Abstract
The aldo-keto reductase superfamily contains 173 proteins which are present in all phyla. Examination of the human and mouse genomes has identified that in some instances a single AKR gene can give rise to alternatively spliced mRNA variants which in some cases can give rise to more than one protein isoform. This is currently well documented in the AKR6A subfamily which contains the β-subunits of the voltage-gated potassium ion channels. With the emergence of second generation sequencing it is likely that the occurrence of transcript variants and protein isoforms from a single AKR gene may become common place. To deal with this issue we recommend that the Ensembl data-base nomenclature be used to annotate the transcript variants from a single AKR gene. However, since multiple transcript variants could give rise to either the same or multiple protein isoforms from the same AKR gene we also propose to expand the nomenclature of the AKR protein superfamily, so that when a protein isoform is shown to be expressed and is functional it would be assigned the standard AKR name followed by a “period or full-stop” and a number for that unique isoform. Numbers will be assigned chronologically and linked to the respective transcripts annotated in Ensembl e.g. AKR6A5.1 (Kvβ2.1) (AKR6A5-001, -006 and -201), followed by AKR6A5.2 (Kvβ2.2) (AKR6A5-002,-202). This nomenclature is expandable and it enables multiple protein isoforms to be assigned to their respective transcripts when they arise from the same AKR gene or for a single protein isoform to be assigned to multiple transcripts when the transcripts encode the same AKR protein.
Keywords: Aldo-Keto Reductase, alternative splicing, nomenclature, transcript variants, protein isoform
1. Introduction
The Aldo-Keto Reductase (AKR) protein superfamily contains more than 173 annotated enzymes and proteins from all kingdoms of life. The universally adopted nomenclature for these proteins was introduced at the 9-th International Meeting on Molecular Biology and Enzymology of Carbonyl Metabolism in Deadwood, South Dakota, and is now accepted by all major databases including NCBI and Ensembl, and by the human and mouse gene nomenclature committees HUGO (HGNC) and MGI organizations, respectively. In addition, the list of existing AKR members is maintained separately by the laboratory of Dr. Penning at the University of Pennsylvania website at http://www.med.upenn.edu/akr/. The official names of AKR family members include the root AKR followed by the family designation (a number), a letter indicating the subfamily and an individual number for the protein, such as AKR1A1 [1, 2]. The corresponding genes are indicated in italics, e.g. AKR1A1. In compliance with the recommendations of the HGNC and MGI gene nomenclature committees, human genes are designated with all upper-case letters, whereas rodent genes are designated with an upper-case first letter followed by lower-case letters. This nomenclature is expandable and new members are added chronologically. The nomenclature makes no assumptions about orthologs across species.
Additional diversity of aldo-keto reductases can be generated by alternative splicing, a phenomenon which allows for several mRNA transcripts and protein polypeptides to be expressed from a single AKR gene. The primary function of alternative splicing is to increase the diversity of mRNA expressed from the limited number of genes [3]. DNA microarray analysis showed that as of 2005, 74% of human genes are alternatively spliced, and on average, a human gene generates two to three transcripts [4]. Alternative splicing patterns constantly change under physiological conditions, allowing an organism to respond to changes in the environment. Additional diversity of the transcriptome may arise from the use of alternative promoters, which also generate alternative transcript variants, with either divergent 5’-untranslated regions or coding sequences. We noticed that the present nomenclature does not include provisions for distinguishing between different protein products generated by a single gene. As discovery of such phenomena is greatly accelerated due to the efforts of large-scale sequencing and availability of large datasets to researchers worldwide, the need for the system to name transcript variants and their corresponding protein isoforms systematically clearly exists. In this report we review the information available on the alternative splicing in the AKR superfamily, existing systems for naming alternatively spliced variants (transcripts and protein isoforms) in other families of proteins, and propose a nomenclature system for the AKRs.
2. Methods
Ensembl and NCBI Genbank databases were searched for the gene names of existing AKR members from the University of Pennsylvania AKR database, and information on the known transcripts was extracted. Following that, a literature search was conducted in PubMed with the keywords “AKR” and “splicing”, and obtained information was manually curated and included in this report.
3. Results and Discussion
3.1. Description of known splice variants in human AKRs
First, we examined how widespread the alternative splicing is in the AKR superfamily. Two types of alternative splicing were found for the AKR genes: 1) events leading to different protein sequences, and 2) events affecting exclusively untranslated regions of mRNA. Of the latter, alternative splicing in the 5’-untranslated region (UTR) is much more widespread than that in the 3’-UTR, where mostly variations in its length are found.
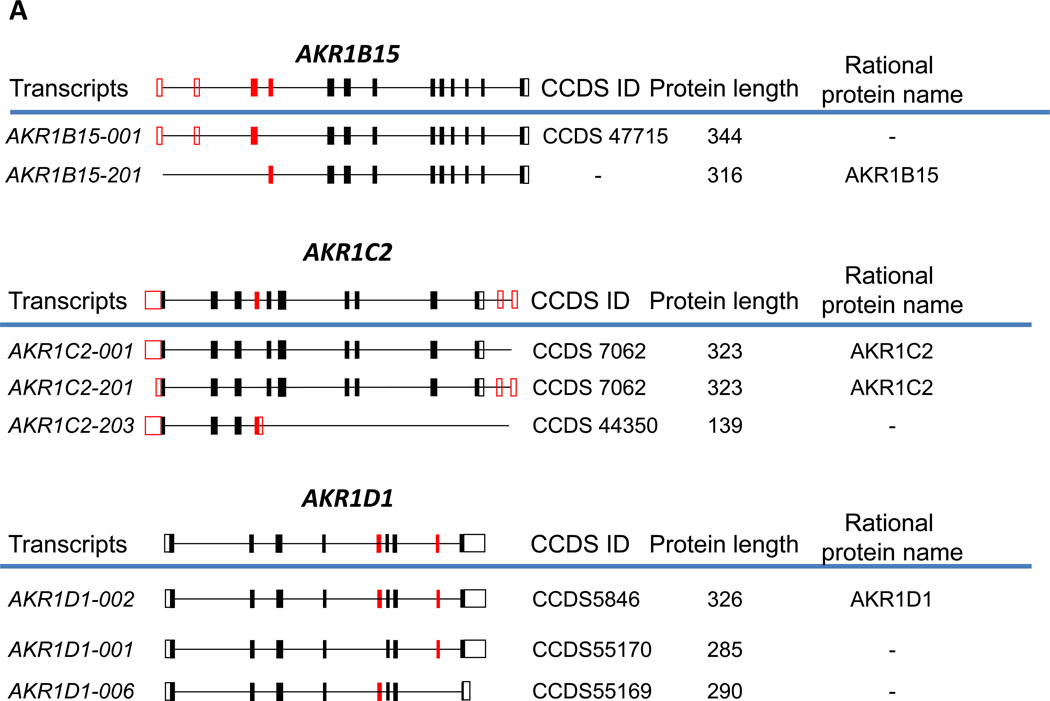
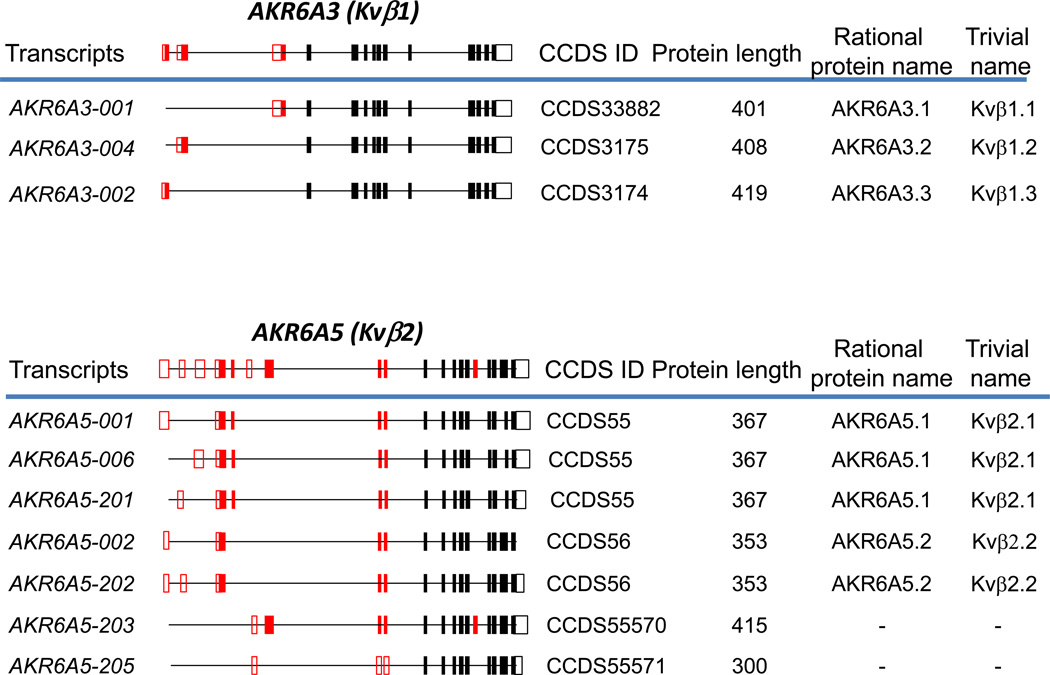
The richest information on splice variants is available for the human genome due to advances in its annotation. As shown in Table 1, more than one transcript has been discovered for all human AKR genes with the exception of AKR7A3. In ten of these genes, the alternatively spliced transcripts contain open reading frames (ORFs) and have the potential to produce protein isoforms in addition to the classically assigned ones. However, the existence of many of these proteins has not been conclusively proven. The high fidelity sequences either described in peer-reviewed literature, or generated by high-throughput sequencing projects and manually curated at the European Bioinformatics Institute (EBI), National Center for Biotechnology Information (NCBI), Wellcome Trust Sanger Institute (WTSI), and University of California, Santa Cruz (UCSC) are assigned a Consensus Coding Sequence Set (CCDS) ID and represent reliable protein coding sequences. The CCDS set includes coding regions that are annotated as full-length (with an initiating ATG and valid stop-codon), can be translated from the genome without frameshifts, and use consensus splice-sites [5]. To this end, 4 human AKR genes are identified as being potentially translated into multiple protein isoforms which have been assigned CCDS ID. The most important belong to the β-subunits of the potassium voltage-gated ion-channel which are members of the AKR6A subfamily. These include AKR6A5, trivially known as Kvβ2, which has 4 predicted protein isoforms; and AKR6A3 (Kvβ1) which has three predicted protein isoforms (which are discussed more fully in section 3.2.). The structure of human and mouse AKR genes, the predicted transcript variants, CCDS IDs and names for the potential protein isoforms they encode are shown in Fig. 1.
Table 1.
Alternative splicing in human AKR genes
| Gene | CCDS IDs (protein length, aa) |
Number of transcripts assigned to CCDS ID |
Number of additional ORFs |
Predicted protein length of additional ORFs (aa) |
Total number of transcripts |
|---|---|---|---|---|---|
| AKR1A1 | CCDS523 (325) | 2 | 1 | 147 | 13 |
| AKR1B1 | CCDS5831 (316) | 1 | 0 | 15 | |
| AKR1B10 | CCDS5832 (316) | 1 | 0 | 4 | |
| AKR1B15 | CCDS47715 (344) | 1 | 1 | 316 | 3 |
| AKR1C1 | CCDS7061 (323) | 2 | 2 | 248, 205 | 6 |
| AKR1C2 | CCDS7062 (323) CCDS44350 (139) |
2 1 |
1 | 297 | 5 |
| AKR1C3 | CCDS7063 (323) | 1 | 1 | 204 | 5 |
| AKR1C4 | CCDS7064 (323) | 2 | 0 | 3 | |
| AKR1D1 | CCDS5846 (326) CCDS55169 (290) CCDS55170 (285) |
1 1 1 |
1 | 96 | 6 |
| AKR1E2 | CCDS31134 (320) | 1 | 6 | 263, 250,222, 199, 122, 56 |
13 |
| AKR6A3 | CCDS33882 (401) CCDS3175 (408) CCDS3174 (419) |
1 1 1 |
4 | 390, 372, 177, 129 |
14 |
| AKR6A5 | CCDS55 (367) CCDS56 (353) CCDS55570 (415) CCDS55571 (300) |
3 2 1 1 |
6 | 254, 253, 173, 173,95,52, |
19 |
| AKR6A9 | CCDS33882 (404) | 1 | 0 | 6 | |
| AKR7A2 | CCDS194 (359) | 1 | 2 | 314, 205 | 5 |
| AKR7A3 | CCDS193 (331) | 1 | 0 | 1 |
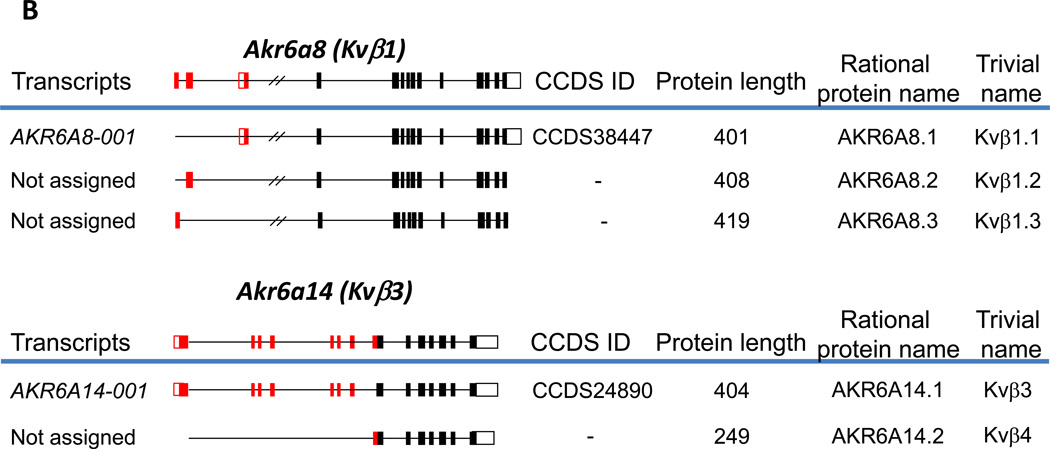
Fig. 1. Alternative splicing of human (A) and murine (B) AKR genes.
Genes with multiple CCDS IDs assigned to them, or genes for which multiple protein isoforms have been reported in the peer-reviewed literature are shown. Constitutive exons are shown in black, and alternatively spliced exons are in red. Solid boxes indicate coding regions, and open boxes indicate untranslated regions. The complete gene structures are shown above the blue lines; below the blue lines are processed transcripts showing only exons included in a particular transcript, along with systematic transcript name, CCDS ID, and the length of the encoded protein. The rational name of the protein isoform encoded by the corresponding alternatively spliced transcript according to the new nomenclature is shown to the right of that transcript. The structure of murine Akr6a8 is drawn according to [15], and that of Akr6a14 is from [20].
Interestingly, the consensus coding sequence CCDS47715 assigned to AKR1B15 gene by the Ensembl and NCBI databases (corresponding to transcript AKR1B15-001) differs at its 5’-end from the sequence published by Salabei et.al [6] which corresponds to the transcript AKR1B15-201 in the Ensembl database. Consequently, the encoded proteins differ at their N-termini. These data suggest that two transcript variants exist and that each may give rise to a different protein isoform.
In the steroid transforming AKRs, AKR1C2 is predicted to have three transcripts and 2 protein isoforms in the CCDS database. Transcripts AKR1C2-001 and AKR1C2-201 encode what is classically known as a 323 amino acid residue -long AKR1C2 protein (type 3 3α-hydroysteroid dehydrogenase/bile-acid binding protein). On the other hand, transcript AKR1C2-203-encodes a protein that utilizes an alternative exon 4 which results in a significantly shorter protein (139 aa) with a different C-terminus. This protein contains the conserved catalytic tetrad found in AKRs but its binding of cofactor is likely to be compromised, and with over half the protein missing it is predicted to be inactive, if expressed.
The AKR1D1 (steroid 5β-reductase) gene consists of 9 exons which could give rise to three different transcripts and three protein isoforms. A well characterized 326 amino acid AKR1D1 enzyme [7, 8] is encoded by an AKR1D1-002 transcript, which includes all 9 exons of the AKR1D1 gene. Transcript AKR1D1-001 does not include exon 5, which is a symmetrical exon, i.e. contains number of nucleotides divisible by 3, so that its omission does not cause a frameshift in the protein. This transcript is conceptually translated into a 285 amino acid protein with identical N- and C-termini to that of full length AKR1D1-002. However, since it is missing some of the residues in the middle of the sequence it is likely this will compromise the β-barrel structure and the protein will not fold properly. On the other hand, AKR1D1-006 mRNA omits exon 8, which causes a frameshift and translates into a 290 amino acid protein with an alternative five C-terminal amino acid residues. However, this protein is missing the C-terminal flexible loop which lines the steroid channel and the last helix in the structure. It is predicted that this isoform may be able to fold correctly but likely losses activity due to a decrease in affinity for steroid substrates. While there is evidence that the AKR6A3 and AKR6A5 transcript variants encode for different protein isoforms that are expressed this is not the case for AKR1C2 and AKR1D1. Only further work will determine whether these protein isoforms are expressed and functionally active.
3.2. Human AKR6 Family Transcript Variants and Protein Isoforms
AKR6 family proteins trivially known as β-subunits of voltage-gated potassium ion channels (Kvβ) are products of a fusion of an AKR domain with a specific N-terminus, which performs functions related to the channel gating. These proteins may have originated in the process of evolution where an ancient AKR enzyme was recruited to serve as a regulator of an ion channel by adding a corresponding N-terminus to the evolutionarily-tested and structurally robust AKR fold. Examination of the AKR family tree supports the assertion that the AKR6 family evolved as a group from a common ancestral protein. In AKR6A3 (Kvβ1) and AKR6A9 (Kvβ3) the N-termini contain a so-called “ball and chain” domain, which functions to impart inactivation on the channel. The N-terminus of AKR6A5 (Kvβ2) does not contain this domain and its function is less clear. Accordingly, AKR6 genes consist of 13 invariable 3’-exons coding for an AKR domain, and one to three 5’- exons coding for a channel-related N-terminal domain. As shown in Fig. 1, in the AKR6A3 gene, only one out of possible three 5’-exons is included in a particular transcript giving rise to three protein isoforms, AKR6A3.1 (Kvβ1.1), AKR6A3.2 (Kvβ1.2), and AKR6A3.3 (Kvβ1.3). These protein isoforms display distinct tissue distribution and channel inactivation properties [9–11], and corresponding mRNA transcripts contain unique 5’-UTRs and are likely transcribed using their own promoters. Although promoters and the regulation of expression of these transcripts have not been studied, AKR6A3 (Kvβ1) represents the best example in the superfamily where alternatively spliced isoforms have been extensively characterized at the protein level.
The splicing situation is more complex in AKR6A5 (Kvβ2). The previously characterized protein isoforms Kvβ2.1 (367 aa) and Kvβ2.2 (353 aa) arise from inclusion of either two (AKR6A5.1) or one (AKR6A5.2) of the most 5’ exons, respectively. These isoforms differ in their effect on inactivation of Kv1.4 [12]. The additional isoform encoded by transcript, AKR6A5-203, utilizes an alternative 5’-exon and includes an additional symmetrical exon inside the AKR domain (Fig. 1). The resultant protein product is predicted to be 415 amino acids in length, has a distinct N-terminus and includes an additional 15 amino acid residues in the middle of its AKR domain without causing a frameshift in the rest of the protein. This insertion is predicted to elongate loop A located between β6 and α4 of the α/β barrel which in other AKRs contains active site residues important for catalytic activity, such as His110 in AKR1B1 (aldose reductase). This loop is shorter in AKR6A5.1 and AKR6A5.2 than that in the enzymes of the AKR1 family [13], and therefore insertion into this loop may have a significant influence on the catalytic activity. Finally, AKR6A5-205 is predicted to encode a 300 amino acid polypeptide with a truncated AKR domain and no N-terminal domain. It is unknown if the last two protein isoforms are expressed and whether they are functional.
3.3. Description of splice-variants in murine AKRs
The murine genome is more complex than the human genome with respect to the AKR superfamily and contains 23 known Akr genes (in comparison with the 15 known human genes). The mouse genome is also predicted to give rise to multiple transcripts and open reading frames for the majority of its Akr genes (Table 2). The information on the alternative splicing in mouse genes contained in public databases is not as rich as in the human genome, and none of those has more than one CCDS ID, despite the fact that for some of those genes, several alternative protein isoforms have been described in the peer reviewed literature. The best example is Akr6a8 (murine Kvβ1), for which, similar to the human gene, 3 protein isoforms have been characterized and even two knockout mouse models have been generated [14, 15]. Despite this fact, the sequences of murine Kvβ1.2 and Kvβ1.3 are found neither in Refseq (NCBI) nor in Ensembl sets. Hence, the relative paucity of reliable information reflects the delays in annotation rather than the actual lack of complexity of the mouse transcriptome, which is evident in the examples cited. The structures of published alternatively spliced transcripts of murine Akr genes extracted from the literature are shown in Fig. 1B. As we will discuss later we prefer to wait for Ensembl to annotate these transcripts but propose a separate nomenclature for the encoded protein isoforms.
Table 2.
Alternative splicing in murine AKR genes.
| Gene | CCDS ID (protein length, aa) |
Number of transcripts assigned to CCDS ID |
Number of additional ORFs |
Predicted protein length of additional ORFs (aa) |
Total number of transcripts |
|---|---|---|---|---|---|
| Akr1a4 | CCDS18514 (325) | 1 | 1 | 204 | 4 |
| Akr1b3 | CCDS19990 (316) | 1 | 1 | 176 | 5 |
| Akr1b7 | CCDS19993 (316) | 1 | 0 | 1 | |
| Akr1b8 | CCDS19991 (316) | 1 | 0 | - | 2 |
| Akr1b16 | CCDS19992 (316) | 1 | 1 | 288 | 5 |
| Akr1c6 | CCDS26223 (323) | 1 | 1 | 175 | 3 |
| Akr1c12 | CCDS26222 (323) | 1 | 0 | 1 | |
| Akr1c13 | CCDS26220 (323) | 1 | 0 | 3 | |
| Akr1c21 | CCDS26225 (323) | 1 | 0 | 2 | |
| Akr1c18 | CCDS26219 (323) | 1 | 1 | 297 | 2 |
| Akr1c14 | CCDS26218 (323) | 2 | 0 | 3 | |
| Akr1c20 | CCDS26224 (262) | 1 | 1 | 323 | 2 |
| Akr1c19 | CCDS26221 (323) | 2 | 0 | 3 | |
| Akr1cl | CCDS48282 (322) | 1 | 2 | 291,156 | 6 |
| Akr1e1 | CCDS26226 (301) | 1 | 1 | 245 | 3 |
| Akr1d1 | CCDS20007 (325) | 1 | 1 | 127 | 3 |
| Akr6a8 | CCDS38447 (401) | 1 | 4 | 366, 347, 295, 178 |
6 |
| Akr6a4 | CCDS19000 (367) | 2 | 5 | 382, 353, 347, 134, 38 |
14 |
| Akr6a14 | CCDS24890 (404) | 1 | 0 | 3 | |
| Akr7a5 | CCDS18844 (367) | 1 | 0 | 2 |
3.4. Alternative splicing in the untranslated region
In addition to variations in the coding sequences, differences in the untranslated regions of multiple transcripts encoding the same protein may reflect differences in mRNA stability, translation rate and susceptibility to silencing by miRNA [16, 17]. These variations might result from true splicing events or from the use of different promoters. The use of cell-specific transcription start sites in combination with alternative splicing in the 5’-untranslated region has been described for human and murine aldehyde reductase AKR1A1 and 1a4 [18]. In addition, multiple 5’-untranslated exons might be present in the AKR6A5 gene.
Many transcripts in the Ensembl database align with AKR genes, but either do not code for a protein, or contain a truncated coding sequence not confirmed experimentally. These sequences may arise from experimental artifacts in cloning, incompletely processed transcripts, inclusion of introns, etc.; thus, the significance of such transcripts remains unclear until further experimental evidence is obtained. In this report we list the number of such transcripts associated with each gene however, interpretation of their biological significance must await further experimental verification.
3.5. Rules for naming mRNA transcripts and protein isoforms from the same AKR gene
The proven existence of multiple transcripts and protein isoforms from the same AKR gene, and an expected increase in information related to the expression and function of such transcripts or isoforms due to advances in sequencing, warrants a nomenclature system for the systematic naming of alternatively spliced transcripts and their protein isoforms. To this end, we reviewed such systems used in other gene families or employed by large databases. NCBI differentiates between mRNA and protein variants by referring to the former as transcript variants and to the latter as (protein) isoforms. Different gene families employ various naming systems including giving roman (perilipin and high mobility group familes) or greek (heat shock protein) letters or a combination of letters and numbers to identify splice variants. The UDP glucuronosyl transferase gene family, where alternative splicing is particularly abundant, utilizes gene name (e.g., UGT1) followed by the unique identifier of the splice variant which corresponds to the name of the 5’-most exon included in the transcript (e.g., UGT1A1) [19]. This nomenclature system is not applicable to the AKR superfamily because names like these are already used to identify different genes/ proteins.
To date, the Ensembl database provides the most comprehensive information on the existing transcript variants in an easy-to-read tabular and graphical format with readily retrievable sequence information. In the Ensembl system, splice variants are identified by a three digit number separated from the gene name by a dash (e.g., AKR1C2-001). Due to its comprehensive coverage and accessibility we recommend adoption of the Ensembl system to name and identify transcript variants in the AKR superfamily. An additional advantage is that when a particular name is used in a publication, readers will be able to obtain desirable information, such as sequence, included and excluded exons, genome position and comparison to other transcripts from the Ensembl web site (http://www.ensembl.org) in an electronic format. The transcript variants characterized-to-date for the human and murine AKR genes and their systematic names according to the Ensembl nomenclature are shown in Fig. 1. In the names of the transcripts arising from the AKR6 family genes the systematic AKR gene name is substituted for the symbol KCNAB (or Kcnab for rodents) which was introduced before the AKR nomenclature for these genes was adopted and is currently used by the Ensembl data base. The three digit Ensembl suffix is retained.
The AKR nomenclature is protein-based. We recommend that when transcript variants give rise to functional protein isoforms they will be assigned the standard AKR name followed by a “period or full-stop” and a number for that isoform. This rule applies to genetically-encoded protein isoforms, and does not include post-translational modifications. Numbers will be assigned chronologically and linked to the respective transcript(s) annotated in Ensembl, e.g. AKR6A5.1 (AKR6A5-001, -006 and -201) and AKR6A5.2 (AKR6A5-002, -202). This nomenclature will enable a single protein isoform to be assigned to multiple transcript(s) when they code for the same protein. The nomenclature system also enables different protein isoforms to be assigned to their respective transcripts when they arise from the same AKR gene. In each instance the protein isoform has its own unique name. The new systematic names for AKR6 family proteins are shown to the right of the respective transcripts in Fig. 1.
3.6. Rules for submission of new sequences
In the event that a researcher discovers a new transcript that codes for a splice variant not included into Ensembl database, we suggest that they submit this information to both Ensembl and the AKR protein superfamily homepage. Ensembl will annotate the transcript according to their open format as described above. The AKR homepage currently maintains a list of potential AKR protein members to which the potential encoded protein isoforms can be added. If the AKR protein isoform has been shown to be expressed as a protein with known function a formal AKR name would then be assigned as follows: AKR6A3.1 (would be the first chronologically annotated protein isoform with validated function); followed by AKR6A3.2, etc, from the AKR6A3 gene. The name for the protein isoform would be linked to the Ensembl database annotation for the respective transcript to maintain consistency.
4. Conclusion
With the unprecedented flow of information fueled mainly by large-scale sequencing projects and ongoing research from individual groups, a large number of previously unknown splice variants of AKR genes are being discovered. An even larger number is expected in the near future as next generation sequencing and annotation of the human genome and genomes of model organisms’ progresses. This influx of information warrants a need for systematic naming of splice variants to provide a common nomenclature for the transcript variants and protein isoforms that they generate. We propose to adopt the system used by Ensembl database to name alternative transcripts in the AKR superfamily. It is important to realize that the AKR nomenclature system is protein-based. Thus, alternative transcripts that give rise to protein isoforms that are functional will be assigned the standard AKR name followed by “period or fullstop” and a number for that isoform. Numbers will be assigned chronologically and linked to the respective transcript annotated in Ensembl e.g. AKR6A5.1 (AKR6A5-001, -006 and -201, currently KCNAB2-001, -006 and -201). This will require researchers to verify the sequence they are using with the databases and annotate the splice variant and protein isoform they are working with accordingly. Using the systematic names will provide readers free access worldwide to full information concerning a particular transcript variant or protein isoform. The procedure proposed is expandable as new spliced variants are discovered.
Acknowledgements
This work was partly supported by NIH grants RR-24489, HL-89372 (to O.A.B.), and R01-DK47015 and P30-ES13508 (to T.M.P.). We thank Dr. Mo Chen for her careful read of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Jez JM, Flynn TG, Penning TM. A new nomenclature for the aldo-keto reductase superfamily. Biochem Pharmacol. 1997;54:639–647. doi: 10.1016/s0006-2952(97)84253-0. [DOI] [PubMed] [Google Scholar]
- 2.Jez JM, Penning TM. The aldo-keto reductase (AKR) superfamily: an update. Chem Biol Interact. 2001;130–132:499–525. doi: 10.1016/s0009-2797(00)00295-7. [DOI] [PubMed] [Google Scholar]
- 3.Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2012 doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salabei JK, Li XP, Petrash JM, Bhatnagar A, Barski OA. Functional expression of novel human and murine AKR1B genes. Chem Biol Interact. 2011;191:177–184. doi: 10.1016/j.cbi.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo KH, Kai MH, Setoguchi Y, Eggertsen G, Sjoblom P, Setoguchi T, Okuda KI, Bjorkhem I. Cloning and expression of cDNA of human delta 4-3-oxosteroid 5 beta-reductase and substrate specificity of the expressed enzyme. Eur J Biochem. 1994;219:357–363. doi: 10.1111/j.1432-1033.1994.tb19947.x. [DOI] [PubMed] [Google Scholar]
- 8.Di Costanzo L, Drury JE, Christianson DW, Penning TM. Structure and catalytic mechanism of human steroid 5beta-reductase (AKR1D1) Mol Cell Endocrinol. 2009;301:191–198. doi: 10.1016/j.mce.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 10.England SK, Uebele VN, Kodali J, Bennett PB, Tamkun MM. A novel K+ channel beta-subunit (hKv beta 1.3) is produced via alternative mRNA splicing. J Biol Chem. 1995;270:28531–28534. doi: 10.1074/jbc.270.48.28531. [DOI] [PubMed] [Google Scholar]
- 11.De Biasi M, Wang Z, Accili E, Wible B, Fedida D. Open channel block of human heart hKv1.5 by the beta-subunit hKv beta 1.2. Am J Physiol. 1997;272:H2932–H2941. doi: 10.1152/ajpheart.1997.272.6.H2932. [DOI] [PubMed] [Google Scholar]
- 12.Akhtar S, McIntosh P, Bryan-Sisneros A, Barratt L, Robertson B, Dolly JO. A functional spliced-variant of beta 2 subunit of Kv1 channels in C6 glioma cells and reactive astrocytes from rat lesioned cerebellum. Biochemistry. 1999;38:16984–16992. doi: 10.1021/bi992114x. [DOI] [PubMed] [Google Scholar]
- 13.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40:553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giese KP, Storm JF, Reuter D, Fedorov NB, Shao LR, Leicher T, Pongs O, Silva AJ. Reduced K+ channel inactivation, spike broadening, and after-hyperpolarization in Kvbeta1.1-deficient mice with impaired learning. Learn Mem. 1998;5:257–273. [PMC free article] [PubMed] [Google Scholar]
- 15.Aimond F, Kwak SP, Rhodes KJ, Nerbonne JM. Accessory Kvbeta1 subunits differentially modulate the functional expression of voltage-gated K+ channels in mouse ventricular myocytes. Circ Res. 2005;96:451–458. doi: 10.1161/01.RES.0000156890.25876.63. [DOI] [PubMed] [Google Scholar]
- 16.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugaut A, Balasubramanian S. 5'-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barski OA, Papusha VZ, Ivanova MM, Rudman DM, Finegold MJ. Developmental expression and function of aldehyde reductase in proximal tubules of the kidney. Am J Physiol Renal Physiol. 2005;289:F200–F207. doi: 10.1152/ajprenal.00411.2004. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 20.Leicher T, Bahring R, Isbrandt D, Pongs O. Coexpression of the KCNA3B gene product with Kv1.5 leads to a novel A-type potassium channel. J Biol Chem. 1998;273:35095–35101. doi: 10.1074/jbc.273.52.35095. [DOI] [PubMed] [Google Scholar]