both syncytin-A and syncytin-B displayed an aggravated phenotype as compared with the syncytin-A single knockout embryos, since they died earlier [55]. Thus, both the ST-I and ST-II syncytial layers cooperate to preserve the structural and functional integrity of the maternal–fetal interface. These mutant mice provided the first demonstration of the critical role played by syncytin genes in placentation. They also constitute a model to investigate further the potential involvement of syncytin gene mutations in placental diseases associated with cell–cell fusion defects.
7. Syncytins are widespread in eutherian mammals
Although they share similar features, the primate and muroid syncytins are clearly not orthologous genes—they are not syntenic in the two species clades—indicating that they are the result of independent gene captures that occurred separately in the genome from ancestors of each lineage. Furthermore, a fifth syncytin gene, syncytin-Ory1, distinct from each of those just listed, has been identified in yet another mammalian lineage, the Leporidae family (rabbit and hare) [59]. The syncytin-Ory1 gene codes for a placenta-specific envelope protein endowed with fusogenic activity and has been conserved for over 12 Myr. Its receptor, ASCT-2, is the same as that of the human syncytin-1 protein. Syncytin-Ory1 expression was detected in the placenta junctional zone, where the invading placental syncytia come into contact with the maternal decidua, suggesting its possible involvement in syncytiotrophoblast formation. More recently, a sixth functional syncytin gene, syncytin-Car1, was found to be present in the 26 species of carnivores investigated [60]. Carnivores are members of the Laurasiatheria superorder, which diverged from the Euarchontoglires more than 80 Ma. This makes syncytin-Car1 the oldest known syncytin gene. Both the dog and cat syncytin-Car1 display fusogenic activity and are specifically expressed in the placenta within the syncytiotrophoblast layer at the maternal–fetal interface [60].
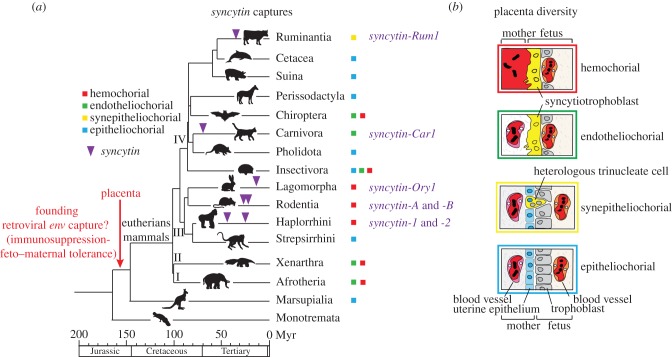
Altogether, these studies provide clear evidence that a number of retroviral env genes have been independently co-opted on multiple occasions in the course of mammalian evolution (figure 5) and may have thus repeatedly prompted the emergence of a syncytial maternal–fetal interface, exemplifying a remarkable phenomenon of evolutionary convergence. Interestingly, comparative anatomy reveals that the placenta is the most variable organ among mammalian species (reviewed in Wooding & Burton [62]; figure 5). Placentae essentially differ in the extent of uterine tissue invasion by the trophoblast cells covering the blastocyst at implantation and by the architecture of the resulting maternal–fetal interface. In the epitheliochorial placenta (horse and pig), the fetal trophoblast is composed of unfused, mononucleated cytotrophoblasts that form a monolayer adjacent to the uterine epithelium. In the three other types of placental interface, the synepitheliochorial (e.g. ruminants), the endotheliochorial (e.g. carnivores) and the hemochorial (e.g. humans, mice and rabbits) types, the degree of invasiveness of the trophoblast increases from one type to the next, respectively, and is accompanied by sequential loss of the intervening maternal cell layers (epithelial and endothelial cells), up to direct contact between the syncytiotrophoblast and maternal blood in the case of the hemochorial placenta (figure 5). However, the various placental structures cannot be predicted merely based on species taxonomy, and their evolutionary pathways are still a matter of debate [63]. Along this line, it seems relevant to ask in what measure, depending on expression pattern, intrinsic fusogenic activity or receptor identity and localization, the syncytin genes present in each species may account for the observed variability in the morphology and organization of the utero–placental interface.
Figure 5.
Multiple syncytin gene captures and diversity of placental structures in eutherian mammals. (a) Phylogenetic tree of mammals, with the four major clades of eutherians: (I) Afrotheria, (II) Xenarthra, (III) Euarchontoglires and (IV) Laurasiatheria (adapted from Meredith et al. [61]). The four basic types of placentation are indicated by coloured squares (the colour code corresponds to that of the boxes that frame the images shown to the right). The time of insertion of the different syncytin genes identified to date is indicated. Branch length is proportional to time (in million years, Myr). (b) Schematic colour-coded representation of the maternal–fetal interface in the four main types of placental structures. Placental types are classified from top to bottom in the order of decreasing extent of syncytialization and invasive properties.
In this context, it is of interest to note that a seventh syncytin gene was recently discovered in the suborder Ruminantia [64], in whose species the placenta lacks an extended syncytium layer, but displays synepitheliochorial placentation, which involves a very limited cell fusion process, with only trinucleate cells (or small multinucleated syncytial plaques in the case of sheep) being formed and with evidence for heterologous fusion between cells of fetal and maternal origin, a feature not found among the other eutherian mammals. This gene, named syncytin-Rum1, was first identified in the cow, where it was shown to be specifically expressed in the placenta. Next, it was found to be also present within the genome of sheep, as well as of 14 other higher ruminant species that were investigated, such as cervids, giraffes and antelopes, indicating that it has been conserved for more than 30 Myr. Both the cow and sheep orthologues were found to display fusogenic activity in ex vivo assays by conferring infectivity on ‘pseudotypes’ (i.e. defective retroviral particles) and triggering cell–cell fusion. In situ hybridization on placenta sections revealed specific expression of syncytin-Rum1 in the trophoblast binucleate cells, consistent with a role in the formation, by heterologous cell fusion with uterine cells, of the trinucleate cells of the cow and the syncytial plaques of the ewe. Syncytin-Rum1 is common to all ruminant species displaying a synepitheliochorial placenta but is not found in any other mammalian species. Therefore, its capture may have contributed to the emergence of this unique placental structure. Altogether, the available data strongly support the notion that the exaptation of syncytin genes has driven major morphological innovations in mammalian placentae, and provides new molecular clues to understand the remarkable transformations that took place during placenta evolution.
Finally, it has to be noted that the involvement of env gene capture in placenta development may not be restricted to the bona fide syncytin genes endowed with fusogenic activity. For instance, in sheep, injection of antisense morpholino-oligonucleotides in utero showed that the enJSRV env genes—acquired through the recent endogenization of an infectious JSRV retrovirus in the Caprinae (sheep/goat) lineage 5–7 Ma (but not found in the cow)—are involved in the control of placental growth and differentiation at early stages of implantation, independently of cell fusion [65]. Another placenta-specific ERV env gene, designated env-Cav1, was discovered within the genome of the guinea pig [66], which is a member of the Caviomorpha rodent suborder, distant from muroids. An orthologous gene with a conserved ORF was found to be present in all the Caviomorpha species that were investigated, consistent with a time of insertion more than 30 Ma. Guinea pig env-Cav1 is specifically expressed at the level of the placenta junctional zone, suggesting a role in the invasion of the uterine tissues by cytotrophoblasts, as hypothesized for syncytin-1 in humans. No fusogenic activity of the env-Cav1-encoded protein could be detected, indicating that this gene may not play a role in placental architecture, as that played by the previously identified classical syncytins, but be involved in invasiveness and/or exert immunosuppressive functions (see §8).
8. Conclusion: a model for retroviral syncytin capture, life and death in the natural history of placental mammals
The determination of an ever increasing number of whole genome sequences from living organisms allows us to reconstruct the evolutionary history of fossil retroviral sequences, some of which have conserved an activity beneficial to their host over millions of years. The development of this emerging field of ‘functional paleovirology’ has led to the retrieval of genes of ancestral retrovirus origin encoding proteins that have been ‘exapted’ to fulfil essential physiological roles, syncytin genes being undoubtedly among the most remarkable examples. Indeed, syncytins are ‘new’ genes encoding proteins derived from the envelope protein of endogenous retroviral elements that have been captured and domesticated on multiple occasions and independently in diverse mammalian species, through a process of convergent evolution. Knockout of syncytin genes in genetically modified mice provided evidence for their absolute requirement for placenta development and embryo survival, via formation by cell–cell fusion of syncytial cell layers at the fetal–maternal interface. Thus, viral genes initially indispensable for the completion of the multiplication cycle of retro-viruses appear to have been converted to genuine ‘cellular genes’ endowed with an essential physiological role.
These genes of exogenous origin and acquired ‘by chance’ raise a paradox, as they are ‘necessary’ to carry out a basic function common to placental mammals, and yet they appear to have been acquired lately. Indeed, although the capture of some of them dates back to up to 80 Myr, the presently identified syncytins can still be considered as relatively recent genes by comparison with the date of emergence of a primitive placenta in a mammalian ancestor (approx. 150 Ma). To resolve this paradox, an evolutionary model can be proposed which is consistent with our present knowledge on the paleontology of retroviral elements, and the functional properties identified for the env genes, either from present-day infectious retroviruses or from anciently captured ERVs.
The model states that a pivotal event in the emergence of placental mammals has been the capture of a founding retroviral env gene, but that this ancestral env gene has been subsequently replaced in the diverse lineages emerging in the course of the mammalian radiation upon successive and independent germline infections by new retroviruses and co-optation of their env gene, each new gene providing its host with a positive selective advantage (figure 5). Such a hypothesis would account for the evolutionary transition from egg-laying to placental animals as well as for the diversity in both the nature and time of insertion of the captured syncytins that can be currently found. It would also rather simply account for the multiplicity of placental structures that have emerged, concomitantly with the diversity of the captured env genes. A consequence of this model is that evidence should exist for ‘lost syncytins’ in eutherian mammals, and this is precisely what was found in a recent study of another human envelope protein gene, belonging to a HERV-V provirus, named envV, which is also specifically expressed within the human placenta, and whose putative role in human placentation remains to be investigated since it was found to be non-fusogenic [67]. Actually, this env gene entered the primate genome concomitantly with syncytin-2, that is, more than 45 Ma as inferred from its present status in primates. Interestingly, it was recently shown that the gene is fusogenic in the Old World monkeys where it most probably still behaves as a bona fide syncytin, whereas its fusogenic activity appears to have been lost in higher primates (including humans) and in some New World monkeys [68]. This situation well illustrates how a captured retroviral gene may undergo distinct evolutionary fates depending on the evolution of the host itself, in a complex reciprocal interplay. Other env genes, such as ERV3, might also correspond to degenerating syncytins, on their way to be replaced by new emerging syncytins, in a continuous evolutionary process whereby incoming retroviruses would play the role of ‘generators of diversity’.
A question still pending on the primary role of syncytins in the emergence of placental mammals concerns the function of the ancestral env gene that was originally co-opted. Indeed, we now know from the data reported above that Env proteins are not only involved in membrane fusion (a property necessary for syncytiotrophoblast formation) but also are bona fide immunosuppressive effectors. As mentioned above, the TM subunit of the Env protein from infectious retroviruses contains an ISD endowed with an immunosuppressive function critical for the propagation of the virus in vivo, in normal immunocompetent animals. It turns out that in all the species where they have been found, syncytins have conserved this feature from their ancestral infectious progenitor, at least one of them carrying a functional ISD in the case of primates and muroids where syncytins are present as pairs. This syncytin attribute is likely to be—or to have been—involved in maternal immune tolerance towards the semi-allogeneic feto–placental unit which expresses histo-incompatible paternal antigens. In fact, this feature could even constitute the primordial function of syncytins, prior to their fusogenic activity.
Indeed, the emergence of a primitive placenta in an oviparous ancestor may have been made possible thanks to the co-optation of a founding env gene that would have kept the immunosuppressive capacity of its ancient viral progenitor, allowing ‘grafting’ of a fetus exhibiting ‘foreign’ antigens from the father within the mother's body (figure 5). Along this line, it will be of interest to investigate the presence of syncytin-like genes in the epitheliochorial placenta (found in horse and pig), a tissue where cell–cell fusion does not occur. If the above hypothesis is correct, such genes are also expected there, which would have lost their fusogenic activity or their cognate receptor, and whose primary role would be immune protection of the fetus. Evidence for such a scenario could be provided by experiments—in progress—with genetically modified ‘knock-in’ mice in which the immunosuppressive function of the syncytins is specifically invalidated via introduction of appropriate point mutations that do not alter their fusogenic activity.
References
- 1.Tollis M, Boissinot S. 2010. The evolutionary dynamics of transposable elements in eukaryote genomes. Genome Dyn. 7, 68–91 (doi:10.1159/000337126) [DOI] [PubMed] [Google Scholar]
- 2.Makalowski W, Pande A, Gotea V, Makalowska I. 2012. Transposable elements and their identification. Methods Mol. Biol. 855, 337–359 (doi:10.1007/978-1-61779-582-4_12) [DOI] [PubMed] [Google Scholar]
- 3.Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (doi:10.1038/nrg2165) [DOI] [PubMed] [Google Scholar]
- 4.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6, e1001191 (doi:10.1371/journal.pgen.1001191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13, 283–296 (doi:10.1038/nrg3199) [DOI] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (doi:10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 7.Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (doi:10.1038/nature01262) [DOI] [PubMed] [Google Scholar]
- 8.Huang CR, Burns KH, Boeke JD. 2012. Active transposition in genomes. Annu. Rev. Genet. 46, 651–675 (doi:10.1146/annurev-genet-110711-155616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin HL, Moran JV. 2011. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627 (doi:10.1038/nrg3030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host–virus arms races. Annu. Rev. Genet. 46, 677–700 (doi:10.1146/annurev-genet-110711-155522) [DOI] [PubMed] [Google Scholar]
- 11.Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 10, 395–406 (doi:10.1038/nrmicro2783) [DOI] [PubMed] [Google Scholar]
- 12.Kidwell MG, Lisch DR. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55, 1–24 (doi:10.1111/j.0014-3820.2001.tb01268.x) [DOI] [PubMed] [Google Scholar]
- 13.Volff JN. 2006. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28, 913–922 (doi:10.1002/bies.20452) [DOI] [PubMed] [Google Scholar]
- 14.Alzohairy AM, Gyulai G, Jansen RK, Bahieldin A. 2013. Transposable elements domesticated and neofunctionalized by eukaryotic genomes. Plasmid 69, 1–15 (doi:10.1016/j.plasmid.2012.08.001) [DOI] [PubMed] [Google Scholar]
- 15.Zdobnov EM, Campillos M, Harrington ED, Torrents D, Bork P. 2005. Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res. 33, 946–954 (doi:10.1093/nar/gki236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurka J, Kapitonov VV, Kohany O, Jurka MV. 2007. Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 8, 241–259 (doi:10.1146/annurev.genom.8.080706.092416) [DOI] [PubMed] [Google Scholar]
- 17.Chenais B, Caruso A, Hiard S, Casse N. 2012. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509, 7–15 (doi:10.1016/j.gene.2012.07.042) [DOI] [PubMed] [Google Scholar]
- 18.Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46, 21–42 (doi:10.1146/annurev-genet-110711-155522) [DOI] [PubMed] [Google Scholar]
- 19.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42, 709–732 (doi:10.1146/annurev.genet.42.110807.091501) [DOI] [PubMed] [Google Scholar]
- 20.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26, 291–315 (doi:10.1023/A:1024455415443) [DOI] [PubMed] [Google Scholar]
- 21.Stocking C, Kozak CA. 2008. Murine endogenous retroviruses. Cell Mol. Life Sci. 65, 3383–3398 (doi:10.1007/s00018-008-8497-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varela M, Spencer TE, Palmarini M, Arnaud F. 2009. Friendly viruses: the special relationship between endogenous retroviruses and their host. Ann. NY Acad. Sci. 1178, 157–172 (doi:10.1111/j.1749-6632.2009.05002.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442, 79–81 (doi:10.1038/nature04841) [DOI] [PubMed] [Google Scholar]
- 24.Cosset FL, Lavillette D. 2011. Cell entry of enveloped viruses. Adv. Genet. 73, 121–183 (doi:10.1016/B978-0-12-380860-8.00004-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter E. 1997. Viral entry and receptors. In Retroviruses (eds Coffin JM, Hughes SH, Varmus HE.), pp. 71–119 Plainview, NY: Cold Spring Harbor Laboratory Press; [PubMed] [Google Scholar]
- 26.Cianciolo GJ, Copeland TD, Oroszlan S, Snyderman R. 1985. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Science 230, 453–455 (doi:10.1126/science.2996136) [DOI] [PubMed] [Google Scholar]
- 27.Mangeney M, Heidmann T. 1998. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc. Natl Acad. Sci. USA 95, 14 920–14 925 (doi:10.1073/pnas.95.25.14920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. 2007. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl Acad. Sci. USA 104, 20 534–20 539 (doi:10.1073pnas.0707873105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlecht-Louf G, Renard M, Mangeney M, Letzelter C, Richaud A, Ducos B, Bouallaga I, Heidmann T. 2010. Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc. Natl Acad. Sci. USA 107, 3782–3787 (doi:10.1073/pnas.0913122107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Best S, Le Tissier PR, Stoye JP. 1997. Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol. 5, 313–318 (doi:10.1016/S0966-842X(97)01086-X) [DOI] [PubMed] [Google Scholar]
- 31.Huerta L, Lopez-Balderas N, Rivera-Toledo E, Sandoval G, Gomez-Icazbalceta G, Villarreal C, Lamoyi E, Larralde C. 2009. HIV-envelope-dependent cell–cell fusion: quantitative studies. ScientificWorldJournal 9, 746–763 (doi:10.1100/tsw.2009.90) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77, 10 414–10 422 (doi:10.1128/JVI.77.19.10414-10422.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villesen P, Aagaard L, Wiuf C, Pedersen FS. 2004. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 1, 32 (doi:10.1186/1742-4690-1-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herve CA, Forrest G, Lower R, Griffiths DJ, Venables PJ. 2004. Conservation and loss of the ERV3 open reading frame in primates. Genomics 83, 940–943 (doi:10.1016/j.ygeno.2003.10.003) [DOI] [PubMed] [Google Scholar]
- 35.de Parseval N, Heidmann T. 1998. Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J. Virol. 72, 3442–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74, 3321–3329 (doi:10.1128/JVI.74.7.3321-3329.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mi S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (doi:10.1038/35001608) [DOI] [PubMed] [Google Scholar]
- 38.Blaise S, de Parseval N, Benit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA 100, 13 013–13 018 (doi:10.1073pnas.2132646100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esnault C, Priet S, Ribet D, Vernochet C, Bruls T, Lavialle C, Weissenbach J, Heidmann T. 2008. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl Acad. Sci. USA 105, 17 532–17 537 (doi:10.1073pnas.0807413105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl Acad. Sci. USA 101, 1731–1736 (doi:10.1073pnas.0305763101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Parseval N, Diop G, Blaise S, Helle F, Vasilescu A, Matsuda F, Heidmann T. 2005. Comprehensive search for intra- and inter-specific sequence polymorphisms among coding envelope genes of retroviral origin found in the human genome: genes and pseudogenes. BMC Genomics 6, 117 (doi:10.1186/1471-2164-6-117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bischof P, Irminger-Finger I. 2005. The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 37, 1–16 (doi:10.1016/j.biocel.2004.05.014) [DOI] [PubMed] [Google Scholar]
- 43.Gude NM, Roberts CT, Kalionis B, King RG. 2004. Growth and function of the normal human placenta. Thromb. Res. 114, 397–407 (doi:10.1016/j.thromres.2004.06.038) [DOI] [PubMed] [Google Scholar]
- 44.Harris LK. 2010. Review: trophoblast–vascular cell interactions in early pregnancy: how to remodel a vessel. Placenta 31(Suppl.), S93–S98 (doi:10.1016/j.placenta.2009.12.012) [DOI] [PubMed] [Google Scholar]
- 45.Muir A, Lever AM, Moffett A. 2006. Human endogenous retrovirus-W envelope (syncytin) is expressed in both villous and extravillous trophoblast populations. J. Gen. Virol. 87, 2067–2071 (doi:10.1099/vir.0.81412-0) [DOI] [PubMed] [Google Scholar]
- 46.Malassine A, Handschuh K, Tsatsaris V, Gerbaud P, Cheynet V, Oriol G, Mallet F, Evain-Brion D. 2005. Expression of HERV-W Env glycoprotein (syncytin) in the extravillous trophoblast of first trimester human placenta. Placenta 26, 556–562 (doi:10.1016/j.placenta.2004.09.002) [DOI] [PubMed] [Google Scholar]
- 47.Yu C, Shen K, Lin M, Chen P, Lin C, Chang GD, Chen H. 2002. GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 277, 50 062–50 068 (doi:10.1074/jbc.M209316200) [DOI] [PubMed] [Google Scholar]
- 48.Malassine A, Blaise S, Handschuh K, Lalucque H, Dupressoir A, Evain-Brion D, Heidmann T. 2007. Expression of the fusogenic HERV-FRD Env glycoprotein (syncytin 2) in human placenta is restricted to villous cytotrophoblastic cells. Placenta 28, 185–191 (doi:10.1016/j.placenta.2006.03.001) [DOI] [PubMed] [Google Scholar]
- 49.Frendo JL, Olivier D, Cheynet V, Blond JL, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. 2003. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell Biol. 23, 3566–3574 (doi:10.1128/MCB.23.10.3566-3574.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vargas A, Moreau J, Landry S, Lebellego F, Toufaily C, Rassart E, Lafond J, Barbeau B. 2009. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 392, 301–318 (doi:10.1016/j.jmb.2009.07.025) [DOI] [PubMed] [Google Scholar]
- 51.Potgens AJ, Drewlo S, Kokozidou M, Kaufmann P. 2004. Syncytin: the major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum. Reprod. Update 10, 487–496 (doi:10.1093/humupd/dmh039) [DOI] [PubMed] [Google Scholar]
- 52.Malassine A, Frendo JL, Evain-Brion D. 2010. Trisomy 21- affected placentas highlight prerequisite factors for human trophoblast fusion and differentiation. Int. J. Dev. Biol. 54, 475–482 (doi:10.1387/ijdb.082766am) [DOI] [PubMed] [Google Scholar]
- 53.Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl Acad. Sci. USA 102, 725–730 (doi:10.1073pnas.0406509102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl Acad. Sci. USA 106, 12 127–12 132 (doi:10.1073pnas.0902925106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, Pierron G, Heidmann T. 2011. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl Acad. Sci. USA 108, E1164–E1173 (doi:10.1073/pnas.1112304108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Georgiades P, Ferguson-Smith AC, Burton GJ. 2002. Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23, 3–19 (doi:10.1053/plac.2001.0738) [DOI] [PubMed] [Google Scholar]
- 57.Watson ED, Cross JC. 2005. Development of structures and transport functions in the mouse placenta. Physiology 20, 180–193 (doi:10.1152/physiol.00001.2005) [DOI] [PubMed] [Google Scholar]
- 58.Simmons DG, Natale DR, Begay V, Hughes M, Leutz A, Cross JC. 2008. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 135, 2083–2091 (doi:10.1242/dev.020099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. 2009. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit : a new ‘syncytin’ in a third order of mammals. Retrovirology 6, 107 (doi:10.1186/1742-4690-6-107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Veron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl Acad. Sci. USA 109, E432–E441 (doi:10.1073/pnas.1115346109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meredith RW, et al. 2011. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (doi:10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 62.Wooding FB, Burton GJ. 2008. Comparative placentation. Structures, functions and evolution. Berlin, Germany: Springer [Google Scholar]
- 63.Carter AM, Mess A. 2007. Evolution of the placenta in eutherian mammals. Placenta 28, 259–262 (doi:10.1016/j.placenta.2006.04.010) [DOI] [PubMed] [Google Scholar]
- 64.Cornelis G, et al. 2013. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl Acad. Sci. USA 110, E828–E837 (doi:10.1073/pnas.1215787110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. 2006. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl Acad. Sci. USA 103, 14 390–14 395 (doi:10.1073pnas.0603836103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vernochet C, Heidmann O, Dupressoir A, Cornelis G, Dessen P, Catzeflis F, Heidmann T. 2011. A syncytin-like endogenous retrovirus envelope gene of the guinea pig specifically expressed in the placenta junctional zone and conserved in Caviomorpha. Placenta 32, 885–892 (doi:10.1016/j.placenta.2011.08.006) [DOI] [PubMed] [Google Scholar]
- 67.Blaise S, de Parseval N, Heidmann T. 2005. Functional characterization of two newly identified human endogenous retrovirus coding envelope genes. Retrovirology 2, 19 (doi:10.1186/1742-4690-2-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esnault C, Cornelis G, Heidmann O, Heidmann T. 2013. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV ‘syncytin’, captured for a function in placentation. PLoS Genet. 9, e1003400 (doi:10.1371/journal.pgen.1003400) [DOI] [PMC free article] [PubMed] [Google Scholar]