Abstract
Patient motion can cause serious artifacts in diffusion tensor imaging (DTI), diminishing the reliability of the estimated diffusion tensor information. Studies in this field have so far been limited mainly to the correction of miniscule physiological motion. In order to correct for gross patient motion it is not sufficient to correct for misregistration between successive shots; the change in the diffusion-encoding direction must also be accounted for. This becomes particularly important for multishot sequences, whereby—in the presence of motion—each shot is encoded with a different diffusion weighting. In this study a general mathematical framework to correct for gross patient motion present in a multishot and multicoil DTI scan is presented. A signal model is presented that includes the effect of rotational and translational motion in the patient frame of reference. This model was used to create a nonlinear least-squares formulation, from which the diffusion tensors were obtained using a nonlinear conjugate gradient algorithm. Applications to both phantom simulations and in vivo studies showed that in the case of gross motion the proposed algorithm performs superiorly compared to conventional methods used for tensor estimation.
Keywords: diffusion, DTI, motion, parallel imaging
Diffusion-weighted imaging (DWI) has been known for its ability to provide a unique tissue contrast that can be used for early detection of ischemic stroke (1). Diffusion tensor imaging (DTI) is an extension of DWI whereby multiple diffusion-weighted measurements are applied to obtain a diffusion tensor for each pixel (2,3). Because of prolonged scan times and motion sensitivity of diffusion-weighting gradients, countermeasures to prevent motion-induced artifacts in DTI are important for the success of a study.
One way to eliminate the effects of motion is to “freeze” the anatomy being imaged by using snapshot imaging techniques—the most popular of which is single-shot EPI (sshEPI). However, susceptibility artifacts due to the very low bandwidth per pixel in the phase-encode direction and T2*-induced blurring caused by the long EPI readout substantially limit the applicability of this method. To address the limitations of sshEPI, the use of parallel imaging has been suggested (4). Nevertheless, the maximum speedup in the phase encode direction that can be achieved by parallel imaging is limited and, thus, the degree of distortion reduction. Another method employed frequently is to use interleaved k-space acquisitions to traverse faster through k-space and, in this way, to reduce the aforementioned distortions. However, diffusion-weighted multishot sequences are very sensitive to motion (5–8). Each interleaf accrues a different (typically nonlinear) phase in the image domain. Navigator-based methods that utilize 1D navigators (5,8–13), 2D navigators (14–17), or self-navigating trajectories (18–20) have demonstrated variable efficacy to eliminate these motion artifacts.
While the phase-navigation used in DTI is usually focused on minuscule motion (i.e., motion in a range that affects image phase but not pixel position), gross patient motion becomes a concern, especially in the case of uncooperative patients such as children and patients that suffer from a specific medical condition (e.g., stroke) that keeps them from staying stationary. Mere misregistration can be corrected for by advanced registration methods. However, motion exposes the object to a different diffusion-encoding gradient than the desired one (21). Neglecting the change in the diffusion encoding caused by motion can cause erroneous estimations of the diffusion tensor orientation and anisotropy (22) (Fig. 1). The studies on the consequences of gross motion during diffusion encoding are limited, mostly because distortions on a much smaller scale (e.g., brain pulsation and miniscule bulk motion) have often been more of a concern. Only recently, an approach to correct for these deviations from the desired encoding has been suggested for sshEPI (21).
FIG. 1.
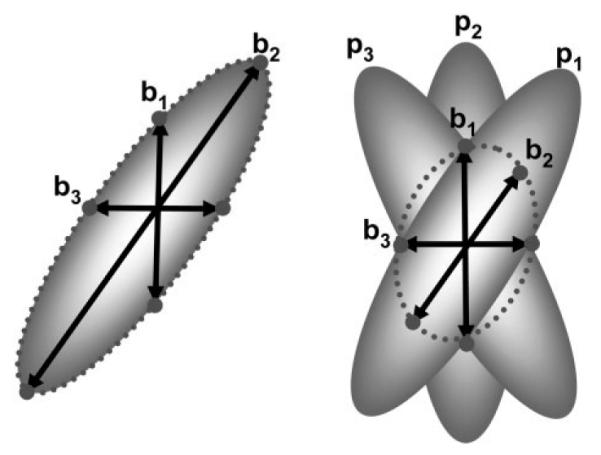
The effect of random subject rotation on tensor estimation. Let bi denote the applied diffusion encoding and let pi represent the tensor orientation, which depends on subject motion. In case of no motion, the estimated diffusion tensor (dotted line) is the same as the actual tensor (left). If there is rotational motion (right), the estimated tensor orientation will be different and the anisotropy will be lower than the actual values. The error in FA and orientation will depend on the severity of the motion and the pattern of orientation changes.
In the case of multishot data the situation is much more complex. Several interleaves, potentially with different actual diffusion-encoding directions, contribute to form a single diffusion-weighted image. Thus, the correction for motion effects on the image data and the diffusion-encoding has to occur simultaneously. Recently, it has been shown that in the absence of diffusion weighting, rigid body motion that occurred in a multishot sequence can be corrected during gridding. This correction was accomplished using parallel imaging techniques to compensate for motion-induced k-space undersampling (23). However, in the presence of diffusion-encoding gradients subject motion not only causes k-space undersampling, but also introduces a different diffusion-weighting matrix for each interleaf. While accounting for motion in DTI is relatively simple for single-shot acquisitions (21), the conventional 2-step method for tensor estimation, that is 1) the reconstruction of individual diffusion-weighted images followed by 2) multivariate regression, cannot be used for multishot acquisition.
In this study we propose a novel single-step algorithm that can be used to estimate diffusion tensors directly from complex k-space data. We start by a very general formulation of the problem for the case of multicoil, multishot arbitrary k-space data including an arbitrary number of diffusion-weighting directions and rigid body motion. Next, a nonlinear conjugate gradient (NLCG) (24)-based algorithm is used to minimize the cost function derived from this model. In both simulations and experiments it is demonstrated that the proposed algorithm is efficient in correcting both small and gross interscan motions.
THEORY
In the general case when there is no motion, the signal equation for a multishot DTI acquisition using a multichannel coil is given by the following expression:
| (1) |
where γ stands for the coil number, δ for the diffusion-weighting direction number, ζ for the interleaf number, κ for k-space point within an interleaf, nρ for the number of image space points (=N2), ρ for image space point, and i,l for tensor indices (i,l = 1,2,3). Here, m(rρ) is the nondiffusion-weighted image and sγ(rρ) is the coil sensitivity. The existence of rigid body motion requires Eq. [1] to be modified in order to account for rigid body rotations and translations. Specifically, the artifacts caused by motion are going to be of a different nature depending on whether they occur during diffusion encoding (intrascan motion) or between shots (interscan motion).
Intrascan Motion
In this section, we will focus on small motion that happens while the strong diffusion-encoding gradients are played out. Small motion (e.g., brain pulsation) during the application of diffusion-encoding gradients results in an additional, unpredictable phase term that varies spatially and for each interleaf, and needs to be removed prior to reconstruction as it conflicts with regular image encoding (5).
The approach taken in this study to correct for this perturbation is similar to the one in Ref. (19): the nonlinear phase accrued for each interleaf is treated as a modification of the underlying image encoding function and in this way it is considered part of the complex coil sensitivity in Eq. [1]. Hence, the coil sensitivity maps in Eq. [1] have to be modified accordingly to reflect the different spatially varying (nonlinear) phase information ϕδ,ζ(rρ) for each diffusion-encoding direction δ and interleaf ζ:
| (2) |
Interscan Motion
In this section we will focus on the effect of (rigid body) motion between shots on the diffusion-weighted images. The correction of interscan rotations and translations is based on defining a patient frame of reference and finding the spatial transformations needed to warp images from a reference frame defined by the scanner frame of reference to the patient frame of reference (23). In other words, in case of motion we are locking on to the patient so that the anatomy under examination is stationary and the scanner (including the RF coil elements and the diffusion-encoding gradients) is “moving” around the patient. Using the Fourier Transform properties, the rotational motion is corrected by counterrotating the k-space trajectories and translational motion is corrected by applying a linear phase to the complex k-space data. In addition, coil sensitivity profiles are also counterrotated and countertranslated to account for the altered coil sensitivity exposure of the anatomy under examination (23). Similarly, the diffusion-encoding direction will also change due to rotation. Thus, in the case when the anatomy under examination is rotated, spins will be exposed to different diffusion-encoding fields that differ from the desired field for each shot.
Following these observations, and assuming a stationary coil geometry, we can rewrite Eq. [1] in the “scanner-frame-of-reference” after adding the effect of rotation and translation:
| (3) |
Here, Rδ,ζ is the rotation matrix and Δrδ,ζ the translation vector from the scanner frame of reference to the patient frame of reference. Substituting in the summation represents a transformation from the scanner frame of reference to patient frame of reference, in which case one gets (after dropping the prime symbol):
| (4) |
Defining and , the final equation for a DTI acquisition in the case of rigid body motion becomes:
| (5) |
Conventional tensor estimation schemes do not consider gross rigid body motion. In these schemes, Eq. [1] is solved for D(rρ) in two steps: 1) Reconstruction of δ diffusion-weighted images m(rρ)e−∑i,l[bδ]i,l[D(rρ)]i,l, 2) calculation of diffusion tensors D(rρ) from diffusion-weighted images using multivariate regression. However, if there is rigid body motion the diffusion-weighted image corresponding to each interleaf is given by m(rρ)e−∑i,l[bδ’,ζ]i,l[D(rρ)]i,l. Since each interleaf has essentially undergone a different diffusion encoding, it becomes unfeasible to reconstruct correct individual diffusion-weighted images. In this study a single-step optimization algorithm to estimate the diffusion tensors D(rρ) from the unity of all k-space data is presented.
Nonlinear Conjugate Gradient Method for Direct Tensor Estimation
In Eq. [5] the unknowns to be determined are the diffusion tensors D(rρ) and the nondiffusion-weighted image m(rρ). To solve for D and m simultaneously, an NLCG-based method is needed to solve the cost function f(D,m) defined in Eq. [6] below. A flow chart showing the iterative reconstruction algorithm to obtain the diffusion tensor information on a per-pixel basis is shown in Fig. 2. In essence, this algorithm tries to minimize the error (in a least-squares sense) between the acquired k-space data d and the corresponding k-space data following our model approach after factoring in motion terms (i.e., phase error, rotation, translation):
| (6) |
In order to minimize the cost function f(D,m), an NLCG method with Polak–Ribiere and Secant 1D line search was used (24; http://www.cs.cmu.edu/~quake-papers/painless-conjugate-gradient.pdf). Inherent to all variants of conjugate gradient (CG) methods is their fast convergence rates, the minimal number of function evaluations, low memory requirements, and the non-necessity of evaluating the exact Hessian matrix—rendering them ideal for solving high dimensional optimization problems. In this sense, CG methods are more memory-efficient and converge faster compared to other methods such as Levenberg–Marquardt or Steepest Descent.
FIG. 2.
Flowchart of the reconstruction algorithm. Evaluation of the derivative of f(D,m) with respect to D(rρ) and m(rρ) can be carried out efficiently by using inverse and forward gridding, represented by FT1 and FT2.
The NLCG approach requires the evaluation of the first derivative vector with respect to the unknown parameters at each iteration step. Since this evaluation is a major time-limiting factor for the reconstruction, a fast algorithm was introduced which is based on forward (25) and inverse gridding (26) to evaluate the first and second derivative of the cost function with respect to the diffusion tensors D and the nondiffusion-weighted image m. By defining a diffusion-weighted image for the γth coil, δth diffusion weighting and ζth interleaf as:
| (7) |
the cost function can be rewritten as:
| (8) |
Here, [●]* denotes the complex conjugate. The first derivative of this cost function with respect to the (i,l)th (i,l = 1,2,3) element of the diffusion tensor at location rρ (ρ= 1…N2) becomes:
| (9) |
The evaluation of Eq. [9] takes a significant amount of effort due to the inner summations over ρ. In fact, these summations represent a discrete Fourier transform (DFT) of the dwi image (Eq. [7]) onto an arbitrary k-space trajectory given by kδ,ζ,κ. Therefore, this DFT operation can be sped up significantly by approximating it by an inverse FFT followed by an inverse gridding operation:
| (10) |
Here, GRID−1δ,ζ denotes the inverse gridding operation of Cartesian DWI data onto the ζth interleaf of the δth diffusion-weighting direction. To be consistent with the notation in Ref. (27), this operation is represented by
| (11) |
Here, FT2 includes roll-off correction, zero padding to a 2x grid, FFT into k-space, and inverse gridding. This way, Eq. [11] becomes:
| (12) |
which eventually evaluates to:
| (13) |
From Eq. [13] it can be seen that the inner summations over all k-space sample points κ represent an inverse discrete Fourier Transform. Again, this inverse DFT can be replaced with forward gridding and FFT to improve efficiency:
| (14) |
Here, GRIDδ,ζ denotes forward gridding and the FT1 operation includes forward gridding on a 2x grid, FFT into image domain, matrix cropping, and roll-off correction. Finally, Eq. [9] can be simplified to:
| (15) |
Here, the Re{●} operation stands for taking the real part of the argument within the curly braces. It is possible to improve the convergence rate of the NLCG method using preconditioners or by improving the line search for each search direction. Both of these are possible by finding an approximation of the Hessian matrix. This approximation includes only the diagonal entries of the Hessian matrix, with the cost of increasing the number of iterations in 1D line searches or imperfect regularization. Using similar methods as described above, the second derivative of the cost function with respect to the tensor elements becomes:
| (16) |
where nκ is the number of k-space points per interleaf.
Using a very similar approach, it is possible to evaluate the first and second derivative of the cost function with respect to the nondiffusion-weighted image:
| (17) |
| (18) |
Because the units of D (s/mm2) and m (a.u.) are different, their scaling will also be different. In this case, preconditioning becomes necessary to bring these parameters to the same scaling in order to speed up convergence. In this study, regularization was accomplished using the diagonal Hessian matrices, as given by Eqs. [16] and [18]. Incorporation of Eqs. [15–18] into the NLCG optimization is explained in Ref. (24) and in http://www.cs.cmu.edu/~quake-papers/painless-conjugate-gradient.pdf.
MATERIALS AND METHODS
Pulse Sequence
For this study a diffusion-weighted spin echo single-shot spiral-in and interleaved spiral-out sequence was used. Specifically, the single-shot spiral-in part was used to acquire a fully sampled low-resolution navigator image for each excitation, while the spiral-out part was used to acquire individual interleaves for the reconstruction of a final high-resolution image (Fig. 3). The motion parameters (rotation and translation) were then obtained by using the low-resolution navigator images and performing image coregistration. In this study, the Pearson correlation coefficient and simplex-based optimization were used to find the motion parameters (23), but other registration methods or positional detection methods could be used without loss of generality. The coil sensitivities were also obtained from these low-resolution navigator images to capture the nonlinear phase accrued by each interleaf. Recently, it has been shown that navigators of 32 × 32 are of sufficient resolution to reliably identify motion of the extent we are trying to correct for with this approach (28).
FIG. 3.

The pulse sequence used for in vivo studies. The readout part consists of a single-shot spiral-in and an interleaved variable-density spiral-out acquisition. The spiral-in readout gives a low-resolution navigator image that can be used to estimate the amount of gross patient motion for each shot. In addition the navigator is used for coil sensitivity estimation, nonlinear phase correction, and the elimination of irreversibly corrupted k-space data. The variable-density spiral out readout makes up one interleaf of the final k-space data to form a high-resolution diffusion-weighted image.
Computer Simulations
All simulations and postprocessing steps were performed on a PC (Dell, Pentium IV 3.4 GHz, 2.5 GB 14 RAM) running IDL 6.1 Student Edition (RSI, Boulder, CO). Synthetic data for 8-interleaf variable-density spiral (pitch factor = 3.0) (29) and 8-interleaf EPI acquisitions were generated using a synthetic DTI dataset containing six tensor element images and one image with no diffusion weighting. The synthetic phantom contained a circular ring and two crossing rods (Fig. 4). The major eigenvalue λ1 was assumed to be 1000 × 10−6mm2/s, while λ2 and λ3 were both 100 × 10−6mm2/s. For the circular ring the tensors were oriented so that the major eigenvector e1 was along the tangent of the ring. For the two crossing rods e1 was oriented along the rods, and for the area where the rods intersect e1 was pointing perpendicular to the plane of the phantom.
FIG. 4.
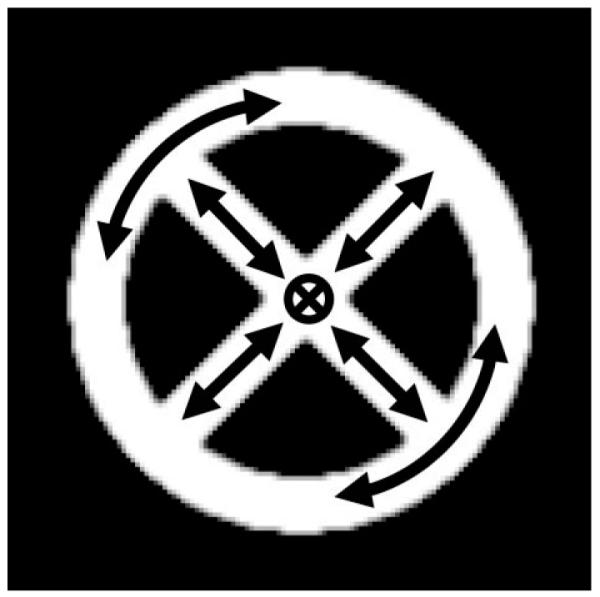
Phantom image used for computer simulations. The software phantom consists of a circular ring in which the major eigenvector of the diffusion tensor is oriented along the ring and four spokes in which the major eigenvector is oriented along the spoke (arrows). In the area where the spokes intersect, the major eigenvector is oriented out of the page. The FA value inside the phantom is 0.88.
Complex k-space data were generated using the model in Eq. [3]. First, the nδ × nζ diffusion-weighted images were obtained with the diffusion gradient directions [(0 0 0)T, (1 1 0)T,(1 0 1)T, (0 1 −1)T, (−1 1 0)T, (0 1 1)T, (1 0 −1)T] and b = 800 s/mm2. Rotation, translation, and random phase in image domain were simulated. In order to simulate the effect of rotation, the diffusion tensors D(rρ) and the image were rotated according to Eq. [3]. Thereafter, diffusion weighting was applied by multiplying the non-diffusion-weighted image by the diffusion-weighting exponential term. Next, the image rotation was established via IDL’s built-in rot function which uses bicubic interpolation to resample the motion simulated image onto a Cartesian grid. Translation of the image was accomplished by applying a linear phase in the Fourier domain. The random phase was simulated by applying a linear phase in the image domain. Next, these images were multiplied by eight coil sensitivities to simulate diffusion-weighted images dwiγ,δ,ζ(rρ) obtained with a phased array coil. After FFT, these nγ × nδ × nζ (= 8 × 7 × 8 = 448) images were inverse gridded onto the prescribed variable-density spiral trajectory to create the synthetic k-space data. The coil sensitivities were also altered to include the effect of random linear phase. The simulations were carried out with 1) no motion; 2) θ=±10° rotation with probability of 1/2 for each angle, Δr =±1.3 pixels translation with probability of 1/2 for each shift, and ψ=±1 pixel shift in k-space, uniformly distributed; and 3) θ=±20° rotation with probability of 1/2 for each angle, Δr =±2.6 pixels translation with probability of 1/2 for each shift, and ψ=±2 pixel shift in k-space, uniformly distributed.
To assess the performance of our motion correction algorithm, the generated data were reconstructed using four methods: A) Reconstruction with no motion correction using conventional gridding: individual diffusion-weighted images were generated by means of gridding reconstruction of simulated k-space data. Thereafter, multivariate regression was performed to estimate the elements of the diffusion tensor in each voxel. B) Reconstruction with phase correction using CG-SENSE: Using the approach in Ref. (19), an augmented iterative SENSE reconstruction was used to obtain diffusion-weighted images. In this method the conventional coil sensitivity was expanded by the phase map that reflects the effect of motion during diffusion-encoding. Again, pixel-wise estimation of the diffusion tensor elements was performed by multivariate regression. C) Reconstruction with phase and motion correction using CG-SENSE: In this approach individual diffusion-weighted images were reconstructed using augmented iterative SENSE reconstruction (see B) (19) and motion correction (23). Similar to A and B, tensor elements were computed by multivariate regression. However, the change in the b matrix due to motion, particularly rotation, was neglected. D) Reconstruction with phase and motion and diffusion-encoding direction correction: Direct estimation of the tensor element maps was performed using NLCG under the consideration of rigid body motion, phase errors due to motion during the presence of diffusion-encoding, and diffusion-encoding errors due to rigid body motion.
In order to eliminate errors that may result from inaccuracies in motion detection, that is, registration inaccuracies, the motion parameters assumed for the simulation (i.e., simulated rotation) were directly used during reconstruction. The coil sensitivities used to perform the simulations were also included directly in the reconstruction after the addition of random linear phase to eliminate any errors that might arise due to the inaccuracies in coil sensitivity estimation. The four reconstructed images were compared with the original reference image. Three measures were used to assess the performance of our algorithm: 1) FA maps; 2) mean diffusivity maps; and 3) the angular deviation between the major eigenvectors of the original image and the reconstructed images in each voxel.
In Vivo Studies
In vivo studies were carried out on four volunteers on a 1.5T whole body system (Signa CVi,; GE, Milwaukee, WI) with a high-performance gradient set (maximum gradient strength = 50 mT/m, maximum slew rate = 150 mT/m/s). Signal reception was accomplished using an 8-channel receive only head array coil (MRI devices, Milwaukee, WI), while signal excitation was performed with the integrated quadrature body coil. All human studies were approved by the internal review board of our institution. Informed written consent was obtained from the subject after the nature of the study was fully explained to them.
The DTI sequence used for this in vivo study employed a single-shot spiral-in and an interleaved variable-density spiral-out readout, where the center of k-space was acquired at the instance when the diffusion-weighted spin echo was formed (Fig. 3). The scan parameters were as follows: TR/TE = 2500/55 ms, six diffusion gradient directions [(1 1 0)T, (1 0 1)T, (0 1 −1)T, (−1 1 0)T, (0 1 1)T, (1 0 −1)T], NEX = 4, matrix size = 128 × 128, navigator matrix size = 32 × 32, 8 interleaves, variable density pitch factor = 3. Two extra scans were also obtained with no diffusion weighting (i.e., with the diffusion-encoding gradients turned off).
During the acquisition of DTI data the subjects were asked to move their head to a varying extent. Specifically, the subject performed a fast head rotation, followed by 20–30 sec in the resting state. The head rotation was approximately around the axis defined by the point where the subject’s head was touching the coil and the line parallel to the B0 field. Using the same parameters an additional reference DTI scan was also obtained where the subject was asked to stay stationary. This acquisition was deemed the gold standard for our comparative evaluations. The four reconstruction methods as described for the Computer Simulations section were used to reconstruct the in vivo data.
To evaluate the quality and performance of the reconstruction algorithms, two regions of interest (ROIs) were selected in the genu and splenium of the corpus callosum. The performance metrics described for the phantom study were used to evaluate the quality of reconstruction in these areas. The image reconstructed with method C (phase and motion correction using iterative SENSE) in the case of no subject motion was used as a reference for all the reconstructed images and for all degrees of motion. In order to guarantee alignment between the reconstructed images and the reference image, registration between the nondiffusion-weighted images and the diffusion tensor maps were accomplished before the comparisons were done.
RESULTS
Computer Simulations
Figures 5 and 6 show the consequences of simulated motion and random shot-to-shot phase on simulated 8-inter-leaf spiral and EPI sequences. Specifically, Fig. 5 shows the FA maps obtained using the four reconstruction methods under different degrees of motion using vd-spiral and EPI trajectories. In the case of no simulated motion, all methods perform similarly (Fig. 5a–d,m–p). In the case of simulated rotational motion the application of motion correction with method C (CG SENSE with motion correction) improves the image quality compared to methods A (conventional gridding) and B (CG SENSE with no motion correction) (Fig. 5g,k,s,w). However, since the change in the diffusion-encoding direction was not accounted for, some residual fluctuations in the FA maps still remain in method C. The application of method D (NLCG) removes these fluctuations and yields a completely uniform FA map, which was very similar to the gold standard (Fig. 5h,l,t,x). Figure 6 shows the angular deviation of the major eigenvectors between the reconstructed and original (motion-free) image. In the absence of simulated motion, all methods performed similarly, as seen by the low angular deviation of the major eigenvectors from the reference orientations (Fig. 6a–d,m–p). In the case of simulated motion the application of methods A and B caused the major eigenvectors to deviate significantly from the true values because of motion-related artifacts (Fig. 6e,f,i,j,q,r,u,v).
FIG. 5.
The FA maps obtained from the computer simulations using the four reconstruction methods under several degrees of rotational motion and using EPI and spiral trajectories. All methods performed similar when no rotational motion was simulated (first and fourth rows). In the case of simulated rotational motion the FA maps reconstructed with methods A and B show misregistration and aliasing artifacts due to motion (e,f,i,j,q,r,u,v). These artifacts are partly reduced by the application of motion correction using method C; however, some residual artifacts still remain due to the uncorrected diffusion-encoding direction (g,k,s,w). These are removed successfully by the application of NLCG (method D), which gave a completely alias-free image (h,l,t,x).
FIG. 6.
Angular deviations of the major eigenvectors between reconstructed images and the reference image. If there was no simulated motion all methods performed similar (first and fourth rows). In the case of simulated motion direct reconstruction with no motion correction led to significant deviation (e,f,i,j,q,r,u,v). After the application of motion correction and reconstruction with CG-SENSE (method C) the orientation error decreased; however, some residual error remained because the change in diffusion-encoding direction was not taken into account (g,k,s,w). Motion and diffusion-encoding direction correction using NLCG significantly reduced orientation errors to ≈0.3° for all level of motion severity (fourth column).
After the application of motion correction with method C, the deviation decreased but some residual error remained. This is due to unaccounted changes in diffusion-weighting directions following conventional motion correction (Fig. 6g,k,s,w). When accounting for the change in the diffusion-encoding direction by using method D, the error significantly decreased (Fig. 6h,l,t,x).
The efficacy of NLCG in eliminating rigid body motion-related artifacts can also be seen in Table 1, where the average angular deviation, FA, and mean diffusivity over the whole phantom are given. When there is no motion all methods perform similarly—as seen by the low angular deviation (≈0.1° maximum) and the closeness of the FA (0.88) and mean diffusivity values (371–373 × 10−6 mm2/sec) to the reference values of FAref = 0.88 and Mean Difref = 371 × 10−6 mm2/sec. In the case of simulated motion the mean angular deviation decreased significantly by the application of NLCG (method D) compared to CG-SENSE (method C) from 2.82 and 8.48 degrees to 0.22 and 0.26 degrees, respectively, for the two degrees of simulated motion in the case of EPI and from 2.10 and 7.09 to 0.18 and 0.19 in case of vd-spiral. On the other hand, application of method D also increased the mean FA slightly for all simulations with motion—consistent with the fact that uncorrected diffusion-encoding directions can cause a decrease in the measured FA value (22). The mean FA values obtained by method D are around 0.88 for all degrees of simulated motion, which is identical to the real value. No significant change in mean diffusivity was observed between methods C and D. This was due to the fact that the mean diffusivity is, by definition, independent of the orientation of the tensor.
Table 1.
Average FA, Mean Diffusivity, and Angular Deviation Values Obtained for the Phantom Experiments
| Method A No Correction (Gridding) |
Method B Phase Correction (CG SENSE) |
Method C Phase & Motion Correction (CG SENSE) |
Method D Phase & Motion & Diff. Enc. Dir. Correction (NLCG) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt Ref. (degrees) |
FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt Ref. (degrees) |
FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt Ref. (degrees) |
FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt Ref. (degrees) |
||
| EPI | No motion | 0.88 | 371.0 | 0.06 | 0.88 | 370.9 | 0.06 | 0.88 | 370.9 | 0.07 | 0.88 | 372.8 | 0.07 |
| Small motiona |
0.96 | 478.8 | 50.13 | 0.92 | 415.9 | 20.23 | 0.86 | 370.2 | 2.82 | 0.88 | 374.4 | 0.22 | |
| Moderate motionb |
0.97 | 453.5 | 51.75 | 0.99 | 461.4 | 34.46 | 0.84 | 365.7 | 8.48 | 0.89 | 370.0 | 0.26 | |
| SPIRAL | No motion | 0.88 | 371.0 | 0.07 | 0.88 | 370.9 | 0.06 | 0.88 | 370.9 | 0.07 | 0.88 | 372.8 | 0.07 |
| Small motiona |
1.07 | 341.0 | 49.35 | 0.92 | 393.4 | 13.07 | 0.86 | 370.9 | 2.10 | 0.88 | 373.9 | 0.18 | |
| Moderate motiona |
1.02 | 402.3 | 43.11 | 0.90 | 377.4 | 21.32 | 0.82 | 367.7 | 7.09 | 0.88 | 373.1 | 0.19 | |
For all datasets, FAREF = 0.88 and meandifREF = 371 × 10−6 mm2/sec.
For simulated small motion, θ = ±10° rotation, Δr = ±1.3 pixels shift in image space, φ = ±1 pixels shift in k-space.
For simulated moderate motion, θ = ±20° rotation, Δr = ±2.6 pixels shift in image space, φ = ±2 pixels shift in k-space.
In Vivo Studies
Figure 7 shows the results of in vivo studies. The 16 images show the reconstructed images with the four methods explained above for the cases of negligible (θ≈±1° rotation, Δr ≈±0.05 pixels translation), small (θ≈±7° rotation, Δr ≈±2 pixels translation), moderate (θ≈±20° rotation, Δr ≈±3 pixels translation), and large (θ≈±50° rotation, Δr ≈±5 pixels translation) subject motion. Without phase correction (method A), all images showed significant artifacts resulting from the random nonlinear phase accrued during the diffusion-encoding part of each interleaf and positional changes (Fig. 7a,e,i,m). In the case of no subject motion, method D performed very similarly to method C, as shown by the similarity in the FA maps (Fig. 7c,d). In the presence of patient motion, method C increases the quality of the FA maps (Fig. 7g,k,o) significantly. Accounting for the altered diffusion-encoding direction using method D introduced improvement in image quality over method C in the case of moderate motion (Fig. 7l). For the case of large subject motion, the improvement in image quality of the FA map derived from method D over C was even more pronounced (Fig. 7p).
FIG. 7.
The FA maps obtained from an in vivo experiment, reconstructed using the four different reconstruction methods. For all degrees of motion, images reconstructed with gridding alone leads to serious artifacts due to motion (a,e,i,m). For the case of negligible motion, reconstructions with motion correction and CG SENSE (method C) (c) and NLCG (method D) (d) give similar results compared to the image reconstructed with no motion correction (method B) (b). In the case of medium motion, motion correction with method C (g,k) removes most of the motion-related artifacts that are otherwise seen on the image reconstructed with method B (f,j). The application of method D (h) gives a similar FA map compared to method C (g) in the case of small motion. For moderate motion, some improvement in the FA map was observed by the application of method D (k,l). In the case of large motion the FA map obtained by the application of method D (p) has higher overall anisotropy compared to method C (o). Clearly, the image quality of CG-SENSE breaks down if no motion correction is applied (n).
Figure 8 shows the angular deviation maps between the major eigenvectors corresponding to the reference and reconstructed tensor maps in the splenium of the corpus callosum. Table 2 reports the average FA, mean diffusivity and angular deviation values in the selected ROIs in the splenium and genu of the corpus callosum. These data show that in the case of no subject motion the average angular deviation between the eigenvectors corresponding to the tensor data reconstructed with methods C and D is around 2°, and the average FA values (0.68, 0.69) are close to each other (Fig. 8c,d). In other words, in the absence of motion the application of NLCG does not introduce additional errors compared to the conventional method. However, in the presence of subject motion correcting for the altered b matrix produces an eigenvector field that is more aligned with the reference orientation (Fig. 8h,l,p). Moreover, after application of the NLCG reconstruction, FA values resembled more closely to the values from the reference dataset than any other method.
FIG. 8.
The angular deviation maps between the major eigen-vectors corresponding to the reference data and to the four reconstruction schemes in the splenium of the corpus callosum. The map of the major eigenvector reconstructed with method C in the absence of motion was chosen to be the reference dataset. In the case of no motion the deviation between the eigenvectors reconstructed with NLCG and reference is on the order of 2° (first row). When there is subject motion, NLCG performs generally better than CG SENSE, as shown by the lower angular deviation from the reference (third and fourth columns).
Table 2.
Average FA, Mean Diffusivity, and Angular Deviation Values Obtained for the In Vivo Experiments in an ROI Containing the Genu and Splenium of Corpus Callosum (CC)
| Method A No Correction (Gridding) |
Method B Phase Correction (CG SENSE) |
Method C Phase & Motion Correction (CG SENSE) |
Method D Phase & Motion & Diff. Enc. Dir. Correction (NLCG) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt ref. (degrees) |
FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt ref. (degrees) |
FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt ref. (degrees) |
FA | Mean Diffusivity (10−6 mm2/sec) |
Angular Deviation wrt ref. (degrees) |
||
| Splenium of CC |
Negligible motiona |
0.67 | 941.4 | 19.28 | 0.68 | 760.1 | 1.18 | 0.68 | 747.9 | 0.0 (Ref) | 0.69 | 750 | 1.47 |
| Small motionb |
0.37 | 1279.3 | 25.6 | 0.66 | 903.6 | 9.56 | 0.67 | 836.0 | 5.16 | 0.69 | 814.2 | 4.43 | |
| Moderate motionc |
0.32 | 1030.6 | 34.25 | 0.48 | 753.5 | 14.64 | 0.57 | 735.9 | 7.83 | 0.64 | 730.2 | 5.22 | |
| Large motiond |
0.32 | 1228.5 | 67.09 | 0.49 | 592.4 | 32.59 | 0.32 | 688.6 | 20.26 | 0.48 | 698.3 | 17.73 | |
| Genu of CC | Negligible motione |
0.51 | 1118.8 | 46.97 | 0.47 | 805.0 | 9.50 | 0.51 | 816.6 | 0.0 (Ref) | 0.52 | 817.4 | 1.43 |
| Small motionf |
0.28 | 1249.1 | 50.1 | 0.40 | 813.6 | 37.93 | 0.50 | 800.8 | 11.80 | 0.52 | 790.0 | 10.65 | |
| Moderate motiong |
0.26 | 1197.3 | 48.27 | 0.26 | 746.5 | 44.47 | 0.44 | 788.3 | 8.07 | 0.52 | 799.6 | 6.31 | |
| Large motionh |
0.32 | 1241.3 | 56.28 | 0.41 | 679.4 | 34.74 | 0.31 | 732.1 | 27.06 | 0.47 | 740.3 | 13.32 | |
θ ≈ ±1°, Δr ≈ ±0.05 pixels, FAREF = 0.68, meandifREF = 748.0 × 10−6 mm2/sec.
θ ≈ ±7°, Δr ≈ ±2 pixels, FAREF = 0.67, meandifREF = 813.8 × 10−6 mm2/sec.
θ ≈ ±25°, Δr ≈ ±3 pixels, FAREF = 0.68, meandifREF = 748.0 × 10−6 mm2/sec.
θ ≈ ±50°, Δr ≈ ±5 pixels, FAREF = 0.68, meandifREF = 748.0 × 10−6 mm2/sec.
θ ≈ ±1°, Δr ≈ ±0.05 pixels, FAREF = 0.51, meandifREF = 817.0 × 10−6 mm2/sec.
θ ≈ ±7°, Δr ≈ ±2 pixels, FAREF = 0.50, meandifREF = 770.0 × 10−6 mm2/sec.
θ ≈ ±25°, Δr ≈ ±3 pixels, FAREF = 0.51, meandifREF = 817.0 × 10−6 mm2/sec.
θ ≈ ±50°, Δr ≈ ±5 pixels, FAREF = 0.51, meandifREF = 817.0 × 10−6 mm2/sec.
Finally, Fig. 9 shows the eight navigator images obtained for a scan with diffusion weighting. It can be seen that significant signal loss was present in the third interleaf as a result of subject motion during the application of diffusion weighting. This particular navigator showed a lower correlation with the template after registration, as shown by the lowered correlation coefficient of 0.61 compared to the average correlation coefficient of ≈0.98. It was possible to detect these interleaves, which were irreversibly corrupt by motion, and eliminate them from reconstruction. In such cases we used the parallel imaging features of our approach to reduce the effect of k-space undersampling.
FIG. 9.

Navigator images corresponding to 8 interleaves in a diffusion-weighted scan. Top row: before registration. Bottom row: after registration to a reference navigator image. The third interleaf shows significant signal loss due to motion during the application of diffusion-encoding gradients. Such k-space data can be detected and eliminated by determining a correlation coefficient value following the registration of the individual navigators with the reference navigator (third row). As a rejection threshold for a particular shot a correlation coefficient lower than the median correlation coefficient (over all navigators) –3 × standard deviations can be used.
DISCUSSION
Due to its prolonged acquisition time, the correction of motion artifacts in DTI is essential to guarantee accurate image quality and assure the accuracy of the diffusion tensors. In the past, attempts to perform motion correction for diffusion tensor imaging have been limited to correction for miniscule motion. While these methods predominantly correct for unwanted phase terms, gross patient motion correction must account for both unwanted phase terms and changes in the effective diffusion-encoding direction. A few DTI studies have proposed correction methods for gross patient motion; so far, however, these methods were limited to single-shot scans. Several software packages offer one the ability to coregister individual diffusion-weighted images prior to computing the tensor. However, as shown in this study, only correcting for altered patient position is insufficient to provide accurate tensor maps. This is because body motion has two effects on the DTI data: 1) positional displacement; and 2) rotation of the effective diffusion-encoding gradient with respect to the patient frame of reference due to rotational motion and, thus, the incorrect estimation of the orientation of the tensors.
The correction for the first of these two effects is straight-forward (23). For single-shot sequences the second effect can be accounted for by rotating the b-matrix according to the positional changes that occurred during the acquisition of each diffusion-weighted image (relative to a reference position) prior to the estimation of the diffusion tensor. For multishot approaches the situation is more complicated. In this case, patient motion, particularly rotation, causes the diffusion-encoding matrix b to vary between different shots. This, in turn, leads to a k-space with inconsistent diffusion encoding and affects the accuracy of tensor information derived from such data. This data in-consistency was the motivation for the introduction of a novel mathematical framework that allows one to solve for diffusion tensor information in each pixel in the presence of both macroscopic and microscopic motion. Despite the fact that only translational and rotational motion were considered in this study, it should be noted that rigid motion can be represented by a set of affine transformations. Thus, it is straightforward to extend the algorithm to account for affine transformations as long as the affine transformation parameters can be obtained from low-resolution navigators or using any other method. It is also possible to extend this algorithm to correct for general rigid and nonrigid motion as long as the displacement field and the effect of nonrigid motion on diffusion tensors are known (34). Unless DTI experiments are performed on a gradient system with noticeable gradient nonlinearities (30), the errors introduced by translational motion on diffusion tensor orientation can be neglected.
In this context, it is important to mention that the mathematical framework presented here also applies to undersampled k-space data, i.e., parallel imaging. This allows one to utilize the methods proposed in this study to perform parallel imaging reconstruction with acceleration factors greater than one. Another important motivation of using parallel imaging techniques in the presence of rigid body motion is that counterrotation of k-space trajectories for rotation correction creates undersampling in k-space and residual artifacts in the final motion-corrected image (23). These gaps can be filled up by parallel imaging.
The performance of our algorithm was evaluated using both computer simulations and in vivo studies. The images reconstructed with no application of gross motion correction showed significant artifacts. The application of motion correction with the standard two-step algorithm (method C) successfully removed most of these artifacts; however, some error remained due to the varying diffusion-encoding directions between successive interleaves—as shown by the lowered FA values and considerable deviation in the orientation of the major eigenvectors relative to reference values. With the application of the proposed NLCG method (method D), the error could be reduced even further. Some residual artifacts remained in the in vivo images, which were due most likely to through-plane motion, misregistration, and inaccuracies in the estimation of coil sensitivities. Through-plane motion and spin history effects are a general limitation of retrospective correction methods.
Our algorithm was also shown to work for a wide range of motions. The algorithm was demonstrated with rotation angles as large as 50°. For the other end of the spectrum of motion, it is also important to point out that in the absence of motion the NLCG (method D) provided similar results as the augmented iterative SENSE (method C). Do note that in order to avoid unwanted blurring introduced by potential slight positional inaccuracies of our motion detection, only motion above a certain threshold (typically the precision of our coregistration method) was considered true motion.
It is clear that in the presence of rigid body motion and when k-space data are obtained with multishot DTI sequences, diffusion tensors can only be reconstructed with nonlinear methods. The benefit that one would get over the conventional linear tensor estimation method by using nonlinear methods certainly depends on the amount of rigid body motion present. The benefit of using the proposed NLCG method increases for larger motion. For smaller degrees of motion, however, the error in eigenvector orientation and FA is mostly dominated by the noise inherent in DTI raw data. Thus, the error resulting from the change in diffusion-encoding direction is less important for smaller positional changes. However, our single-step nonlinear method proposed in this study can still be quite effective in correcting for smaller motion in studies that use a large number of diffusion-encoding gradients and that have high signal-to-noise ratio (SNR). Despite the fact that the range of motion evaluated in this study was above average for adult clinical studies, such considerable positional changes happen quite frequently in DTI studies of unsedated, small infants and in fetal imaging (33).
Due to the direct computation of the tensor information and the more complex cost function, the reconstruction time becomes longer than with regular augmented iterative SENSE. Essentially, for each iteration a forward and inverse gridding over nc × nδ × nζ interleaves must be carried out. In addition to the linear CG, NLCG requires minimization along one dimension after the search direction is determined (24). This causes NLCG to contain two nested iterations, which takes more time. In particular, for nc = 8, nδ = 26 and nζ = 8, method C takes ≈1 hr with 15 iterations per diffusion-weighted image and method D takes ≈4 hr with 12 outer and 4 inner iterations. Nevertheless, it is possible to provide a major speed-up by a factor of 50% using the transfer function approach to replace the forward and inverse gridding steps by a convolution operation (31). Moreover, the entire algorithm can be run in multiple threads on a parallel CPU architecture which can provide further speed-up.
Since the algorithm proposed herein solves for the tensor maps within a single step, one has more control over the optimization procedure. For example, regularization of the reconstruction problem is straightforward and can be performed by adding extra terms to the cost function (Eq. [6]). This regularization can be either a total variation-based constraint or an edge-preserving function (32). A detailed description of the regularization approach that can be used for our NLCG approach is beyond the scope of this article and the subject of a forthcoming study.
CONCLUSIONS
Despite coregistering individual diffusion-weighted images, the accuracy of computed tensor information (e.g., orientation of eigenvectors, FA) can be significantly impaired due to falsely assumed diffusion-encoding directions following motion correction. Particularly for multishot methods, such as diffusion-weighted PROPELLER FSE and EPI or multishot spiral and EPI, k-space data can be generated with inconsistent diffusion-encoding leading to ambiguous tensor information. In this study a new reconstruction method has been proposed that estimates maps of the diffusion tensor elements in a single step. Its efficacy in simultaneously correcting for both microscopic and macroscopic motion was demonstrated in phantom and in vivo experiments. The presented generalized mathematical framework can be applied to any trajectory and can be used in conjunction with parallel imaging. Although the degree of improvement depends, of course, on the severity and pattern of motion, our initial experimental data demonstrate that a considerable improvement in the accuracy of the observed tensor information can be achieved using this novel approach.
ACKNOWLEDGMENTS
We thank Prof. Gary Glover for providing us with his spiral pulse sequence, which we used as a basis for our experiments, and for his support and mentorship. We also thank Dr. Samantha Holdsworth for proofreading the article.
Grant sponsor: National Institutes of Health (NIH); Grant numbers: R01EB002711, R01NS047607, R01NS34866, P41RR09784, K99NS057943-01; Grant sponsor: Lucas Foundation; Grant sponsor: Oak Foundation.
Footnotes
Presented in part at the 15th Scientific Meeting of the ISMRM, Berlin, Germany, 2007.
REFERENCES
- 1.Moseley ME, Cohen Y, Mintorovitch J, Chileuitt L, Shimizu H, Kucharczyk J, Wendland MF, Weinstein PR. Early detection of regional cerebral ischemia in cats: comparison of diffusion and T2-weighted MRI and spectroscopy. Magn Reson Med. 1990;14:330–346. doi: 10.1002/mrm.1910140218. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Res Series B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 3.Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201:637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- 4.Bammer R, Keeling SL, Augustin M, Pruessmann KP, Wolf R, Stollberger R, Hartung HP, Fazekas F. Improved diffusion-weighted single-shot echo-planar imaging (EPI) in stroke using sensitivity encoding (SENSE) Magn Reson Med. 2001;46:548–554. doi: 10.1002/mrm.1226. [DOI] [PubMed] [Google Scholar]
- 5.Anderson AW, Gore JC. Analysis and correction of motion artifacts in diffusion weighted imaging. Magn Reson Med. 1994;32:379–387. doi: 10.1002/mrm.1910320313. [DOI] [PubMed] [Google Scholar]
- 6.Norris DG. Implications of bulk motion for diffusion-weighted imaging experiments: effects, mechanisms, and solutions. J Magn Reson Imag. 2001;13:486–495. doi: 10.1002/jmri.1072. [DOI] [PubMed] [Google Scholar]
- 7.Chenevert TL, Pipe JG. Effect of bulk tissue motion on quantitative perfusion and diffusion magnetic resonance imaging. Magn Reson Med. 1991;19:261–265. doi: 10.1002/mrm.1910190212. [DOI] [PubMed] [Google Scholar]
- 8.Trouard TP, Sabharwal Y, Altbach MI, Gmitro AF. Analysis and comparison of motion-correction techniques in diffusion-weighted imaging. J Magn Reson Imag. 1996;6:925–935. doi: 10.1002/jmri.1880060614. [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, Golay X, Zijl PCM, Mori S. Origin and minimization of residual motion-related artifacts in navigator corrected segmented diffusion-weighted EPI of the human brain. Magn Reson Med. 2002;47:818–822. doi: 10.1002/mrm.10102. [DOI] [PubMed] [Google Scholar]
- 10.Bammer R, Stollberger R, Augustin M, Simbrunner J, Offenbacher H, Kooijman H, Ropele S, Kapeller P, Wach P, Ebner F, Fazekas F. Diffusion-weighted imaging with navigated interleaved echo-planar imaging and a conventional gradient system. Radiology. 1999;211:799–806. doi: 10.1148/radiology.211.3.r99jn15799. [DOI] [PubMed] [Google Scholar]
- 11.Crespigny AJ, Marks MP, Enzmann DR, Moseley ME. Navigated diffusion imaging of normal and ischemic human brain. Magn Reson Med. 1995;33:720–728. doi: 10.1002/mrm.1910330518. [DOI] [PubMed] [Google Scholar]
- 12.Mori S, Zijl PCM. A motion correction scheme by twin-echo navigation for diffusion-weighted magnetic resonance imaging with multiple RF echo acquisition. Magn Reson Med. 1998;40:511–516. doi: 10.1002/mrm.1910400403. [DOI] [PubMed] [Google Scholar]
- 13.Butts K, Crespigny A, Pauly JM, Moseley M. Diffusion-weighted inter-leaved echo-planar imaging with a pair of orthogonal navigator echoes. Magn Reson Med. 1996;35:763–770. doi: 10.1002/mrm.1910350518. [DOI] [PubMed] [Google Scholar]
- 14.Atkinson D, Porter DA, Hill DLG, Calamante F, Connelly A. Sampling and reconstruction effects due to motion in diffusion-weighted echo planar imaging. Magn Reson Med. 2000;44:101–109. doi: 10.1002/1522-2594(200007)44:1<101::aid-mrm15>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 15.Atkinson D, Counsell S, Hajnal JV, Batchelor PG, Hill DLG, Larkman DJ. Nonlinear phase correction of navigated multi-coil diffusion images. Magn Reson Med. 2006;56:1135–1139. doi: 10.1002/mrm.21046. [DOI] [PubMed] [Google Scholar]
- 16.Butts K, Pauly J, Crespigny A, Moseley M. Isotropic diffusion-weighted and spiral-navigated interleaved EPI for routine imaging of acute stroke. Magn Reson Med. 1997;38:741–749. doi: 10.1002/mrm.1910380510. [DOI] [PubMed] [Google Scholar]
- 17.Miller KL, Pauly JM. Nonlinear phase correction for navigated diffusion imaging. Magn Reson Med. 2003;50:343–353. doi: 10.1002/mrm.10531. [DOI] [PubMed] [Google Scholar]
- 18.Nunes RG, Jezzard P, Behrens TEJ, Clare S. Self-navigated multishot echo-planar pulse sequence for high-resolution diffusion-weighted imaging. Magn Reson Med. 2005;53:1474–1478. doi: 10.1002/mrm.20499. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Moseley ME, Bammer R. Simultaneous phase correction and SENSE reconstruction for navigated multi-shot DWI with non-Cartesian k-space sampling. Magn Reson Med. 2005;54:1412–1422. doi: 10.1002/mrm.20706. [DOI] [PubMed] [Google Scholar]
- 20.Pipe JG, Farthing VG, Forbes KP. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn Reson Med. 2002;47:42–52. doi: 10.1002/mrm.10014. [DOI] [PubMed] [Google Scholar]
- 21.Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med. 2004;51:103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 22.Maniega SM, Bastin ME, Armitage PA. Effects of rotation on icosahedral diffusion sampling schmemes in DT-MRI. Proc Joint Annual Meeting ISMRM-ESMRMB; Berlin. 2007. p. 312. [Google Scholar]
- 23.Bammer R, Aksoy M, Liu C. Augmented generalized SENSE reconstruction to correct for rigid body motion. Magn Reson Med. 2007;57:90–102. doi: 10.1002/mrm.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scales LE. Introduction to non-linear optimization. Macmillan; London: 1985. pp. 73–83.pp. 113–118. [Google Scholar]
- 25.Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding. IEEE Trans Med Imag. 1991;10:473–478. doi: 10.1109/42.97598. [DOI] [PubMed] [Google Scholar]
- 26.Rasche V, Proksa R, Sinkus R, Bornert P, Eggers H. Resampling of data between arbitrary grids using convolution interpolation. IEEE Trans Med Imag. 1999;18:385–392. doi: 10.1109/42.774166. [DOI] [PubMed] [Google Scholar]
- 27.Pruessmann KP, Weiger M, Bornert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Magn Reson Med. 2001;46:638–651. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 28.Aksoy M, Liu C, Moseley M, Bammer R. The effect of navigator resolution on registration accuracy in rigid head motion correction. Proc Joint Annual Meeting ISMRM-ESMRMB; Berlin. 2007. p. 654. [Google Scholar]
- 29.Liu C, Bammer R, Kim DH, Moseley M. Self-navigated interleaved spiral (SNAILS): application to high-resolution diffusion tensor imaging. Magn Reson Med. 2004;52:1388–1396. doi: 10.1002/mrm.20288. [DOI] [PubMed] [Google Scholar]
- 30.Bammer R, Markl M, Barnett A, Acar B, Alley MT, Pelc NJ, Glover GH, Moseley ME. Analysis and generalized correction of the effect of spatial gradient field distortions in diffusion-weighted imaging. Magn Reson Med. 2003;50:560–569. doi: 10.1002/mrm.10545. [DOI] [PubMed] [Google Scholar]
- 31.Wajer FTAW, Pruessmann KP. Major speedup of reconstruction for sensitivity encoding with arbitrary trajectories. Proc 9th Annual Meeting ISMRM; Glasgow. 2001. p. 767. [Google Scholar]
- 32.Charbonnier P, Feraud LB, Aubert Gm, Barlaud M. Deterministic edge-preserving regularization in computed imaging. IEEE Trans Image Process. 1997;6:298–311. doi: 10.1109/83.551699. [DOI] [PubMed] [Google Scholar]
- 33.Jiang S, Counsell SJ, Xue H, Allsop JM, Rutherford MA, Rueckert D, Hajnal JV. In-utero 3D high resolution fetal brain diffusion tensor imaging. Proc Joint Annual Meeting of ISMRM-ESMRMB; Berlin. 2007. p. 152. [DOI] [PubMed] [Google Scholar]
- 34.Batchelor PG, Atkinson D, Irarrazaval P, Hill DLG, Hajnal J, Larkman D. Matrix description of general motion correction applied to multishot images. Magn Reson Med. 2005;54:1273–1280. doi: 10.1002/mrm.20656. [DOI] [PubMed] [Google Scholar]







