Abstract
Background
Child abuse is highly prevalent and associated with increased risk for a range of health problems including: cancer, cardiovascular disease, diabetes, psychiatric disorders, and other health problems. Little is currently known about the mechanism by which early adversity confers risk for health problems later in life.
Purpose
To determine if there are epigenetic differences associated with child maltreatment that may help explain association between adverse childhood experiences and later health problems.
Methods
As part of a study examining genetic and environmental factors associated with depression, saliva DNA specimens were collected on 96 maltreated children removed from their parents due to abuse or neglect and 96 demographically-matched control children between 2003 and 2010. In 2011, the Illumina 450K BeadChip was used on stored DNA specimens and analyzed to examine whole-genome methylation differences between maltreated and control children.
Results
After controlling for multiple comparisons, maltreated and control children had significantly different methylation values at 2868 CpG sites (p< 5.0 × 10−7, all sites; average methylation difference per site=17%; range 1%–62%). The gene set contained numerous markers of diseases and biological processes related to the health problems associated with early childhood adversity.
Conclusions
While replication is required, this study suggests that epigenetic mechanisms may be associated with risk for health problems later in life in maltreated children. This study lays the groundwork for future studies examining health and methylation measures to further characterize the role of epigenetic mechanisms in conferring risk for medical problems in individuals with histories of early adversity.
Introduction
Child abuse occurs at epidemic rates, with nearly 700,000 substantiated reports of child maltreatment each year,1 many reported cases of actual abuse referred to protective services that are not verified,2 and countless other cases that never come to the attention of authorities.2–4 Child maltreatment and other adverse childhood experiences are nonspecific risk factors for multiple psychiatric disorders5–7 and several health risk behaviors including smoking, overeating, and excessive alcohol and drug use.8–10 Above and beyond the effect of these risk behaviors, however, adverse childhood experiences predict ischemic heart disease,9,11,12 stroke,9 respiratory problems,13,14 diabetes,9,12 and cancer.9,15
How do experiences of early adversity confer risk for later psychiatric and medical problems? Epigenetics has been proposed as one possible mechanism.16–18 Epigenetics refers to functionally relevant modifications to the genome that do not involve a change in DNA nucleotide sequence.19 These modifications regulate gene activity and play a role in acute regulation of genes in response to changes in the environment.20,21
DNA methylation is one of the most studied epigenetic mechanisms.22 In mammalian cells, DNA methylation occurs mainly at discrete CpG sites in the genome, regions where cytosine nucleotides occur next to guanine nucleotides.23,24 Although gene regulation is influenced by DNA methylation in other regions of the genome, the impact of methylation in promoters is best understood; it can lead to gene silencing.
The effect of a history of childhood abuse on methylation has been examined previously in candidate gene studies. In two reports, a history of childhood abuse was associated with increased methylation in lymphoblast DNA in the promoter region of the serotonin transporter (SLC6A4), a gene implicated in several neuropsychiatric disorders.25,26 A third report found childhood abuse associated with differences in methylation at four CpG sites in nonpromoter regions of SLC6A4, but these did not withstand correction for multiple comparisons.27 Methylation in the glucocorticoid receptor (NR3C1), a gene involved in the stress response and implicated in several neuropsychiatric disorders has also been examined, and two independent research groups reported that a history of childhood abuse was associated with altered methylation of NR3C1 in hippocampal-derived DNA.28,29
With the advent of methodologies to complete high-throughput methylation profiling of the entire genome,30 novel epigenetic modifications associated with early adversity can also be identified. Two small-scale studies used the Illumina 27K BeadChip, which assays 27,578 CpG promoter-associated sites across the genome to examine methylation differences associated with early adversity. The first study included 28 children: 14 reared from birth in institutions and 14 reared with their parents.31 The second study included 100 adults: 25 who met criteria for Posttraumatic Stress Disorder (PTSD) secondary to childhood abuse; 25 who met criteria for PTSD secondary to other traumas; 25 adults with histories of childhood abuse without PTSD; and 25 adults without PTSD and without histories of childhood abuse.32 The first study reported methylation differences between institution- and family-reared children in numerous CpG sites in genes involved in immune response and cellular signaling systems.31 These findings were not replicated in the second study with adults which used more-rigorous controls for multiple comparisons and contained adults with PTSD secondary to other traumas in the non-abused control group.32
The present investigation examines methylation profile differences in a sample of maltreated and nontraumatized comparison children using the new Illumina 450K BeadChip, which examines a broader range of CpG sites involved in gene regulation. In addition to examining promoter-associated CpG sites, this array also assays CpG sites involved in gene regulation located on the gene body, 3′ untranslated regions (3′UTR), 5′UTRs, and intergenic regions.30 The goal of the current study is to identify novel pathways and mechanisms by which child abuse and other adverse early experiences may confer risk for a range of health problems later in life.
Methods
Participants included 192 children recruited between 2003 and 2010: of these, 96 were maltreated children removed from their parents' care due to reports of abuse and/or neglect within the 6 months of study enrollment, and 96 were controls with no history of maltreatment or exposure to intrafamilial violence. All maltreated children in this investigation also participated in a prior published report of genetic and environmental factors associated with depression33; the cohort of controls was expanded for this current investigation. The 192 children were from 136 families with various numbers of siblings and half-siblings (range: 1–4) in each family.
Children ranged in age from 5 years to 14 years, with a mean age of 10.2 years. The sample was approximately evenly divided by gender (42% male) and was of mixed racial origin (17% European-American, 38% Hispanic, 30% African-American, and 15% biracial). Maltreated and control children did not differ in terms of age (t=0.2, ns), gender (χ2=0.1, ns), or race (χ2=3.3, ns).
Inclusion Criteria
All children were: (1) English-speaking; (2) of normal intelligence (IQ≥70); and (3) had no physical handicaps or medical problems that would interfere with participation in the day camp program where most of assessments for the gene-environment study were collected. Inclusion criteria for maltreated children additionally required recent removal from parental care due to verified report of abuse or neglect. Additional criteria for the control group included: (1) annual household income <$30,000; (2) no contact with protective services and no history of abuse, neglect, or exposure to domestic violence based on mothers' and children's report and verified by the state protective services records; and (3) no lifetime history of psychiatric illness verified using standard research assessments.33–36 IRBs at Yale University and the Connecticut Department of Children and Families approved the present investigation.
Maltreatment History
Multiple informants and data sources (e.g., parents, children, and protective services case records) were used to obtain a best estimate of a child's maltreatment history.37 Before the maltreated children's removal from their parents' care, the children in this study had a mean of three substantiated reports of abuse or neglect (range: 1–7). In addition, 92% experienced more than one type of maltreatment: 65% were physical abused, 24% sexually abused, 83% neglected, 65% emotionally abused, and 70% witnessed domestic violence.
Many experienced the most extreme forms of these categories of maltreatment; 31% of physically abused children required medical attention for injuries; 83% of sexually abused children experienced genital fondling, oral–genital contact, or vaginal or anal intercourse; 37% of children who witnessed domestic violence observed episodes in which a weapon was used or the parent received an injury requiring medical care; and 16% of the emotionally abused children were actually abandoned—either left unattended for several days while a parent was on a drug binge or forsaken by a parent in favor of a partner who sexually abused the child. Although the sample is heterogeneous in terms of number and type of maltreatment experiences, it is homogenous in terms of having a recent experience of maltreatment of sufficient severity to warrant out-of-home placement, and having a recent removal, a substantial stressor common among all the maltreated children in the study.
DNA Specimens
Saliva was collected for DNA extraction, with specimens collected from maltreated children at time of ongoing and extreme stress; within 6 months after an incident of maltreatment of sufficient severity to warrant out-of-home placement, and during a time of multiple ongoing stressors (e.g., frequent changes in placements, separation from siblings, irregular contact with birth parents). Specimens were obtained at the research day camp where many of the assessments for the depression study were obtained.33,35,36,38 Ten ml of Scope mouthwash was dispensed into 50-ml tubes; children swished for 45 seconds, then spit back into the tube. Specimens were refrigerated within 2 hours of collection and DNA extracted using Puregene kits.
To prepare specimens for methylation analyses, 500 ng of genomic DNA was treated with bisulfite reagents included in an EZ-96 DNA methylation kit according to manufacturer's protocol. Unmethylated cytosines were converted to uracils; methylated cytosines remained unchanged. Bisulfite-converted DNA samples were then used in the array-based DNA methylation assay. Methylation levels generated using salivary DNA show excellent test–retest reliability (r =0.99) and high correlation with methylation levels obtained from blood DNA (r =0.97; supplemental supportive data are shown in Appendixes A–E, available online at www.ajpmonline.org).
Array-Based Genome-Wide DNA Methylation
Illumina 450K Methylation BeadChip analyses were completed in 2011 at Keck Biotechnology Laboratory at Yale using standard procedures. This BeadChip interrogates >485,000 CpG sites per sample at single-nucleotide resolution. It covers most designable (96%) RefSeq genes, with 41% of CpG sites in promoter regions, 31% in the gene body, 3% UTRs, and 25% in intergenic regions. GenomeStudio software was used to generate beta values for each CpG site, with beta values ranging from 0.0–1.0 quantifying percentage methylation at each CpG site. GenomeStudio normalizes data using various internal and background probe controls on a HumanMethylation450 BeadChip. Standard quality-control tests were run, and CpG sites that had detection p-value >0.001 were removed to ensure only high-confidence probes were included in subsequent analysis (30/485,578 CpG sites removed, 0.006%).
Data Analysis
In order to take familial correlations into consideration while modeling the effects of maltreatment status on methylation values, data were analyzed using a linear mixed-effects model (LME), which addresses familial correlations by assigning a random effect to each family39 using the “nlme” command in the R software environment (www.cran.r-project.org/web/packages/nlme//index.html). To normalize residuals, an indicator variable to differentially specify random effects for families with one child and those with more than one child was also used. Demographic variables age, gender, and race were also included in analyses as covariates.
There were noteworthy differences in the SDs of methylation beta values for CpG sites with low (<0.2), medium (0.2–0.8), and high (>0.8) mean methylation values (Appendix A, available online at www.ajpmonline.org). Given these differences and the heteroscedasticity of beta values, as recommended by Du and colleagues,40 M-values (logit transformation in log2 scale) were used in all analyses. To correct for multiple comparison testing, the significance threshold for analyses was set to 5.0 × 10−7, consistent with the level recommended by Raykan and colleagues.41
Results
Group Differences in Methylation
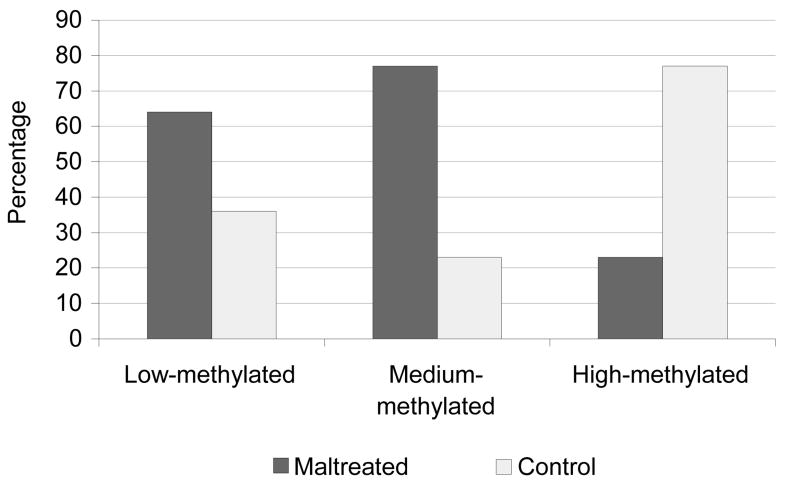
After controlling for demographic factors (e.g., race, gender, age), and multiple comparisons, maltreated and control children had significantly different methylation values at a total of 2868 CpGs sites of the 485K sites included on the array (p<5.0 × 10−7, all sites). On average, methylation values of maltreated and control children differed by 17% (range: 1%–62%). Important CpG sites were identified on all 23 chromosomes; 2113 (74%) at sites with low methylation values (<0.2), 288 (10%) at sites with mid-range methylation values (0.2–0.8), and 467 (16%) at sites with high-range methylation values (>0.8). A consistent pattern emerged: maltreated children generally had elevated methylation values at sites with low- and mid-range methylation values, and reduced methylation at sites with high-methylation values (X2=310, p<0.0001; Figure 1).
Figure 1. Pattern of methylation differences at low, medium, and high-methylated CpG sites.
Forty-five (2%) of the noted CpG sites were in 3′UTR regions, and 213 (7%) in 5′UTR regions. In all, 848 were promoter-associated (30%); 524 were in intergenic regions (18%), and 1238 were on the gene body (43%). Illumina IDs and summary data for each of the noteworthy CpG sites are included in Appendix B (available online at www.ajpmonline.org). As some of the CpG sites are associated with more than one gene, and multiple CpG sites were detected within some genes, an alphabetic listing of associated genes and the number of noteworthy CpG sites within each gene is included in Appendix C (available online at www.ajpmonline.org). Although the majority of genes (88%) had significant methylation differences in only one CpG site, 256 genes had significant methylation differences in two CpG sites, and 27 genes had significant methylation differences in three CpG sites. Eight genes had significant methylation differences in four or more CpG sites: CCDC85C; FANK1; FRG1; TMED2; WNT3A; PTPRN; SLC29A4; and PTPRN2.
Replication of Prior Candidate Gene Findings
As the Illumina 450K Beadchip does not include CpG sites in the promoter regions of SLC6A4 or NR3C1, direct comparisons with prior published candidate gene studies are not feasible. Methylation at one CpG site on the body of the NR3C1gene (cg04111177), however, was significantly lower in maltreated children after controlling for demographic factors and multiple comparisons (p=2.19 × 10−7).
Disease Biomarker-Gene Associations
To identify affiliated diseases, the complete list of genes associated with noteworthy CpG sites was analyzed using GeneGo Metcore® software. The MetCore software tests for significance of markers using a variation of Fisher's exact test with Benjamini-Hochberg False Discovery Rate (FDR) corrections. A substantial number of genes associated with lung, colorectal, prostatic, breast, colon, and ovarian neoplasms were contained in the gene set showing differential methylation among maltreated and comparison children (Table 1).
Table 1. Disease biomarkers significantly associated with genes that are differentially methylated in maltreated and control children.
| Disease Biomarkers | p-value | A | B |
|---|---|---|---|
| Lung Neoplasms_Cell cycle | 7.114E-04 | 16 | 43 |
| Colorectal Neoplasms_Regulation of progression through cell cycle | 4.523E-03 | 20 | 68 |
| Breast neoplasm_GPCR pathway regulation | 4.642E-03 | 23 | 82 |
| Prostatic Neoplasms_Regulation of progression through cell cycle | 6.484E-03 | 20 | 70 |
| Prostatic Neoplasms_Anti-apoptosis | 7.111E-03 | 12 | 35 |
| Breast neoplasm_Cell-cell signaling | 8.090E-03 | 26 | 100 |
| Breast neoplasm_Endothelins | 8.946E-03 | 17 | 58 |
| Prostatic Neoplasms_Transcription | 1.069E-02 | 27 | 107 |
| Breast neoplasm_Cellmotility | 1.097E-02 | 15 | 50 |
| Breast neoplasm_Gene transcription | 1.097E-02 | 15 | 50 |
| Colonic Neoplasms_Cell cycle | 1.514E-02 | 17 | 61 |
| Lung Neoplasms_Regulation of progression through cell cycle | 1.570E-02 | 11 | 34 |
| Ovarian Neoplasms (core network 1) | 1.941E-02 | 28 | 117 |
| Breast neoplasm_CREB1 | 2.132E-02 | 14 | 49 |
| Breast neoplasm_Transcription regulation | 2.364E-02 | 34 | 150 |
Note: Column A includes the number of genes in the disease biomarker set, and Column B represents the number of genes within the set that had methylation differences in the maltreated and control children.
Biological Pathways-Gene Associations
The GeneGo Metcore® software was also used to identify biological pathways and processes affiliated with the set of genes showing differing patterns of methylation in maltreated and control children. Genes associated with a wide array of biological processes relevant to many of the diseases associated with early adversity were identified (Appendix E, available online at www.ajpmonline.org).
Discussion
Child maltreatment was associated with widespread differences in methylation across the entire genome. After rigorously controlling for demographic factors and multiple comparisons, group differences in methylation values emerged at a total of 2868 CpGs sites of the 485K sites included on the array. A general pattern emerged, such that maltreated children had elevated methylation values at CpG sites with methylation values in the low to mid range, and reduced methylation values at CpG sites with methylation values in the high range.
Approximately 20% of noteworthy sites were in intergenic regions. Many CpG islands in intergenic regions are enriched for factor-binding sites and are involved in the three-dimensional organization of the genome and gene regulation.42,43 Transcription factor-binding sites and chromatin insulators within intergenic regions are believed to mediate intra- and inter-chromosomal interactions, affecting gene expression at both proximal and distal locations. There are numerous instances where intergenic genetic variation is associated with disease risk,44 and methylation in intergenic regions has been implicated in neuropsychiatric diseases,45 cardiovascular disease and obesity,46 and a variety of cancers.47–51 As less than 2% of the more than three billion DNA base pairs in human genome code for proteins, it is not surprising that a role in gene regulation and disease risk is emerging for intergenic regions of DNA.
The majority of noteworthy CpG sites, however, were within intragenic regions, with many of the genes with the greatest number of CpG sites associated with diseases previously linked to early childhood adversity.11,13,15,52 Of the genes with the greatest number of noteworthy CpG sites, CCDC85C is involved in cortical development,53PTPRN2 is associated with risk for depression and substance dependence,54 as well as insulin-dependent diabetes mellitus55; FANK1 is linked to asthma,55 and WNT3A is implicated in cancer.56
A substantial number of genes implicated in lung, colorectal, prostatic, breast, colon, and ovarian neoplasms were contained in the set of genes showing differential methylation among maltreated and comparison children (Table 1). In multiple studies, experiences of childhood adversities in general9,15,57—and child abuse in particular57,58—have been associated with increased risk for lung, prostate, breast, and other cancers. The increased risk for cancer associated with adverse early experiences may be due to the health risk behaviors, such as smoking, obesity, and excessive alcohol consumption, that are associated with early adversity,58 and it may, as this study suggests, also be due to epigenetic changes in key genes implicated in cancer.
The gene set showing differential methylation between the maltreated and comparison children also contained genes involved in biological processes relevant in psychiatric and substance use disorders (e.g., neurogenesis, axonal guidance), heart disease (e.g., cardiac development), stroke (development of blood vessel morphogenesis), respiratory disease (e.g., interleukin regulation), diabetes (e.g., leptin signaling), and cancer (e.g., WNT signaling, NOTCH signaling; Appendix A, available online at www.ajpmonline.org). Multiple networks involved in inflammation (e.g., IL-2 signaling) and gene regulation (e.g., translation initiation, regulation of telomere length) were also identified.
Although there is controversy in the field about the use of peripheral DNA methylation markers to study tissue-specific disease processes, there are emerging research findings across multiple areas of medicine documenting the utility of peripheral DNA methylation measures.59–63 A recent review of studies that examined breast cancer methylation markers in plasma and serum concluded that these peripheral methylation markers are potentially a very attractive additive method for early tumor detection.57 Methylation profiling of DNA specimens derived from saliva specimens have also identified epigenetic differences between diabetic patients with end stage renal disease and diabetics without nephrology56; and methylation patterns in DNA derived from postmortem brain tissue, white blood cells, and germlines were found to be different in bipolar patients and controls, with white blood cell and sperm DNA methylation levels mirroring most of the brain findings.59 While additional research is required to better understand the extent and circumstances for similarities and differences in DNA methylation across tissue types, there is a growing body of research suggesting a role for peripheral DNA methylation measures in understanding disease pathology and deriving biomarker sets to predict risk, diagnosis, and prognosis.
The role of epigenetic mechanisms in disease is an evolving area of medicine. Much is unknown about the epigenetic regulation of genes involved in normal development and various disease processes. Epigenetic changes in some genes, such as those involved in establishing ocular dominance and visual acuity, are responsive to environmental manipulations only during certain sensitive periods. Within a narrow window of development, experiences of monocular deprivation can lead to long-term changes in brain development and visual processing that are mediated through epigenetic mechanisms. These changes were previously thought to be permanent, but recent research has shown that they can be reversed with pharmacologic interventions and experiences of environmental enrichment later in development, allowing complete function to be restored.64
The current study provides preliminary support for the notion that epigenetic mechanisms may be involved in conferring risk for a host of health problems later in life among individuals with a history of child maltreatment. The study is limited by its modest sample size, the absence of cross-sectional and longitudinal health data, and failure to obtain gene expression data. This study lays the groundwork, however, for future studies to further explore the role of epigenetic mechanisms in conferring risk for health problems in individuals with histories of early adversity.
Nonreplication of genetics findings has been a problem across multiple areas of medicine.65,66 Future larger-scale studies can employ split-half designs and pyrosequencing to validate methylation findings. In addition, emerging findings suggest the need to examine possible confounds such as lead exposure,67 as well as genetic polymorphisms which may moderate methylation effects.68
Child abuse is a preventable risk factor associated with a range of health problems,69 and a history of child abuse is often associated with poor treatment outcome.70–72 Further research in this area will help to identify novel prevention and intervention strategies to reduce the burden associated with stress-related health problems. Epigenetic changes are often long-lasting, but they need not be permanent.64
Supplementary Material
Acknowledgments
The authors thank the children and families who participated in this research, and the administration of the Connecticut Department of Children and Families for their collaboration on this effort. This work was supported by a Brain and Behavior Research NARSAD Young Investigator award (B-ZY); the Leon Levy Foundation (NW); the NIH—K01 DA24758 (B-ZY), K99/R00 DA022891 (HZ), T32 MH067763 (NW), DA12849 (JG), DA12690 (JG), AA017535 (JG), AA11330R01 (JG), and MH077087 (JK); the National Center for Posttraumatic Stress Disorder-Veterans Affairs Connecticut (HD-P, JG, JK); and the Veterans Affairs Depression Research Enhancement Award Program (Veterans Affairs Connecticut; JG, JK).
Footnotes
No financial disclosures were reported by the authors of this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
References
- 1.DHHS. Child Maltreatment 2009. Washington, DC: DHHS, Administration for Children and Families, Administration on Children, Youth and Families, Children's Bureau; 2010. [Google Scholar]
- 2.Sedlak AJ, Mettenburg J, Basena M, et al. Executive Summary. Washington DC: DHHS, Administration for Children and Families; 2010. Fourth National Incidence Study of Child Abuse and Neglect (NIS–4): Report to Congress. [Google Scholar]
- 3.Wolfner GD, Gelles RJ. A profile of violence toward children: a national study. Child Abuse Negl. 1993;17(2):197–212. doi: 10.1016/0145-2134(93)90040-c. [DOI] [PubMed] [Google Scholar]
- 4.Dowd M. Penn State's Culpability. New York Times. 2011 Nov 8;A:30. [Google Scholar]
- 5.Kendler KS, Bulik CM, Silberg J, Hettema JM, Myers J, Prescott CA. Childhood sexual abuse and adult psychiatric and substance use disorders in women: an epidemiological and cotwin control analysis. Arch Gen Psychiatry. 2000;57(10):953–959. doi: 10.1001/archpsyc.57.10.953. [DOI] [PubMed] [Google Scholar]
- 6.Molnar BE, Buka SL, Kessler RC. Child sexual abuse and subsequent psychopathology: results from the National Comorbidity Survey. Am J Public Health. 2001 May;91(5):753–760. doi: 10.2105/ajph.91.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufman J. Lewis' Child and Adolescent Psychiatry: A Comprehensive Textbook. Fourth. Baltimore, MD: Lippincott Williams & Wilkins; 2007. Child Abuse. [Google Scholar]
- 8.Burke Nj, Hellman JL, Brandon SG, Weems CF, Carrion VG. The impact of adverse childhood experiences on an urban pediatric population. Child Abuse and Neglect. 2011;35:408–413. doi: 10.1016/j.chiabu.2011.02.006. 20110621 DCOM- 20111031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felitti VJ, Anda RF, Nordenberg D, et al. Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. The Adverse Childhood Experiences (ACE) Study. Am J Prev Med. 1998;14(4):245–258. doi: 10.1016/s0749-3797(98)00017-8. [DOI] [PubMed] [Google Scholar]
- 10.Anda RF, Croft JB, Felitti VJ, et al. Adverse childhood experiences and smoking during adolescence and adulthood. JAMA. 1999;282(17):1652–1658. doi: 10.1001/jama.282.17.1652. [DOI] [PubMed] [Google Scholar]
- 11.Dong M, Giles WH, Felitti VJ, et al. Insights into causal pathways for ischemic heart disease: adverse childhood experiences study. Circulation. 2004;110(13):1761–1766. doi: 10.1161/01.CIR.0000143074.54995.7F. [DOI] [PubMed] [Google Scholar]
- 12.Romans S, Belaise C, Martin J, Morris E, Raffi A. Childhood abuse and later medical disorders in women. An epidemiological study. Psychother Psychosom. 2002;71(3):141–150. doi: 10.1159/000056281. [DOI] [PubMed] [Google Scholar]
- 13.Anda RF, Brown DW, Dube SR, Bremner JD, Felitti VJ, Giles WH. Adverse childhood experiences and chronic obstructive pulmonary disease in adults. Am J Prev Med. 2008;34(5):396–403. doi: 10.1016/j.amepre.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dube SR, Fairweather D, Pearson WS, Felitti VJ, Anda RF, Croft JB. Cumulative childhood stress and autoimmune diseases in adults. Psychosom Med. 2009;71(2):243–250. doi: 10.1097/PSY.0b013e3181907888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown DW, Anda RF, Felitti VJ, et al. Adverse childhood experiences are associated with the risk of lung cancer: a prospective cohort study. BMC Public Health. 2010;10(20):20. doi: 10.1186/1471-2458-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danese A, Caspi A, Williams B, et al. Biological embedding of stress through inflammation processes in childhood. Mol Psychiatry. 2011;16(3):244–246. doi: 10.1038/mp.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62(1):3–12. doi: 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shonkoff JP, Boyce WT, McEwen BS. Neuroscience, molecular biology, and the childhood roots of health disparities: building a new framework for health promotion and disease prevention. JAMA. 2009;301(21):2252–2259. doi: 10.1001/jama.2009.754. 20090603 DCOM- 20090608. [DOI] [PubMed] [Google Scholar]
- 19.Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Annu Rev Psychol. 2010;61:439–466. C431–433. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]
- 20.Nestler EJ. Epigenetic mechanisms in psychiatry. Biol Psychiatry. 2009;65(3):189–190. doi: 10.1016/j.biopsych.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Rutten BP, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr Bull. 2009;35(6):1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nestler E, Hyman SE, Malenka RC. Molecular Neuropharmacology: A Foundation for Clinical Neuroscience. Second. New York, N.Y: McGraw-Hill Professional; 2008. [Google Scholar]
- 23.Szyf M. The early life environment and the epigenome. Biochim Biophys Acta. 2009;1790(9):878–885. doi: 10.1016/j.bbagen.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Champagne FA, Curley JP. Maternal regulation of estrogen receptor alpha methylation. Curr Opin Pharmacol. 2008;8(6):735–739. doi: 10.1016/j.coph.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA. Methylation at SLC6A4 is linked to family history of child abuse: an examination of the Iowa Adoptee sample. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):710–713. doi: 10.1002/ajmg.b.31028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA. Methylation at 5HTT mediates the impact of child sex abuse on women's antisocial behavior: an examination of the Iowa adoptee sample. Psychosom Med. 2011;73(1):83–87. doi: 10.1097/PSY.0b013e3181fdd074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vijayendran M, Beach SR, Plume JM, Brody GH, Philibert RA. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front Psychiatry. 2012;3(55):55. doi: 10.3389/fpsyt.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labonte B, Yerko V, Gross J, et al. Differential glucocorticoid receptor exon 1(b), 1(c), and 1(h) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72(1):41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 29.McGowan PO, Sasaki A, D'Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009 Mar;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011 Oct;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. Epub 2011 Aug 2012. [DOI] [PubMed] [Google Scholar]
- 31.Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, Grigorenko EL. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Dev Psychopathol. 2011 Nov 29;29:1–13. doi: 10.1017/S0954579411000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AK, Conneely KN, Kilaru V, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011 Jun 28;28(10):31212. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman J, Yang BZ, Douglas-Palumberi H, et al. Brain-Derived Neurotrophic Factor-5- HTTLPR Gene Interactions and Environmental Modifiers of Depression in Children. Biological Psychiatry. 2006;59:673–680. doi: 10.1016/j.biopsych.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Grasso D, Boonsiri J, Lipschitz D, et al. Posttraumatic stress disorder: the missed diagnosis. Child Welfare. 2009;88(4):157–176. [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufman J, Yang BZ, Douglas-Palumberi H, et al. Social supports and serotonin transporter gene moderate depression in maltreated children. Proc Natl Acad Sci USA. 2004;101(49):17316–17321. doi: 10.1073/pnas.0404376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weder N, Yang BZ, Douglas-Palumberi H, et al. MAOA genotype, maltreatment, and aggressive behavior: the changing impact of genotype at varying levels of trauma. Biol Psychiatry. 2009;65(5):417–424. doi: 10.1016/j.biopsych.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman J, Jones B, Steiglitz E, Vitulano L, Mannarino A. The use of multiple informants to assess children's maltreatment experiences. J Family Violence. 1994;9:227–248. [Google Scholar]
- 38.Kaufman J, Yang BZ, Douglas-Palumberi H, et al. Genetic and environmental predictors of early alcohol use. Biol Psychiatry. 2007;61(11):1228–1234. doi: 10.1016/j.biopsych.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 39.McCulluch CE, Neuhaus JM. General Linear Mixed Models. In: Colton PAT, editor. Encyclopedia of Biostatistics. Hoboken, NJ: Wiley; 2005. [Google Scholar]
- 40.Du P, Zhang X, Huang C, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodges E, Molaro A, Dos Santos CO, et al. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell. 2011 Oct 07;44(1):17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Corces VG. Chromatin insulators: a role in nuclear organization and gene expression. Adv Cancer Res. 2011;110:43–76. doi: 10.1016/B978-0-12-386469-7.00003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cotnoir-White D, Laperriere D, Mader S. Evolution of the repertoire of nuclear receptor binding sites in genomes. Mol Cell Endocrinol. 2011;334(1-2):76–82. doi: 10.1016/j.mce.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 45.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson M, Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD. Developmental plasticity and developmental origins of non-communicable disease: theoretical considerations and epigenetic mechanisms. Prog Biophys Mol Biol. 2011;106(1):272–280. doi: 10.1016/j.pbiomolbio.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Dedeurwaerder S, Fumagalli D, Fuks F. Unravelling the epigenomic dimension of breast cancers. Curr Opin Oncol. 2011;23(6):559–565. doi: 10.1097/CCO.0b013e32834bd481. [DOI] [PubMed] [Google Scholar]
- 48.Ogoshi K, Hashimoto S, Nakatani Y, et al. Genome-wide profiling of DNA methylation in human cancer cells. Genomics. 2011;98(4):280–287. doi: 10.1016/j.ygeno.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem. 2010;52:145–167. doi: 10.1016/s0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- 50.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775(1):138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Yegnasubramanian S, Wu Z, Haffner MC, et al. Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences. BMC Genomics. 2011;12(313):313. doi: 10.1186/1471-2164-12-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dube SR, Anda RF, Felitti VJ, Edwards VJ, Croft JB. Adverse childhood experiences and personal alcohol abuse as an adult. Addict Behav. 2002 Sep-Oct;27(5):713–725. doi: 10.1016/s0306-4603(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 53.Mori N, Kuwamura M, Tanaka N, et al. Ccdc85c Encoding a Protein at Apical Junctions of Radial Glia Is Disrupted in Hemorrhagic Hydrocephalus (hhy) Mice. Am J Pathol. 2012 Jan;180(1):314–327. doi: 10.1016/j.ajpath.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 54.Yang BZ, Han S, Kranzler HR, Farrer LA, Gelernter J. A genomewide linkage scan of cocaine dependence and major depressive episode in two populations. Neuropsychopharmacology. 2011;36(12):2422–2430. doi: 10.1038/npp.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremblay K, Lemire M, Potvin C, et al. Genes to diseases (G2D) computational method to identify asthma candidate genes. PLoS One. 2008;3(8):e2907. doi: 10.1371/journal.pone.0002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev. 2007;3(1):30–38. doi: 10.1007/s12015-007-0006-6. [DOI] [PubMed] [Google Scholar]
- 57.Morton PM, Schafer MH, Ferraro KF. Does childhood misfortune increase cancer risk in adulthood? J Aging Health. 2012;4:4. doi: 10.1177/0898264312449184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuller-Thomson E, Brennenstuhl S. Making a link between childhood physical abuse and cancer: results from a regional representative survey. Cancer. 2009;115(14):3341–3350. doi: 10.1002/cncr.24372. [DOI] [PubMed] [Google Scholar]
- 59.Brennan K, Garcias-Closas M, Orr N, et al. Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res. 2012;28:28. doi: 10.1158/0008-5472.CAN-11-3157. [DOI] [PubMed] [Google Scholar]
- 60.Sapienza C, Lee J, Powell J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6(1):20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 61.Suijkerbuijk KP, van Diest PJ, van der Wall E. Improving early breast cancer detection: focus on methylation. Ann Oncol. 2011;22(1):24–29. doi: 10.1093/annonc/mdq305. [DOI] [PubMed] [Google Scholar]
- 62.Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20(24):4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaminsky Z, Tochigi M, Jia P, et al. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Mol Psychiatry. 2011;7:7. doi: 10.1038/mp.2011.64. [DOI] [PubMed] [Google Scholar]
- 64.Weder N, Kaufman J. Critical periods revisited: implications for intervention with traumatized children. J Am Acad Child Adolesc Psychiatry. 2011 Nov;50(11):1087–1089. doi: 10.1016/j.jaac.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Redden DT, Allison DB. Nonreplication in genetic association studies of obesity and diabetes research. J Nutr. 2003;133(11):3323–3326. doi: 10.1093/jn/133.11.3323. [DOI] [PubMed] [Google Scholar]
- 66.Tan EC, Chong SA. Identification of genes for schizophrenia susceptibility. Ann Acad Med Singapore. 2000 May;29(3):305–312. [PubMed] [Google Scholar]
- 67.Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010 Jun;118(6):790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.vanIJzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R. Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry. 2010;68(5):405–407. doi: 10.1016/j.biopsych.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eckenrode J, Ganzel B, Henderson CR, Jr, et al. Preventing child abuse and neglect with a program of nurse home visitation: the limiting effects of domestic violence. JAMA. 2000;284(11):1385–1391. doi: 10.1001/jama.284.11.1385. [DOI] [PubMed] [Google Scholar]
- 70.Boxer GH, Carson J, Miller BD. Neglect contributing to tertiary hospitalization in childhood asthma. Child Abuse Negl. 1988;12(4):491–501. doi: 10.1016/0145-2134(88)90066-x. [DOI] [PubMed] [Google Scholar]
- 71.Nanni V, Uher R, Danese A. Childhood maltreatment predicts unfavorable course of illness and treatment outcome in depression: a meta-analysis. Am J Psychiatr. 2012;169(2):141–151. doi: 10.1176/appi.ajp.2011.11020335. [DOI] [PubMed] [Google Scholar]
- 72.Nemeroff CB, Heim CM, Thase ME, et al. Differential responses to psychotherapy versus pharmacotherapy in patients with chronic forms of major depression and childhood trauma. Proc Natl Acad Sci USA. 2003;100(24):14293–14296. doi: 10.1073/pnas.2336126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.