Abstract
Bracoviruses represent the most complex endogenous viral elements (EVEs) described to date. Nudiviral genes have been hosted within parasitoid wasp genomes since approximately 100 Ma. They play a crucial role in the wasp life cycle as they produce bracovirus particles, which are injected into parasitized lepidopteran hosts during wasp oviposition. Bracovirus particles encapsidate multiple dsDNA circles encoding virulence genes. Their expression in parasitized caterpillars is essential for wasp parasitism success. Here, we report on the genomic organization of the proviral segments (i.e. master sequences used to produce the encapsidated dsDNA circles) present in the Cotesia congregata parasitoid wasp genome. The provirus is composed of a macrolocus, comprising two-thirds of the proviral segments and of seven dispersed loci, each containing one to three segments. Comparative genomic analyses with closely related species gave insights into the evolutionary dynamics of bracovirus genomes. Conserved synteny in the different wasp genomes showed the orthology of the proviral macrolocus across different species. The nudiviral gene odv-e66-like1 is conserved within the macrolocus, suggesting an ancient co-localization of the nudiviral genome and bracovirus proviral segments. By contrast, the evolution of proviral segments within the macrolocus has involved a series of lineage-specific duplications.
Keywords: polydnavirus, bracovirus, parasitoid wasp, obligatory mutualism, comparative genomics
1. Introduction
Bracoviruses (BVs) are symbiotic viruses associated with tens of thousands of braconid wasp species [1]. They have atypical virus life cycles that require two separate host species. The primary hosts are parasitoid wasps, in which the virus particles are produced. The secondary host are the lepidopteran larvae parasitized by the wasp, in which the virus is expressed in infected cells (reviewed in [1]). BV particles are produced from endogenous viral elements (EVEs) integrated in the wasp genomes, and contain multiple dsDNA circular molecules. BVs are produced in specialized cells of the wasp ovaries and constitute the major component of the fluid injected with the eggs into the parasitized caterpillar host during wasp oviposition. The wasps use BVs as gene-transfer agents to express virulence factors that manipulate the immune defences of the lepidopteran host [2]. BVs are essential for the survival and development of the wasp eggs and larvae, which would otherwise be encapsulated in a cellular sheath of haemocytes and killed by the potent immune system of the caterpillar hosts.
Bracovirus-associated wasps form a monophyletic group, which evolved approximately 100 million years ago (Ma) [3]. Their common ancestor integrated in its germline the genome of a virus belonging to nudiviruses: a sister group of the Baculoviridae [4,5]. All BVs associated with contemporary wasps originated from this unique evolutionary event: the capture of a nudivirus genome. Half of the nudiviral genes identified within the genome of the braconid wasp Cotesia congregata are still localized in a 17 kb region referred to as the nudiviral cluster. This EVE corresponds to the major remnant of the nudivirus genome captured by the wasp ancestor [4,6,7], whereas other nudiviral genes have been dispersed in the wasp genome. Nudiviral genes encode the viral RNA polymerase, BV particle structural components and envelope proteins [6,8,9]. However, they are not packaged in BV particles, which instead contain multiple dsDNA circular molecules, called ‘circles’. BV circles are produced from ‘proviral segments’. They encode virulence factors involved in the manipulation of the host [10] and contain conserved regulatory sequences (termed direct repeat junctions, DRJs) involved in their production. As no nudiviral genes are present in the DNA of the particles, BVs cannot replicate in parasitized caterpillars, such as free viruses would do. Consequently, the BV genomes (nudiviral EVEs and proviral segments) are exclusively transmitted vertically as parts of the wasp genome.
Co-options of single EVE genes by cells in order to perform specific physiological functions have been described. For example, different mammalian lineages have independently acquired retroviral genes that are known to be involved in placental development [11]. In the case of BVs, parasitoid wasps have co-opted a nudiviral genome to ensure virus particle production. The BV particles and the DNA they enclose act together to ensure wasp survival in the host. This essential functional role has protected BV genome sequences from the mutation load generally incurred by non-functional EVEs [1,5].
Previous studies using molecular approaches [12,13] and in situ hybridization on wasp chromosomes [14] showed that the proviral segments analysed were clustered together. This led to the hypothesis that all proviral segments might be organized in the wasp genome in tandem arrays constituting a macrolocus [14,15]. However, a more complex picture emerged from the first extensive genomic analysis of proviral segments in two wasp species belonging to the genus Glyptapanteles (G. indiensis and G. flavicoxis). A large majority of proviral segments (75% corresponding to 21 segments) were indeed located in a single region within the wasp genome constituting the so-called macrolocus. However, contrary to the prediction, seven segments were dispersed in five localizations (designated as dispersed loci) [16]. Moreover, although proviral segments and nudiviral genes are believed to originate from the ancestral nudivirus genome, no physical link could be identified between them at the time.
Here, we present an extensive analysis of the C. congregata bracovirus (CcBV) proviral segments found within the wasp genome based on the BAC inserts sequencing approach. Five new proviral segments were identified, which together with de novo annotation of all proviral segments led to an increase in the total number of predicted genes in CcBV particles. We also performed extensive analyses of DRJ regulatory sequences involved in circle production. Moreover, we identified, for the first time, a physical link between a nudiviral gene and the proviral segments. To determine whether BV organization was evolutionarily conserved or whether viral sequences were mobile in the wasp genome, we compared the results obtained on C. congregata with data from the closely related Cotesia sesamiae and from the Glyptapanteles spp. [16]. The comparisons highlighted the striking conservation of bracovirus genomic organization over the approximately 17 Myr period since the separation of both genera. Most proviral segments were localized at homologous positions in all four parasitoid wasp genomes. By contrast, the evolution of proviral loci contents involved numerous rearrangements. In particular, the macrolocus was shaped by successive large lineage-specific duplications, each creating a series of new circles encoding similar genes.
2. Material and methods
(a). Insects and DNA extraction
The gregarious wasp C. congregata (Braconidae) was reared under laboratory conditions on host larvae, Manduca sexta (Sphingidae) maintained on artificial diet at 27°C, under a 16 L : 8 D photoperiod [17]. Virus particles were purified from 200 C. congregata ovaries by SpinX filtration (Costar, France), and the DNA packaged in the particles was extracted as previously described [17]. Genomic DNA used for PCR approaches was extracted from over 80 wasps (50 mg) using the Easy-DNA kit (Invitrogen, France).
(b). Isolation of proviral and flanking sequences within the wasp genome
High molecular weight DNA suitable for BAC library preparation was extracted from C. congregata larvae nuclei in agarose plugs and partially digested with HindIII. DNA fragments of selected size (50 kb) isolated using pulsed field gel electrophoresis were cloned into the pBeloBAC11 vector [18]. Clones (18 432) were selected and spotted onto nylon membranes in duplicate. The filters were then screened by hybridization in high stringency conditions using specific 35-mer oligonucleotide probes (GC% > 50) designed based on each previously sequenced viral circle [19]. Positive clones were further confirmed by PCR using primers located in a different part of the circles in order to provide high screening specificity.
Three successive steps of chromosome walking were performed to extend proviral segment flanking regions. Most of the macrolocus sequence was obtained from overlapping BAC inserts. The gap between the proviral locus 1 and 2 (of the macrolocus) was filled using a PCR approach, and primers designed based on the alignments of conserved wasp genes from Glyptapanteles spp. and C. sesamiae present in this region. Sequencing of overlapping PCR fragments was also used for assembly verifications. Primer sequences are reported in the electronic supplementary material: tables S1 and S2 show how each piece of genomic sequence (BAC and PCR fragments) was obtained and used in the assembly. For amplification of fragments under 3 kb, a 35-cycle PCR was performed (94°C, 60 s; 58 or 60°C, 60 s; 72°C, 120 or 240 s; depending on fragment length) using 50 ng of wasp genomic DNA, 30 pmol of each primer, 2.5 mM MgCl2, 0.2 mM dNTP and one unit of Goldstar Taq polymerase (Eurogentec, France). Larger fragments were obtained using long-range PCR of 35 cycles (20 min extension plus 15 s added at each cycle from the 20th cycle), performed using 50–250 ng of wasp genomic DNA, 20 pmol of each primer, 0.4 mM dNTP and one unit of LA Taq polymerase (Takara, France).
(c). Sequence assembly, proviral segments identification and circle junction PCR
Thirty-four C. congregata BAC inserts and 13 PCR fragment sequences were assembled (see the electronic supplementary material, table S2) and annotated. Proviral segments were identified by comparison with circle sequences [19] and by the MEME/MAST program suite [20], which allowed extensive search of conserved segment extremities (DRJ) that have been shown to terminate bracovirus proviral segments [12]. The genuine presence of newly identified circles in the particles (S16, S24, S27, S28 and S29) was assessed by circle junction PCR tests, as each proviral segment extremity is joined in the circle. These PCRs were performed using 50 ng of DNA extracted from purified virus particles and primers designed in opposite orientation at the extremities of proviral segments, allowing fragment amplification from circles (see the electronic supplementary material, table S3).
The end of the CcBV macrolocus (figure 1) contains unusually short spacers separating segments in the same orientation (see the electronic supplementary material, table S4), and we hypothesized that this could interfere with circle production. The occurrence of larger circles containing the sequence of smaller circles (a feature previously described as ‘nesting’ in symbiotic viruses associated with ichneumonid wasps [21,22]) was assessed by 35-cycle PCR using primers in opposite orientation designed at the extremities of the putative composite proviral segment (3F and 29R for S29/3, 27F and 28R for S28/27, 24F and 32R for S32/24; see the electronic supplementary material, table S3). All PCR fragments (circle junction PCR and nesting) were purified and sequenced to confirm amplification accuracy.
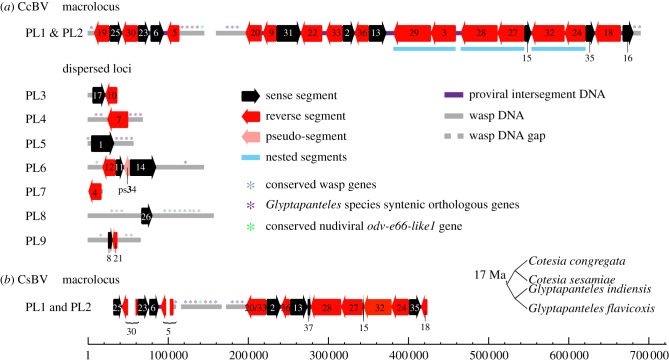
Figure 1.
Structural organization of proviral loci within (a) Cotesia congregata and (b) Cotesia sesamiae. Proviral segments are represented as black or red arrows depending on their orientation. CcBV proviral segments have been given the same number as their corresponding circles packaged in virus particles, whereas CsBV segments were numbered based on their CcBV homologues, except for CsBV S20/33 and S37 specific for CsBV. Only partial sequences of CsBV S25, S5 and S18 could be identified from available data. Loci were named based on those previously characterized in G. indiensis and G. flavicoxis [16] except for PL8 and PL9 (specific for C. congregata). The small tree on the right is a schematic of phylogenetic relationships between wasp species indicating the estimated time since the separation of Cotesia and Glyptapanteles lineages. Note the wasp genes in flanking sequences that are conserved in orthologous positions in Glyptapanteles spp. (purple stars) and the gene of the nudiviral machinery involved in particle production (green star) within the macrolocus. Scale is expressed in basepairs. For detailed analysis of proviral loci flanking region synteny, see figure 2, table 2 and electronic supplementary material, S5.
Cotesia sesamiae sequences were retrieved from NCBI (EF710626–EF710643) and assembled with Geneious Pro assembly software [23]. Newbler Mapper [24] was used to map viral circle sequences onto wasp genomic sequences [25].
(d). Annotation and direct repeat junction regulatory sequences analysis
For both Cotesia spp., gene predictions were performed using a combination of FGENESH and FGENESH+ software from the SoftBerry platform with the Apis mellifera training set (http://linux1.softberry.com/all.htm) and from the EMBL-EBI platform using Wise2 algorithms (http://www.ebi.ac.uk/Tools/Wise2/index.html). Four criteria were used to guide the annotation choice: (i) orthologous gene prediction in previously published BVs or insect sequences, (ii) clustering based on conserved domains, (iii) intron/exon structure prediction in other genes of the family, and (iv) mRNA sequences reported in the literature. Final annotation was conducted using the ARTEMIS software [26]. CcBV-annotated sequences have been deposited at EMBL (accession numbers HF586472–HF586480), and annotation was added to C. sesamiae sequences (EF710626–EF710643 [25]). Sequence coding density was measured as the ratio between the number of bases in coding DNA sequences (CDS) over the total number of bases.
For DRJ analyses, approximately 200 bp surrounding the DRJ highly conserved core were analysed. Sequences upstream of S10 and downstream of S4 (containing S10 5′DRJ and S4 3′DRJ, respectively) were lacking and a former 3′DRJ from S28 (S28*; figure 7) was used. Thus, a total of 34 5′DRJ, 35 3′DRJ and 35 circle junction sequences were aligned using MULTIALIN (http://multalin.toulouse.inra.fr/multalin). Consensus motifs were generated using the MEME program suite [27] and visualized with WebLogo [28]. Proviral segment clustering for the 5′ and 3′DRJ proviral sequences was performed using maximum likelihood on the Phylogeny platform (http://www.phylogeny.fr/version2_cgi/alacarte.cgi) with PhyML v. 3.0, SH-like test and the most adapted substitution model (GTR for the 5′DRJ dataset and HYK85 for the 3′DRJ dataset).
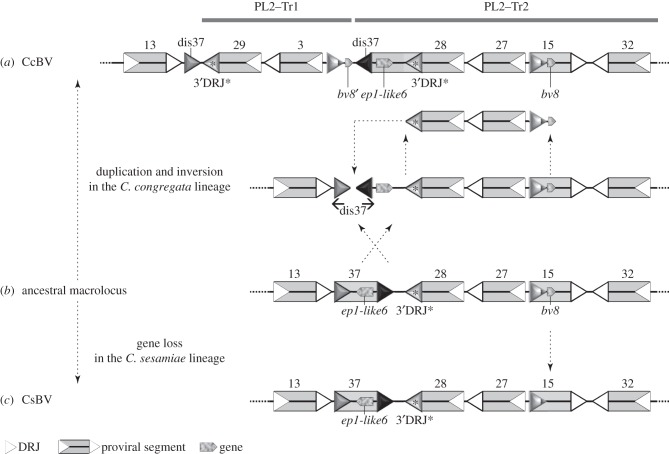
Figure 7.
A proposed parsimonious scenario for the complex rearrangement that may have produced PL2-Tr1 based on the analysis of duplications among Cotesia spp. (a) Cotesia congregata and (c) C. sesamiae macrolocus sequences were used to infer the putative organization of this region in their common ancestor (b). In the lineage leading to C. sesamiae, the bv8 gene was lost (or this gene was acquired specifically in the C. congregata lineage). In the lineage leading to C. congregata, a complex rearrangement occurred resulting in inversion and duplication of proviral segment sequences. Inversion: the segment S37 was inverted and its ep1-like6 gene and regular 3′DRJ (black triangle) were incorporated into an enlarged S28. The regular 3′DRJ became that of S28, replacing the former S28 DRJ readily identified by its particular sequence (3′DRJ*, grey triangle). Duplication: the region encompassing S28, S27 and a part of S15 was duplicated and inserted within S37 that was dismantled (dis37). It should be noted that inversion, duplication and dismantlement might have been produced by a single complex rearrangement caused by errors during replication (fork stalling and template switching model). DRJs are indicated by white triangles to delimit the segments.
(e). Comparative genomic analyses
Glyptapanteles spp. sequences were retrieved from NCBI (accession numbers AC191960 and EF710652–EF710658 for G. indiensis and EF710644–EF710650 for G. flavicoxis) and concatenated to allow macroloci dot plot analyses (543 890 bp for GiBV and 554 319 bp for GfBV). Comparisons of regions within the C. congregata macrolocus and between C. congregata and Glyptapanteles macroloci were performed using MULTIALIN, MAFFT v. 6.8.11 (http://mafft.cbrc.jp/alignment/server/index.html) and DIALIGN-TX (http://dialign-tx.gobics.de/submission?type=dna), and the series of blastn tools available at NCBI. The graphical tool WebACT (http://www.webact.org/WebACT/home) was used to display results.
3. Results
(a). Global proviral segment organization is conserved
Genomic analyses of BV proviral regions from closely related parasitoid wasp species G. indiensis and G. flavicoxis have previously shown that 75% of BV proviral segments were localized within an approximately 550 kb long macrolocus [16,29]. This macrolocus comprised two regions named PL1 and PL2 (for proviral locus 1 and 2) separated by a region containing wasp genes. In addition, several proviral loci, containing one or two proviral segments, were found dispersed in the wasp genome [16]. Recently, circles were reported to integrate in vivo into parasitized host DNA [30], and sequences resembling reintegrated circles were identified within the genome of the wasp C. sesamiae [31]. This raised the question as to whether proviral segments stayed integrated at conserved loci or were mobile within the wasp genome. To understand how BVs evolve within the wasp genome, we characterized the proviral sequences of C. congregata and C. sesamiae, which belong to a genus that separated from Glyptapanteles approximately 17 Ma [3]. We assembled CcBV proviral segments and their flanking regions from C. congregata DNA using genomic BAC libraries, chromosome walking and sequencing of overlapping PCR fragments. Altogether, over 1.2 Mb of C. congregata chromosomal regions, including CcBV proviral segments, was annotated (figure 1a). In parallel, non-annotated sequences available for C. sesamiae were characterized (figure 1b).
CcBV proviral segments were generally clustered and separated by spacers of variable length (114 bp to greater than 10 kb; see electronic supplementary material, table S4). As found in Glyptapanteles [29], a single region corresponding to the macrolocus contained the majority of proviral segments (68%). This macrolocus was larger in C. congregata (approx. 700 kb) but had a similar organization with two parts (PL1 and PL2) linked by a region containing wasp genes. The other proviral segments were dispersed in seven distinct loci (PL3–PL9) each comprising one to three segments (figure 1a). The flanking regions of the proviral loci encoded either wasp genes or remnants of mobile elements. Wasp genes present in the flanking regions were highly conserved between Glyptapanteles and Cotesia spp. (table 1 and electronic supplementary material, S5) indicating that the proviral segments are inserted in homologous genomic regions in these species. We identified that the macrolocus (PL1–PL2) and three isolated loci (PL4, PL5 and PL6) were orthologous within the wasp genomes of G. flavicoxis, G. indiensis, C. sesamiae and C. congregata. Most proviral segments have therefore remained at the same localization in braconid wasp genomes since the separation of the Cotesia and Glyptapanteles lineages approximately 17 Ma. By contrast, some proviral loci appeared to be lineage-specific, such as PL8 and PL9 within C. congregata (for PL3 and PL7, we obtained limited and inconclusive data on flanking regions).
Table 1.
Cotesia congregata conserved wasp genes in proviral flanking regions (for more details, see electronic supplementary material, table S5). No C. congregata wasp gene has been identified in PL3 and PL7 flanking regions. Gene locus tags are displayed for G. indiensis or G. flavicoxis when not available. Cs, C. sesamiae; G. spp., Glyptapanteles spp. +, Present; n.a. not available.
| region |
locus tag | gene name | Cs | G. spp. | |
|---|---|---|---|---|---|
| PL1 | 5′ | 003 | nt5-like1 | n.a. | GIP_L1_050 |
| 3′ | 036 | CcPL1.036 | + | GIP_L1_060 | |
| 037 | nmt | + | GIP_L7_700 | ||
| 038 | hyal | + | GIP_L7_710 | ||
| 039 | hyal-like | + | GIP_L8_010 | ||
| 040 | odv-e66-like1a | + | GIP_L8_020 | ||
| PL2 | 5′ | 001 | nt5-like2 | + | GIP_L8_080 |
| 002 | nt5-like3 | + | GIP_L8_090 | ||
| 003 | nt5-like4 | + | GIP_L8_100 | ||
| 3′ | 179 | CcPL2.179 | n.a. | GIP_L6_040 | |
| 180 | CcPL2.180 | n.a. | GIP_L6_030 | ||
| PL4 | 5′ | 001 | CcPL4.001 | + | GFP_L4_260 |
| 002 | CcPL4.002 | + | GIP_L4_010 | ||
| 3′ | 008 | chits | + | GIP_L4_170 | |
| 009 | slit1 | + | GIP_L4_180 | ||
| 010 | iqca | + | GIP_L4_190 | ||
| PL5 | 5′ | 001 | mtsa | n.a. | GIP_L5_030 |
| 3′ | 013 | kif3 | n.a. | GIP_L5_140 | |
| 014 | prpc | n.a. | GIP_L5_150 | ||
| 015 | pka-C1 | n.a. | GIP_L5_160 | ||
| 016 | ros | n.a. | GIP_L5_170 | ||
| PL6 | 3′ | 028 | ari | n.a. | GIP_L2_110 |
aConserved nudiviral gene.
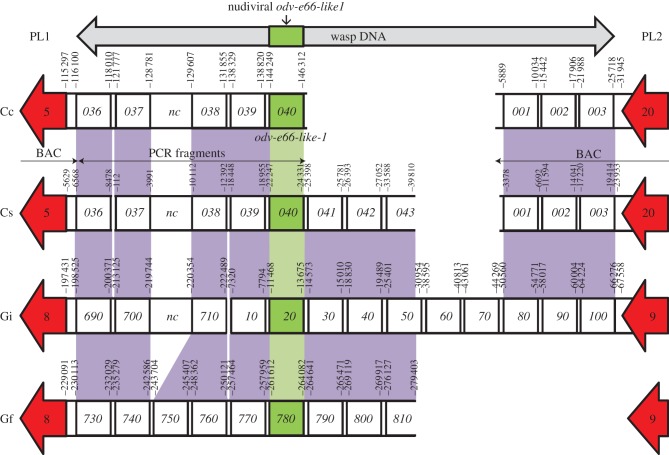
To date, no nudiviral genes involved in the production of particle components have been found in the genomic regions containing the proviral segments [4]. Analyses of these regions in Glyptapanteles and Cotesia spp. revealed that the odv-e66-like1 nudiviral gene encoding a bracovirus particle component [8] was localized between PL1 and PL2 within the macrolocus (figures 1 and 2). For C. congregata, this region was obtained by sequencing overlapping fragments isolated by PCR with primers designed based on conserved Glyptapanteles and C. sesamiae genes (figure 2 and table 1; electronic supplementary material, S5). We thus showed the macrolocus is an EVE composed of both nudiviral and proviral segment sequences, which were already present at this chromosomal location before the separation of the Glyptapanteles and Cotesia lineages.
Figure 2.
Synteny in wasp genes-containing region joining PL1 to PL2 (macrolocus). This region includes the conserved nudiviral odv-e66-like1 gene. Genes are indicated by squares and numbers are those given in GenBank. Their positions on the DNA sequences (following the numbering in GenBank) are indicated above the squares. Gene synteny is highlighted in purple and the nudiviral gene is coloured green. Interruptions in the black lines indicate gaps in the sequence (non-overlapping BACs). White areas correspond to non-homologous sequence or to a lack of data for one species. Proviral segments flanking this region corresponding to the extremities of PL1 and PL2 are shown in red, with arrows indicating their orientation. CcBV sequences were obtained either from overlapping BAC sequencing or PCR fragments as indicated below. Cc, C. congregata; Cs, C. sesamiae; Gi, G. indiensis and Gf, G. flavicoxis; nc, non-coding sequences. CsPL1 and region containing wasp genes (accession number EF710629); CsPL2 (EF710635); GiPL1 (AC191960); GiPL2 (EF710657); GfPL1 (EF710644) and GfPL2 (EF710648).
(b). New Cotesia congregata bracovirus segments were identified within Cotesia congregata bracovirus PL2
In addition to the 30 circles previously reported [19], we were able to predict five new CcBV circles from PL2 proviral segments (S16, S24, S27, S28 and S29) that correspond to duplicated copies of previously reported circles. Specific PCR assays (circle junction PCR) confirmed their presence in BV particles. Thus, the CcBV packaged genome is an assortment of 35 different circles. Unexpectedly, we also detected larger molecules made of two smaller segments from PL2 (S29 + S3, S28 + S27 and S32 + S24; figure 1a). The ‘nesting’ of small circles within large circles shown in ichnoviruses [21,22] therefore also exists in BVs.
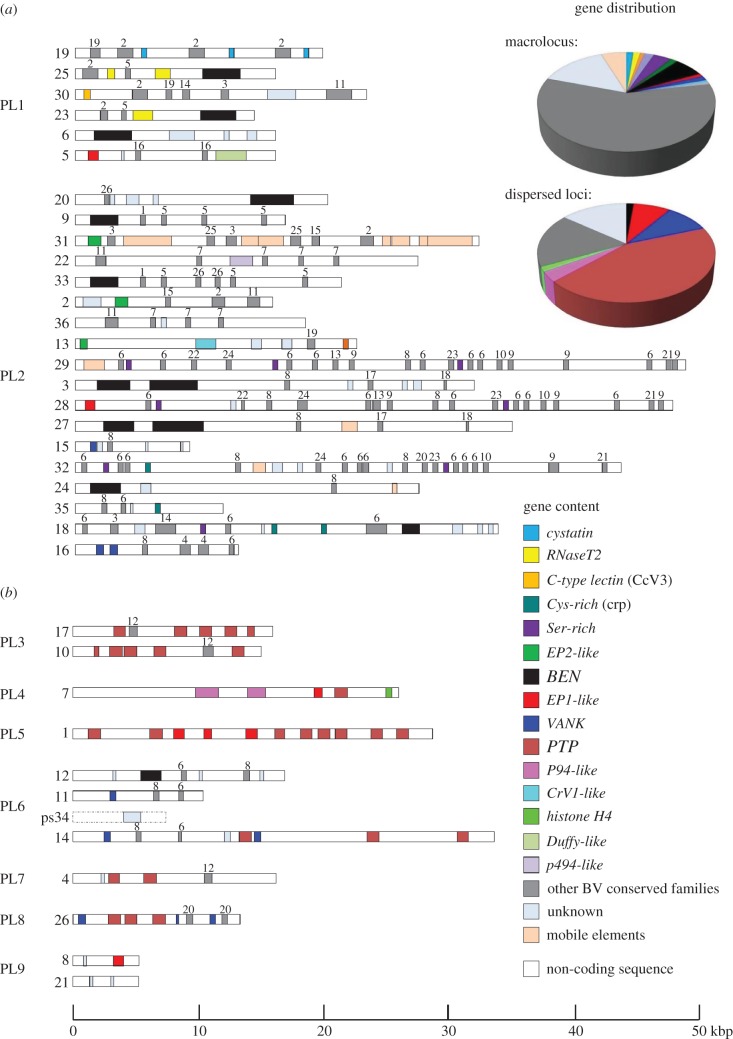
In silico de novo annotation predicted more packaged genes than previously reported [19], with now 222 CDS, 29 putative pseudo-genes and 11 remnants from mobile elements identified within CcBV proviral segments (figure 3). Bracovirus genomes feature numerous gene families: 183 CcBV genes and 26 predicted pseudo-genes belong to 37 families (table 2 and figure 3). Seven of these gene families encoded proteins containing eukaryotic-conserved domains (PTP, VANK, cystatin, RNaseT2, BEN, Cys-rich, C-type lectin), one family codes for a P94-like baculovirus protein and 29 families are specific to BVs (EP1-like, EP2-like, Ser-rich and BV families 1–26). In contrast to nudiviral genes that do not contain introns, 60% of genes present in proviral segments were predicted to contain introns, like cellular genes (see also [19,29]).
Figure 3.
Gene content of CcBV proviral segments within (a) macrolocus and (b) dispersed loci displayed by coloured boxes (macrolocus, 260 genes; dispersed loci, 62 genes). CcBV contains 37 gene families: seven encode proteins with described conserved domains (cystatin, RNaseT2, C-type lectin, Cys-rich, BEN, VANK and ptp) representing approximately 23.5% of the genes and one encoded a baculovirus homologue protein (p94-like). Twenty-nine gene families representing 57% of the genes encode proteins of unknown function conserved in BVs associated with wasps of the Cotesia and Glyptapanteles genera (Ser-rich, EP2-like, EP1-like and BV1–BV26 represented by grey boxes, with the number identifying the family indicated above the boxes). Some other previously identified BV gene families are only represented by one member in CcBV (CrV1, histone H4, Duffy and p494). Other genes of unknown function are unique (approx. 14.5%) and some coding DNA sequences are identified as remnants of genes from mobile elements (approx. 4.25%). Note that the ptp gene family constitutes the major part of the genes from isolated loci (see gene distribution pie charts on the right). Unlike in GiBV, no ptp genes are found in the CcBV macrolocus. Other genes such as cystatin, RNaseT2, C-type lectin or cys-rich are found only in the macrolocus, which contains a majority of BV specific genes (table 2). ps34 shown by a dashed line corresponds to a former proviral segment mutated in the 3′DRJ core homologous to CvBV S30 (accession number HQ009553) but no longer producing a circle.
Table 2.
Cotesia congregata bracovirus gene families segment localization. The BV-specific families BV1–B26 are not reported here. Newly identified segments are italicized. RNaseT2, ribonuclease T2-type; C-type lec, protein with C-type lectin domain (CcV3); Cys-rich, cysteine-rich protein (CRP); EP2-like, homologues of early-expressed protein 2; Ser-rich, serine-rich protein; BEN, proteins with BEN domain; EP1-like, homologues of early-expressed protein 1; VANK, viral ankyrin; PTP, protein tyrosine phosphatase; P94-like, related to P94 baculovirus protein; macr., macrolocus; disp., dispersed loci.
| family coding | Cystatin | RNaseT2 | C-type lec. | Cys-rich | EP2-like | Ser-rich | BEN | EP1-like | VANK | PTP | P94-like |
|---|---|---|---|---|---|---|---|---|---|---|---|
| no. of genes | 3 | 3 | 2 | 4 | 3a | 8 | 14 | 7 | 9 | 27 | 2 |
| segments | 19 | 25, 23 | 30, 13 | 32, 35, 18 | 31, 2, 13 | 29, 28, 24, | 25, 23, 6, 20, | 5, 28, 7, | 15, 16, 11, | 17, 10, 7, | 7 |
| 18 | 9, 33, 3, 27, | 3, 8 | 14, 26 | 1, 14, 4, 26 | |||||||
| 24, 18, 12 | |||||||||||
| proviral locus | 1 | 1 | 1, 2 | 2 | 2 | 2 | 1, 2, 6 | 1, 2, 4, 5, 9 | 2, 6, 8 | 3, 4, 5, 6, 7, 8 | 4 |
| type | macr. | macr. | macr. | macr. | macr. | macr. | macr., disp. | macr., disp. | macr., disp. | disp. | disp. |
aThree other ep2-like genes are present in a duplicated region not producing a circle.
(c). Proviral segment extremities are conserved
All proviral segments from all BVs analysed to date are terminated by direct repeats at both extremities, termed DRJs [12,13,16,29,30,32]. Bracovirus circles contain a unique sequence (circle junction) produced from a recombination event between these DRJs [12,33]. Site-specific tyrosine recombinases identified in the nudiviral machinery (VLF-1a or VLF-1b) were proposed to perform this recombination [7] based on functional homology with the homologous baculovirus protein VLF-1 [34]. VLF-1 has been demonstrated to be a nucleocapsid component and we therefore hypothesize that a VLF-1 complex could bind DRJs terminating a segment and resolve the circles, following encapsidation of BV DNA [7].
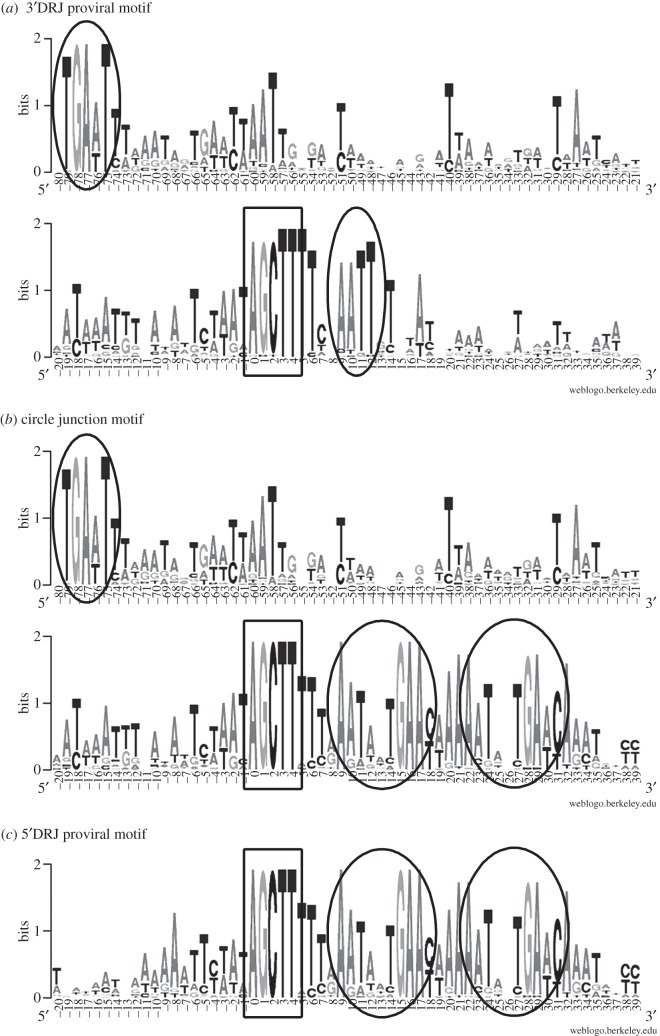
We performed comprehensive sequence analyses of CcBV segment extremities and of their corresponding circle junctions (figure 4). The alignments led to the identification of a perfectly conserved 5 bp direct sequence motif (AGCTT), which constitutes the DRJ core also found in the other wasp species [35]. Less-conserved sequences extend from this core to form a total DRJ of approximately 120 bp (see electronic supplementary material, figure S1). Different conserved motifs were found in the 5′ and 3′DRJ (except for the core), which were subsequently analysed separately (figure 4).
Figure 4.
DRJ sequence motifs within C. congregata proviral segments and CcBV circles visualized using WebLogo. Each logo consists of stacks of bases, with one stack for each position in the sequence. The height of the stack at a position indicates the sequence conservation, whereas the height of a base indicates the relative frequency of this base at this position. Note that the circle junction sequence (b) corresponds to a recombined form of the two DRJs within the perfectly conserved DRJ core shown in the black box. Sequences characterizing (a) 3′DRJ and (c) 5′DRJs are circled in black. A 30 bp sequence containing a 13 bp repeat (TTtnAatantGAAyaaAAatnntGAwcAaa) following the 5′DRJ core was found to be conserved, whereas the sequence following the core in 3′DRJ was smaller (TTcnAATTgt). A highly conserved motif (TGAa/tT) was also identified 80 bp upstream of the 3′DRJ core. These graphical representations were generated from independent alignments of 34 5′DRJ, 35 3′DRJ and 35 circle junction sequences (see electronic supplementary material, figure S1).
We newly identified a highly conserved motif (TGAa/tT) 80 bp upstream of the 3′DRJ core (figure 4a and electronic supplementary material, figure S1a). We also found a conserved 30 bp sequence containing a repeat downstream of the 5′DRJ core (figure 4c and electronic supplementary material, figure S1c). Alignment of circle junctions displayed both the 3′DRJ highly conserved upstream motif and the 5′DRJ repeat-containing sequences following the core (figure 4b and electronic supplementary material, S1b). The most conserved sequences are thus present in the circles and might interact with BV particle components. Comparison between proviral segments and circle sequences indicated recombination occurs within the DRJ core (AGCTT) for all circles (see electronic supplementary material, figure S1b).
As both DRJs of each segment were located either on the plus or minus strand, we were able to determine the orientation (5′–3′ or 3′–5′) of the proviral segments on the wasp chromosome (arrows in figure 1 indicate this orientation). The segments were always separated by spacer sequences more than 114 bp long. Strikingly, segments separated by small spacers (less than 500 bp) were in opposite orientation (13 of 16) except for those involved in nesting (see electronic supplementary material, table S4). As each nucleocapsid contains only one circle [36,37], this might reflect the physical constraints imposed during encapsidation for incorporating the DNA of adjacent segments into different nucleocapsids.
(d). The Cotesia congregata bracovirus macrolocus contains a series of duplications
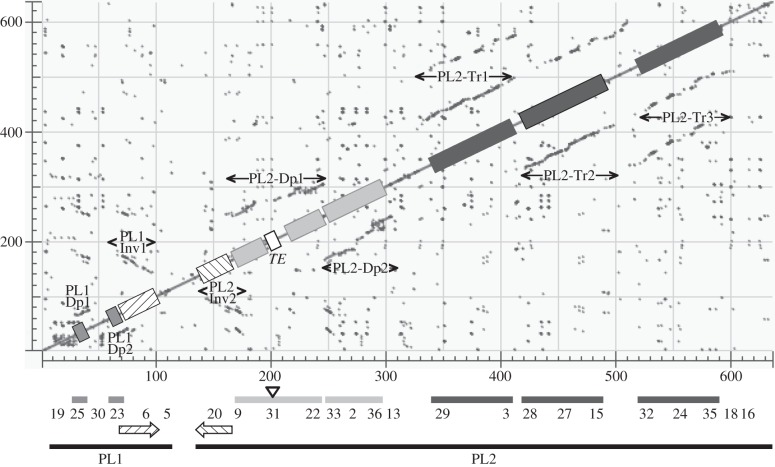
Sequencing of the packaged genome [19] revealed that some CcBV circles were strikingly similar to each other. For example, circles 31 and 2 were 96% identical and only differed by the insertion of retroelements [38] and of a large Maverick DNA transposon [39]. Two hypotheses had been invoked to explain this similarity. First, both circles could correspond to a single polymorphic proviral segment in the C. congregata laboratory strain. Or second, both circles could correspond to two proviral segments (S31 and S2) formed by duplication and fixed in the wasp population. Here, we found the second hypothesis to be true. We further identified a series of duplicated regions of different sizes within the macrolocus by BLASTN analysis of the CcBV macrolocus against itself with dot matrix (figure 5 and electronic supplementary material, table S6). These duplicated regions represent almost three-quarters of the macrolocus.
Figure 5.
Similarity matrix of the C. congregata proviral macrolocus compared with itself. The main diagonal represents sequence alignment with itself; dotted lines (grey) identify duplicated regions within the sequence analysed. Those parallel to the diagonal correspond to duplicated regions in the same orientation; those antiparallel correspond to inverted sequences (striped arrows). Scale is expressed in kilobase pair. Relative positions of proviral loci 1 and 2 and of each CcBV proviral segment forming the macrolocus are indicated below the dot plot matrix. PL1, proviral locus 1; PL2, proviral locus 2; TE, transposable element. Grey boxes: duplication PL1–Dp1/PL1–Dp2 within PL1; striped boxes: inverted duplication PL1–Inv1/PL2–Inv2; light grey boxes: duplication PL2–Dp1/PL2–Dp2; dark grey boxes: triplication PL2–Tr1/PL2–Tr2/PL2–Tr3. Nucleotide positions of duplication extremities are indicated in the electronic supplementary material, table S6.
The first PL1 proviral segment (S19) contained a repetition of three highly similar sequences containing cystatin genes [40]. Downstream, S25 and S23 also constituted duplicated sequences (PL1–Dp1 and PL1–Dp2). Moreover, the second half of PL1 (containing S23 and S6) and the first part of PL2 (S20 and S9) were inverted duplicated regions (PL1–Inv1 and PL2–Inv2). At the beginning of the second proviral locus, PL2–Dp1 (S9, S31, S22 and an incomplete copy of S13) and PL2–Dp2 (S33, S2, S36 and S13) were found to form another large duplicated region. Finally, the second part of PL2 harboured a large triplicated region: PL2–Tr1 (S29, S3), PL2–Tr2 (S28, S27 and S15) and PL2–Tr3 (S32, S24, S35 and S18).
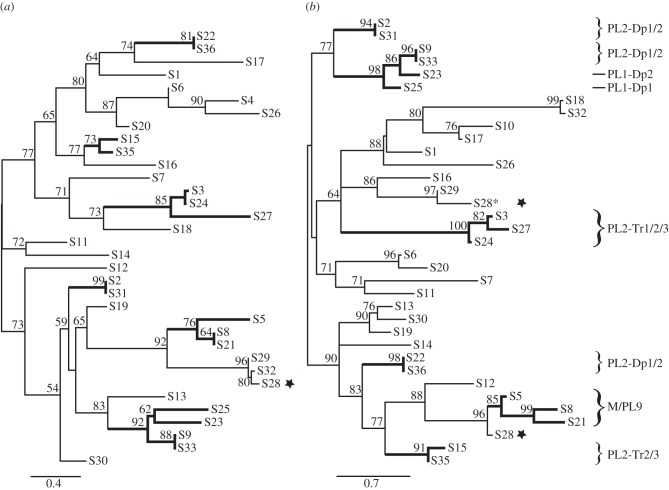
The history of viral segment production was also inferred through 5′DRJ and 3′DRJ maximum-likelihood phylogenies (figure 6). For 16 segments present in the duplicated regions, both DRJs evolved in co-phylogeny (figure 6), but for other segments, 5′ and 3′DRJs had different histories (figure 6), which indicated the formation of mosaic segments (as described in figure 7).
Figure 6.
Proviral segment clustering based on (a) 5′ and (b) 3′DRJ sequences. The trees were obtained from maximum-likelihood phylogenetic inferences based on the alignments of approximately 200 bp sequences of 5′ and 3′DRJs including the DRJ core (‘extended DRJs’). Only SH-like branch values above 50 are indicated. Thick branches highlight 5′DRJs in co-phylogeny with the 3′DRJ of the same segment, produced by complete duplication of proviral segments including their DRJs (duplicated regions containing the DRJs are indicated on the right). The stars indicated the three S28 DRJs. In this segment, the ancestral 3′DRJ S28* (still functional in C. sesamiae) was replaced by a new 3′DRJ recruited during the rearrangement that produced PL2-Tr1. This resulted in a mosaic S28 segment (figure 7).
(e). Macrolocus evolution involved duplications
As BV sequences share a common origin, duplication histories can be tentatively reconstituted by comparing duplicated regions in related species using parsimonious interpretations. To identify the most recent rearrangements, we have analysed the C. sesamiae proviral segments available in GenBank. BAC inserts corresponding to PL1 and PL2 were identified corresponding to a large part of the CsBV macrolocus (figure 1b). Proviral segment boundaries were identified (DRJs), and CsBV segment gene contents were annotated [25]. Orthology between both Cotesia spp. was readily identified, because gene order was mostly conserved. CsBV segments were therefore annotated and numbered based on the CcBV orthologues (figure 1a) except for CsBV S20/33, corresponding to a fusion of CcBV S20 and S33 specific for C. sesamiae (in C. vestalis BV, the segments are separated [41]), and for CsBV S37, which is dismantled in CcBV (figure 7).
Comparison with Glyptapanteles spp. gave insights into evolutionary events dating back to 17 Ma. The macrolocus structure (PL1 region containing wasp genes PL2) was also conserved between Glyptapanteles and Cotesia spp., but the number of segments and their gene content were different. However, in many cases, it was still possible to trace evolutionary relationships between segments of the different genera based on conserved homologous DRJ sequences and of nucleotide similarities remaining between homologous segments. The comparison of CcBV and GiBV macroloci is shown in table 3. Strikingly, homologous segments were in the same orientation even if gene content was not conserved.
Table 3.
Correspondence between CcBV and GiBV segments within the macrolocus. Correspondences were established according to DRJ sequences and gene content by analyses of homologous sequences detected by TBLASTX. Segment orientation is displayed (>sense> and <reverse<). Similarity is given by the percentage of homologous sequences common between the corresponding CcBV and GiBV segments: 10% < * < 35%, 35% < ** < 45%, *** >45%; inv, inversed sequence; °<, conserved DRJ in redesigned segment; —, redesigned segment; n.a. absent; dis37, dismantled segment 37. $, in that last case the percentage of common homologous sequences is low (13%) between CcBV and GiBV, but high (46%) between the intact S37 present in CsBV and the 9p from GiBV.
| macrolocus | homologous segments |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CcBV | PL1 | °<19<° | >25> | <30< | >23> | >6> | <5< | |||||||
| GiBV | PL1 | °<1p< | >2p> | <3p<° | >4p> | <5p< | >6p> | >7p> | <8p< | |||||
| similarity range | * | * | *** | * | *** | *** | *** | |||||||
| CcBV | PL2 | — | <20< | <33/(9)< | — | — | >2/(31)> | <36/(22)< | >13> | |||||
| GiBV | PL1inv | >8p> | <7p< | <6p< | >5p> | <4p< | >3p> | <2p< | >1p> | |||||
| similarity range | ** | *** | * | * | ** | |||||||||
| CcBV | PL2 | >dis37> | <28< | <27< | >15> | <32< | <24< | >35> | °<18 | n.a. | n.a. | <° | n.a. | >16> |
| GiBV | PL2 | >9p> | <10p< | <11p< | >12p> | <13p< | <14p< | >15p> | °<16p< | <17p< | >18p> | <19p<° | >20p> | >21p> |
| similarity range | *$ | ** | *** | * | ** | *** | ** | ** | * | *** | ||||
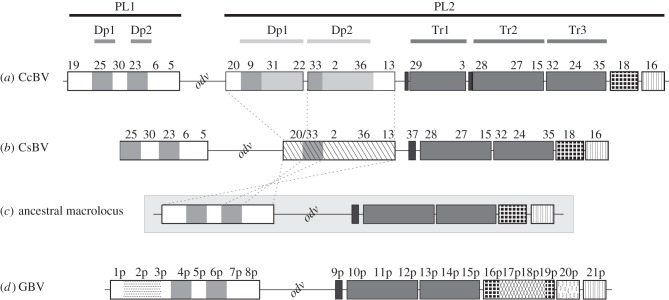
The evolutionary dynamics of macrolocus content has involved duplications that could be traced back to (i) before the separation of the Cotesia and Glyptapanteles lineages (17 Ma), (ii) before the separation of C. congregata and C. sesamiae, or (iii) after this separation (figure 8). Before the separation of Cotesia and Glyptapanteles lineages, the proviral form was already organized in a macrolocus composed of two proviral regions separated by several wasp genes (figure 8c). Two duplications present in the ancestor of the four BVs remain detectable: PL1–Dp1, PL1–Dp2, PL2–Tr2 and PL2–Tr3. Specific events occurred in the Glyptapanteles lineage: (i) the formation of 1p–2p–3p, (ii) that of 17p and 18p, associated with the capture of sugar transporter genes from the wasp genome [16] and (iii) the formation of 20p, by re-integration of a dispersed proviral segment in the macrolocus [31] (figure 8d). Between 17 Ma and the separation of Cotesia spp., an inverted duplication of PL1 sequences (PL1–Inv1) occurred in the PL2 anterior part (PL2–Inv2) of the Cotesia lineage (figure 8b). Finally, in the lineage leading to C. congregata, two main events occurred. A complex duplication in the posterior part of PL2 produced PL2–Tr1 (figure 7). Another duplication also occurred upstream in PL2, leading to the formation of PL2–Dp1 and PL2–Dp2 (figure 8a). The later duplication occurred relatively recently as judged from the very high similarity between the duplicated regions.
Figure 8.
A proposed parsimonious scenario for macrolocus proviral genome evolution in Cotesia and Glyptapanteles lineages based on the analysis of duplications among wasp species. Sequences from C. congregata (a), C. sesamiae (b), G. indiensis and G. flavicoxis (d) were used to infer the putative organization of an ancestral macrolocus (c) containing two proviral regions that would have existed before the separation of the Cotesia and Glyptapanteles lineages over 17 Ma. In the lineage leading to Glyptapanteles spp., new segments 1p–2p–3p, 17p, 18p and 20p (re-integrated) were formed. In the lineage leading to Cotesia spp., inverted duplications of PL1 sequences (hashed boxes) resulted in modifications of the anterior PL2 region. Subsequent rearrangements in the lineage leading to C. sesamiae lead to the fusion of segments S20 and S33 (S20/33) and the loss of segment S15 gene content. In the lineage leading to C. congregata, duplication in the posterior region of PL2 produced PL2–Tr1 and a larger S28. In addition, duplications upstream of PL2 lead to the formation of PL2–Dp1 and PL2–Dp2. Names and box colours for duplications and inversion are the same as in figure 5.
(f). Cotesia congregata bracovirus has two specific dispersed loci
In C. congregata, the seven dispersed proviral loci each contained only one to three segments. However, duplications, such as mirror duplications (S17/S10 on PL3 and S8/S21 on PL9), have also occurred in these regions. Most dispersed segments encode ptp genes, which are involved in complex functional interactions with the caterpillar hosts [31]. It is noteworthy that S1 (PL5), S7 (PL4), S17–S10 (PL3) and S26 (PL8) in addition to ptp genes have a common 3′DRJ with a one base deletion before the DRJ core (see electronic supplementary material, figure S1a). This mutation is also found in GiBV segments 20p (PL2) and 25p (PL5), and in Microplitis demolitor BV (MdBV) segments H, J and M, and could reflect their common origin.
The DRJs of the PL9 segments (S8 and S21) are closely related to those of PL1 S5 (figures 1a and 6; electronic supplementary material, figure S1). PL9 segments are separated from the macrolocus but are known to localize on the short arm of chromosome 5, like the macrolocus [14]. Therefore, PL9 segments could originate from a duplication of S5 followed by a translocation. The unique segment within PL8 (S26) appears to correspond to the integration of a sequence originally present in locus PL4 at a new localization in the wasp genome (see electronic supplementary material, figure S2). No PL8 or PL9 homologues have been identified in Glyptapanteles spp. Therefore, PL8 and PL9 are new proviral loci and could constitute an exception to proviral sequences stability in the wasp genome (unless they have yet to be isolated in Glyptapanteles spp.).
4. Discussion
In this study, we report the characterization of bracovirus proviral segments in the genomes of the wasps C. congregata and C. sesamiae. Comparative genomics with Glyptapanteles proviruses gave further insights into the evolutionary history of BVs. The presence of common hymenopteran genes in flanking regions of most proviral sites indicated that the localizations of bracovirus segments in the wasp genomes have remained the same since the separation of the Cotesia and Glyptapanteles lineages approximately 17 Ma. The proviral sequences were organized in a macrolocus comprising over two-thirds of the proviral sequences completed by seven smaller dispersed loci, each with one to three segments. The macrolocus comprised two proviral loci (PL1 and PL2) joined by a region containing wasp genes.
In C. congregata, the dispersed PL4 and PL9 loci and the macrolocus had previously been visualized on the short arm of chromosome 5 by in situ hybridization [14]. Homologues of wasp genes either flanking or within the macrolocus and PL4 (see electronic supplementary material, table S5) belong to the same linkage group in the genome of A. mellifera [42]. This might suggest PL4 and PL9 were originally a part of the macrolocus, from which they were later separated by chromosomal rearrangements. By contrast, homologues of PL8 wasp genes were located in a different linkage group in A. mellifera and Nasonia vitripennis genomes, implying that this proviral locus might be located on a different chromosome.
The identification of homologous sequences to all proviral loci of Glyptapanteles spp. suggests data concerning the CcBV proviral segment is relatively complete. However, because the BAC clones were identified by hybridization to sequences from previously known segments, we cannot exclude that other proviral segments in dispersed loci have remained unidentified. Most of the packaged circles of C. vestalis bracovirus (CvBV), recently sequenced using a high-throughput approach [41], have CcBV homologues (including the newly identified CcBV circles) except CvBV C19, C31, C32 and C35 that could be specific for CvBV or still missing from our analysis. However, CvBV C35 lacks the DRJ that are characteristic of BV circles. Furthermore, CvBV C35 encodes a helicase and shows high nucleotide similarity with different arthropod genomes, suggesting it could correspond to a mobile element.
We found one nudiviral gene within CcBV proviral locus: the odv-e66-like1 gene, which had been shown to encode a particle component of Chelonus inanitus BV (CiBV) [8]. We showed that this gene is present within the macrolocus of the four species examined (figure 2). We suppose this reflects the ancient presence of the nudiviral genome and proviral macrolocus at the same locus. Forthcoming C. congregata whole genome sequencing should reveal whether the nudiviral cluster is localized on chromosome 5, like the macrolocus.
No nudiviral genes are present in the packaged genome; however, the DRJs terminating the BV proviral segments are probably a component of the nudiviral machinery. Indeed, based on what is known for baculovirus replication, BV DRJs involved in the dsDNA circle production could constitute the binding sites of a nudiviral site-specific recombinase that would ensure the excision of the circles from larger amplified molecules during DNA encapsidation [7]. In baculoviruses, the VLF-1 protein is a tyrosine recombinase localized at one extremity of the nucleocapsid. VLF-1 is involved in the resolution of the baculovirus DNA molecules, which are amplified as genome concatemers during replication [34] and separated as individual genome monomers during encapsidation. Two nudiviral genes related to vlf-1 homologues were expressed in braconid wasp ovaries and could encode the DRJ binding recombinase [7,9].
The DRJ sequences were conserved in all BVs studied (CcBV, CsBV, GiBV, GfBV and MdBV). In CiBV, some variations occurred on the first and last bases of the core DRJ (consensus (A/c)GCT(T/c)), the 13 bp repeat after the 5′DRJ core was absent, and the sequence downstream of the 3′DRJ core showed variation with the CcBV consensus (CiBV consensus: TCnAATt). This could reflect the greater phylogenetic distance of Chelonus inanitus (belonging to Cheloninae subfamily) compared with the relatively closely related Cotesia, Glyptapanteles and Microplitis spp. (all classified in the Microgastrinae subfamily). Still, the high conservation of DRJ motifs underlines their deep phylogenetic relationships going back to the common origin of BVs. In the ancestral nudivirus, a DRJ-related sequence might have been used to separate genome concatemers produced during viral DNA replication such as in baculoviruses. Strong selective constraints linked to the interaction with the nudiviral machinery may have led to high DRJ sequence conservation in comparison with other proviral sequences.
Proviral segments were oriented on the wasp chromosome as both DRJs of a segment are always located either on the positive or negative strand. Proviral segments were separated by spacer sequences more than 114 bp long that are not packaged in the particles [43], and surprisingly segments separated by short spacers (less than 500 bp) were often in opposite directions. This particular organization could derive from physical constraints as adjacent segments are amplified together in the same molecule [43] before their separation and circularization by recombination of the DRJs, which is likely coupled with viral DNA entry into the nucleocapsids. When spacers are short, opposite orientation of segments may permit capsid access to the amplified DNA molecule from opposite sides, whereas segments in the same orientation will induce capsid competition for space. According to this hypothesis, adjacent segments in the same orientation and separated by small spacers could fail to be resolved (figure 1a, blue lines). We were indeed able to observe this phenomenon resulting in large circles containing the sequences of two segments in the particles, a situation similar to circle ‘nesting’ observed in ichnoviruses [44]. Given that the length of BV nucleocapsids is correlated with the size of the dsDNA molecule they contain [36,37], a large range of DNA circle sizes can be encapsidated thus allowing nesting.
As proviral loci of different species are orthologous, it is possible to compare their content and to reconstruct their history. Stability of proviral segment localizations within wasp genomes contrasted sharply with the evolutionary dynamics of their content that has involved duplications. The macrolocus evolved mainly by duplications within segments (S19) or comprising one (PL1-Dp1/PL1-Dp2) to a series of segments (PL2 triplication). Duplication boundaries do not correspond to those of the segments (see electronic supplementary material, table S6). Thus, in most cases, duplications do not appear to have been produced by a viral mechanism (circle re-integration for example). Gene amplifications have been reported to be involved in mosquito resistance to insecticides, but the duplications have not been sequenced, and the mechanism of their production is unknown [45]. In human genetic diseases, complex rearrangements involving duplications have been sequenced and attributed to DNA replication errors [46]. According to this model, unusual genome architecture, such as the presence of repetitive sequences, may confuse the DNA replication machinery. This results in replication fork stalling, causing the DNA polymerase to switch from one template to another. Template switching can occur several times forwards or backwards on the molecule used as master sequence before replication resumes on the original DNA template. This could potentially simultaneously cause both inversions and duplications, such as those described for PL2–Tr1 (figure 7). DRJs, being repeated sequences, could be involved in such a mechanism.
Beyond the hypothetical mechanism leading to their production, the reason duplications are evolutionarily conserved is probably because of the antagonistic coevolution between caterpillar hosts and parasitoid wasps, which resulted in complex evolutionary arms races [1,25,47]. The packaged genes expressed in infected caterpillar tissues produce virulence factors involved in manipulating host physiology and altering host immunity or development [48,49]. The selection of particular beneficial alleles and accelerated mutation accumulation in duplicated genes should provide new weapons against host targets [25], and we found the triplicated cystatin genes (S19) to undergo strong positive selection [50]. In this context, the localization of most proviral sequences within a macrolocus might have been maintained because it readily allowed the production of new circles. Indeed, the large duplications resulted in new proviral sequences with two DRJs that are very likely to produce new circles, whereas re-integrated circles were found to contain a single DRJ and to be dysfunctional [31].
Acknowledgements
We thank Cindy Ménoret for taking care of the insects and Germain Chevignon for helpful discussion. The Cotesia congregata proviral locus study was performed as a part of the projects Evparasitoid and Paratoxose supported by the agencies ANR and CNRS (GDR 2153, GDR 2157 and IFR 136) with the collaboration of the Genoscope (Evry, France) and the European Research Council starting grant GENOVIR (205206). This work has been carried out with the technical support of the Genomics Department (PPF ASB) - University F. Rabelais (Tours).
References
- 1.Herniou EA, Huguet E, Thézé J, Bézier A, Periquet G, Drezen J-M. 2013. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Phil. Trans. R. Soc. B 368, 20130051 (doi:10.1098/rstb.2013.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupuy C, Huguet E, Drezen J-M. 2006. Unfolding the evolutionary story of polydnaviruses. Virus Res. 117, 81–89 (doi:10.1016/j.virusres.2006.01.001) [DOI] [PubMed] [Google Scholar]
- 3.Murphy N, Banks JC, Whitfield JB, Austin AD. 2008. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol. Phylogenet. Evol. 47, 378–395 (doi:10.1016/j.ympev.2008.01.022) [DOI] [PubMed] [Google Scholar]
- 4.Bézier A, et al. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323, 926–930 (doi:10.1126/science.1166788) [DOI] [PubMed] [Google Scholar]
- 5.Théze J, Bézier A, Periquet G, Drezen J-M, Herniou EA. 2011. Paleozoic origin of insect large dsDNA viruses. Proc. Natl Acad. Sci. USA 108, 15 931–15 935 (doi:10.1073/pnas.1105580108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bézier A, Herbinière J, Lanzrein B, Drezen J-M. 2009. Polydnavirus hidden face: the genes producing virus particles of parasitic wasps. J. Invertebr. Pathol. 101, 194–203 (doi:10.1016/j.jip.2009.04.006.) [DOI] [PubMed] [Google Scholar]
- 7.Drezen J-M, Herniou EA, Bézier A. 2012. Evolutionary progenitors of bracoviruses. In Parasitoid viruses symbionts and pathogens (eds Beckage NE, Drezen J-M.), pp. 15–31 San Diego, CA: CRC Press-Elsevier [Google Scholar]
- 8.Wetterwald C, et al. 2010. Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J. Gen. Virol. 91, 2610–2619 (doi:10.1099/vir.0.022699-0) [DOI] [PubMed] [Google Scholar]
- 9.Burke GR, Strand MR. 2012. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor bracovirus. J. Virol. 86, 3293–3306 (doi:10.1128/JVI.06434-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuy C, Gundersen-Rindal D, Cusson M. 2012. Genomic and replication of polydnaviruses. In Parasitoid viruses symbionts and pathogens (eds Beckage NE, Drezen J-M.), pp. 47–61 San Diego, CA: CRC Press-Elsevier [Google Scholar]
- 11.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Veron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl Acad. Sci. USA 109, E432–E441 (doi:10.1073/pnas.1115346109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savary S, Beckage NE, Tan F, Periquet G, Drezen J-M. 1997. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastrinae) at potential recombinase binding sites. J. Gen. Virol. 78, 3125–3134 [DOI] [PubMed] [Google Scholar]
- 13.Annaheim M, Lanzrein B. 2007. Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers and abundance of proviral and excised segments. J. Gen. Virol. 88, 450–457 (doi:10.1099/vir.0.82396-0) [DOI] [PubMed] [Google Scholar]
- 14.Belle E, Beckage NE, Rousselet J, Poirié M, Lemeunier F, Drezen J-M. 2002. Visualization of polydnavirus sequences in a parasitoid wasp chromosome. J. Virol. 76, 5793–5796 (doi:10.1128/JVI.76.11.5793-5796.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasquier-Barre F, Dupuy C, Huguet E, Monteiro F, Moreau A, Poirié M, Drezen J-M. 2002. Polydnavirus replication: the EP1 segment of the parasitoid wasp Cotesia congregata is amplified within a larger precursor molecule. J. Gen. Virol. 83, 2035–2045 [DOI] [PubMed] [Google Scholar]
- 16.Desjardins CA, et al. 2008. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9, R183 (doi:10.1186/gb-2008-9-12-r183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beckage NE, Tan F, Schleifer KW, Lane RD, Cherubin LL. 1994. Characterization and biological effects of Cotesia congregata polydnavirus on host larvae of the tobacco hornworm, Manduca sexta. Arch. Insect Biochem. Physiol. 26, 165–195 (doi:10.1002/arch.940260209) [Google Scholar]
- 18.Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, Kang HL, Simon MI, Shizuya H. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34, 213–218 (doi:10.1006/geno.1996.0268) [DOI] [PubMed] [Google Scholar]
- 19.Espagne E, Dupuy C, Huguet E, Cattolico L, Provost B, Martins N, Poirié M, Periquet G, Drezen J-M. 2004. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306, 286–289 (doi:10.1126/science.1103066) [DOI] [PubMed] [Google Scholar]
- 20.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–208 (doi:10.1093/nar/gkp335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu D, Stoltz DB. 1993. Polydnavirus genome segment families in the ichneumonid parasitoid Hyposoter fugitivus. J. Virol. 67, 1340–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui L, Webb BA. 1997. Homologous sequences in the Campoletis sonorensis polydnavirus genome are implicated in replication and nesting of the W segment family. J. Virol. 71, 8504–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (doi:10.1093/bioinformatics/bts199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (doi:10.1038/nature03959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jancek S, et al. 2013. Adaptive selection on bracovirus genomes drives the specialization of Cotesia parasitoid wasps. PLoS ONE 8, e64432 (doi:10.1371/journal.pone.0064432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 (doi:10.1093/bioinformatics/16.10.944) [DOI] [PubMed] [Google Scholar]
- 27.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (doi:10.1093/bioinformatics/16.10.944) [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (doi:10.101/gr.849004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desjardins CA, et al. 2007. Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus. BMC Microbiol. 7, 61 (doi:10.1186/1471-2180-7-61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck MH, Zhang S, Bitra K, Burke GR, Strand MR. 2011. The encapsidated genome of Microplitis demolitor bracovirus integrates into the host Pseudoplusia includens. J. Virol. 85, 11 685–11 696 (doi:10.1128/JVI.05726-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serbielle C, Dupas S, Perdereau E, Hericourt F, Dupuy C, Huguet E, Drezen J-M. 2012. Evolutionary mechanisms driving the evolution of a large polydnavirus gene family coding for protein tyrosine phosphatases. BMC Evol. Biol. 12, 253 (doi:10.1186/1471-2148-12-253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruber A, Stettler P, Heiniger P, Schumperli D, Lanzrein B. 1996. Polydnavirus DNA of the braconid wasp Chelonus inanitus is integrated in the wasp genome and excised only in later pupal and adult stages of the female. J. Gen. Virol. 77, 2873–2879 (doi:10.1099/0022-1317-77-11-2873) [DOI] [PubMed] [Google Scholar]
- 33.Savary S, Drezen J-M, Tan F, Beckage NE, Periquet G. 1999. The excision of polydnavirus sequences from the genome of the wasp Cotesia congregata (Braconidae, Microgastrinae) is developmentally regulated but not strictly restricted to the ovaries in the adult. Insect Mol. Biol. 8, 319–327 (doi:10.1046/j.1365-2583.1999.83130.x) [DOI] [PubMed] [Google Scholar]
- 34.Vanarsdall AL, Okano K, Rohrmann GF. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 80, 1724–1733 (doi:10.1128/JVI.80.4.1724-1733.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gundersden-Rindal D, Dupuy C, Huguet E, Drezen J-M. 2013. Parasitoid polydnaviruses: evolution, pathology and applications. Biocontrol Sci. Technol. 23, 1–61 (doi:10.1080/09583157.2012.731497) [Google Scholar]
- 36.Albrecht U, Wyler T, Pfister-Wilhelm R, Gruber A, Stettler P, Heiniger P, Kurt E, Schumperli D, Lanzrein B. 1994. Polydnavirus of the parasitic wasp Chelonus inanitus (Braconidae): characterization, genome organization and time point of replication. J. Gen. Virol. 75, 3353–3363 (doi:10.1099/0022-1317-75-12-3353) [DOI] [PubMed] [Google Scholar]
- 37.Beck MH, Inman RB, Strand MR. 2007. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 359, 179–189 (doi:10.1016/j.virol.2006.09.002) [DOI] [PubMed] [Google Scholar]
- 38.Drezen J-M, Bézier A, Lesobre J, Huguet E, Cattolico L, Periquet G, Dupuy C. 2006. The few virus-like genes of Cotesia congregata bracovirus. Arch. Insect Biochem. Physiol. 61, 110–122 (doi:10.1002/arch.20108) [DOI] [PubMed] [Google Scholar]
- 39.Dupuy C, Periquet G, Serbielle C, Bézier A, Louis F, Drezen J-M. 2011. Transfer of a chromosomal Maverick to endogenous bracovirus in a parasitoid wasp. Genetica 139, 489–496 (doi:10.1007/s10709-011-9569-x) [DOI] [PubMed] [Google Scholar]
- 40.Espagne E, et al. 2005. A virus essential for insect host-parasite interactions encodes cystatins. J. Virol. 79, 9765–9776 (doi:10.1128/JVI.79.15.9765-9776.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YF, Gao F, Ye XQ, Wei SJ, Shi M, Zheng HJ, Chen XX. 2011. Deep sequencing of Cotesia vestalis bracovirus reveals the complexity of a polydnavirus genome. Virology 414, 42–50 (doi:10.1016/j.virol.2011.03.009) [DOI] [PubMed] [Google Scholar]
- 42.The Honeybee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949 (doi:10.1038/nature05260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louis F, Bézier A, Periquet P, Ferras C, Drezen J-M, Dupuy C. In press. Bracovirus DNAs and the nudivral cluster involved in particle production are amplified in several replication units. J. Virol. (doi:10.1128/JVI.00886-13) [Google Scholar]
- 44.Kroemer JA, Webb BA. 2004. Polydnavirus genes and genomes: emerging gene families and new insights into polydnavirus replication. Annu. Rev. Entomol. 49, 431–456 (doi:10.1146/annurev.ento.49.072103.120132) [DOI] [PubMed] [Google Scholar]
- 45.Alout H, Labbe P, Berthomieu A, Pasteur N, Weill M. 2009. Multiple duplications of the rare ace-1 mutation F290 V in Culex pipiens natural populations. Insect Biochem. Mol. Biol. 39, 884–891 (doi:10.1016/j.ibmb.2009.10.005) [DOI] [PubMed] [Google Scholar]
- 46.Lee JA, Carvalho CM, Lupski JR. 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (doi:10.1016/j.cell.2007.11.037) [DOI] [PubMed] [Google Scholar]
- 47.Dupas S, Dubuffet A, Carton Y, Poirié M. 2009. Local, geographic and phylogenetic scales of coevolution in Drosophila-parasitoid interactions. Adv. Parasitol. 70, 281–295 (doi:10.1016/S0065-308X(09)70011-9) [DOI] [PubMed] [Google Scholar]
- 48.Strand MR. 2012. Polydnavirus gene products that interact with the host immune system. In Parasitoid viruses symbionts and pathogens (eds Beckage NE, Drezen J-M.), pp. 149–161 San Diego, CA: CRC Press-Elsevier [Google Scholar]
- 49.Beckage NE. 2012. Polydnaviruses as endocrine regulators. In Parasitoid viruses symbionts and pathogens (eds Beckage NE, Drezen J-M.), pp. 163–168 San Diego, CA: CRC Press-Elsevier [Google Scholar]
- 50.Serbielle C, Chowdhury S, Pichon S, Dupas S, Lesobre J, Purisima EO, Drezen J-M, Huguet E. 2008. Viral cystatin evolution and three-dimensional structure modelling: a case of directional selection acting on a viral protein involved in a host–parasitoid interaction. BMC Biol. 6, 38 (doi:10.1186/1741-7007-6-38) [DOI] [PMC free article] [PubMed] [Google Scholar]