Abstract
Thyroid hormones (THs) play an important role in vertebrate development; however, the underlying mechanisms of their actions are still poorly understood. Zebrafish (Danio rerio) is an emerging vertebrate model system to study the roles of THs during development. In general, the response to THs relies on closely related proteins and mechanisms across vertebrate species, however some species-specific differences exist. In contrast to mammals, zebrafish has two TRα genes (thraa, thrab). Moreover, the zebrafish thraa gene expresses a TRα isoform (TRαA1) that differs from other TRs by containing additional C-terminal amino acids. C-terminal extensions, called “F domains”, are common in other members of the nuclear receptor superfamily and modulate the response of these receptors to hormones. Here we demonstrate that the F-domain constrains the transcriptional activity of zebrafish TRα by altering the selectivity of this receptor for certain coactivator binding motifs. We found that the F-domain of zebrafish TRαA1 is encoded on a separate exon whose inclusion is regulated by alternative splicing, indicating a regulatory role of the F-domain in vivo. Quantitative expression analyses revealed that TRαA1 is primarily expressed in reproductive organs whereas TRαB and the TRαA isoform that lacks the F-domain (TRαA1-2) appear to be ubiquitous. The relative expression levels of these TRα transcripts differ in a tissue-specific manner suggesting that zebrafish uses both alternative splicing and differential expression of TRα genes to diversify the cellular response to THs.
Keywords: Thyroid Hormone, Thyroid hormone receptor, Isoforms, Danio rerio, F-domain
1. Introduction
Thyroid hormones (THs) are essential for normal development, differentiation, and metabolic balance of vertebrates (Yen, 2001). In amphibians and some fish THs are the key regulators of metamorphosis, whereas in mammals hypothyroidism leads to cretinism, mental retardation, and deafness (Pitt-Rivers and Tata, 1959; DeLong, 1996; Power et al., 2001). Although the general mechanisms of TH physiological actions are known, the roles of these hormones during development remain largely elusive (Chan and Kilby, 2000; Tata, 1999). Recently, zebrafish has been introduced as a novel non-mammalian model system to facilitate the manipulation, dissection and genetic analysis of TH activities during development (Brown, 1997; Essner et al., 1997; Essner et al., 1999; Liu et al., 2000; Liu and Chan, 2002; Lam et al., 2005). Manipulation of TH levels provided evidence that THs are of particular importance during the embryonic-larval and larval-juvenile transitions (Brown, 1997; Liu and Chan, 2002). Chemical disruption of TH production in zebrafish embryos and larvae results in stunted growth, retarded head cartilage development, paired fin elongation, curled tails, changes in the distribution and structure of melanophores, and impairs the development of the thymus (Brown, 1997; Liu and Chan, 2002; Lam et al., 2005; Elsalini and Rohr, 2003). Zebrafish embryos that have been treated with exogenous THs do not develop swimbladders, have smaller and bent bodies, severely retarded gastrointestinal system development and fewer and smaller melanophores (Liu and Chan, 2002). Complete absence or high concentrations of TH during zebrafish embryogenesis is lethal (Liu and Chan, 2002).
The cellular activity of THs is exerted by two thyroid hormone receptors, TRα and TRβ (Lazar, 1993; Marchand et al., 2001). Differential promoter usage and alternative splicing enables the TRα and β genes to give rise to multiple isoforms (Flamant and Samarut, 2003). The temporal and regional expression of these isoforms constitutes an important mechanism for the stage- and tissue-specific regulation of cellular responses to THs (Liu et al., 2000; Yamano and Miwa, 1998; Chassande et al., 1997; Buchholz et al., 2006). TRs are hormone regulated transcription factors that belong to the nuclear receptor superfamily (Aranda and Pascual, 2001). Like all nuclear receptors, TRs have a modular structure and contain highly conserved DNA (DBD) and ligand binding (LBD) domains as well as a less conserved N-terminal domain (Zhang and Lazar, 2000). Depending on the type of thyroid hormone response element (TRE), TRs can bind DNA as monomers, homodimers, or heterodimers with retinoid X receptors (RXRs) (Lazar et al., 1991; Bugge et al., 1992; Rastinejad et al., 1995).
THs control the activities of TRs by inducing conformational changes that regulate the interaction of the TR LBD with corepressors and coactivators (Renaud and Moras, 2000; Privalsky, 2004). In the absence of hormone, TRs interact with corepressors that are released upon hormone binding and replaced by coactivators (Glass and Rosenfeld, 2000). The hormone-dependent transition of TRs from transcriptional repressors to transcriptional activators plays a decisive role during development (Mai et al., 2004). The dual activity rationalizes why TRs are usually expressed prior to the onset of fetal TH production. Moreover, this mechanism also explains that the absence of TRs, which prevents the active repression and activation of TR target genes, is physiologically less detrimental than reduced levels of THs, which result in constitutive repression of TR target genes.
The general mechanisms of TH actions appear to be similar across vertebrates; however, some species-specific variations have been identified. One of these differences concerns the onset of zygotic TH production and the availability of THs before this point. Although there is evidence that mammalian embryos are exposed to maternal THs and that in transgenic mice TH-dependent reporters can be activated before the onset of zygotic TH production, TH levels are considered to be low during early mammalian development (Chassande et al., 1997; deEscobar et al., 2004). In contrast, oocytes from fish and other non-mammalian vertebrates can contain large amounts of maternal THs (Power et al., 2001). It is still unclear whether these maternal THs are available to activate TRs during embryogenesis. Another species-specific difference is the number and expression of the TR genes and details in the structure of TRs. Due to ancestral gene duplication events, some non-mammalian vertebrate species such as the African clawed frog (Xenopus laevis) and the Japanese flounder (Paralichthys olivaceus) have several TRα-encoding genes (Yaoita and Brown, 1990; Yamano et al., 1994; Yamano and Inui, 1995). Similarly, the genome sequencing project (http://www.sanger.ac.uk) recently identified an additional TRα-encoding gene in zebrafish (Genebank accession number XM_702123). Based on available cDNAs, both zebrafish TRα genes appear to be expressed and give rise to at least two TRα products (TRαA1; TRαB)*.
A seemingly unique feature of zebrafish TRαA1 is the extension of α-helix H12 at the TRα C-terminus by 17 amino acids (Essner et al., 1997; Marchand et al., 2001). C-terminal extensions, called “F domains”, are common in other members of the nuclear receptor superfamily. With the exception of some steroid hormone receptors, for most nuclear receptors the sequence and size of F-domains are highly variable and can differ substantially even for closely related receptors. Thus far, the structures of only a few steroid hormone receptor F-domains have been solved (Williams and Sigler, 1998; Bledsoe et al., 2002; Kauppi et al., 2003) (Fig. 1A). Consistent with these structures, mutational studies indicate that F-domains regulate the response of steroid hormone receptors to ligands by modulating the interactions with coactivators and corepressors (Montano et al., 1995; Schwartz et al., 2002; Peters and Kahn, 1999). Mutational analyses suggested that F-domains of other nuclear receptors have similar functions (Suaud et al., 1999; Sladek et al., 1999; Ruse et al., 2002; Farboud and Privalsky, 2004).
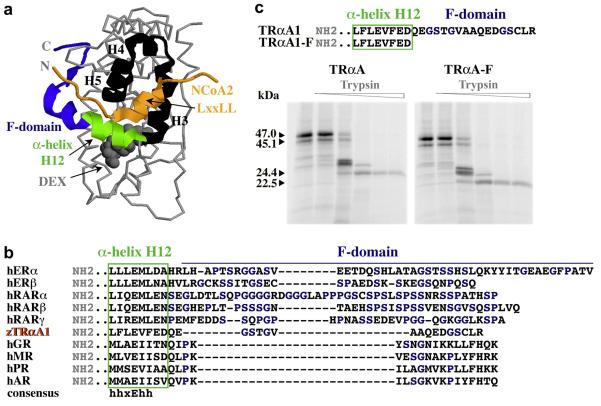
Fig. 1. Comparison of the F-domains of TRαA1 and other nuclear receptors.
A, Structure of the glucocorticoid receptor LBD bound to dexamethasone (grey space-filled) (Bledsoe et al., 2002). The F-domain (blue) extents α-helix H12 (green), which is a hormone-regulated structural switch. In the presence of hormone, the GR α-helix H12 completes a hydrophobic groove formed by α-helices H3, H4, and H5 (all black), which enables the interaction of conserved “LxxLL” motifs in NCoA2 (orange) and other coactivators with this groove. Sequences N-terminal of the LxxLL motifs are located in close proximity to the F-domain. B, Sequence alignment of α-helix H12 and the F-domains of the human estrogen receptor α (P03372), estrogen receptor β (Q92731), retinoic acid receptor α (P10276), retinoic acid receptor β (P10826), retinoic acid receptor γ (P13631), zebrafish thyroid hormone receptor αA1 (Q98867), and the human glucocorticoid receptor (P04150), mineralcorticoid receptor (P08235), progesterone receptor (P06401), and androgen receptor (P10275). Although the F-domains of these receptors display no significant sequence homologies, they are all very rich in glycines, serines and prolines (blue). C, Sequence of the C-termini and proteolytic digest patterns of TRαA1 and zebrafish TRαA1-F. Shown are the digestion patterns of TRαA1 (47 kDa) and TRαA1-F (45.1 kDa) after incubation with various concentrations of trypsin (0, 0.3, 3, 30, 150, 300 μg/ml) at 25°C for 25 min in the presence of 20 μM T3. The 24.4 kDa (TRαA1-F, 22.5 kDa) trypsin fragment is characteristic for hormone-bound TRα.
In this study we investigated whether the F-domain of zebrafish TRαA1 plays a role in regulating the activity of zebrafish TRα and contributes to the stage- and tissue-specific regulation of cellular responses to THs.
2. Materials and methods
2.1 Animal and embryos
Adult zebrafish were maintained as described by (Westerfield, 2000). Embryos were generated by natural crosses and maintained at 28°C in embryo medium (15 mM NaCl, 0.5 mM KCl, 1 mM CaCl2 H2O, 1 mM MgSO4•7H20, 0.15 mM KH2PO4, 0.05 mM Na2HPO4•2H2O, 0.8 mM NaHCO3, 10 mM Hepes). 2 ml/l penicillin-streptomycine solution (Sigma) was added following injections. All described animal experimentations were conducted in accord with accepted standards of humane animal care and approved by the Institutional Animal Care and Use Committee of the University of Oregon.
2.2 Molecular cloning
Zebrafish TRαA1 (TRα1) and TRβ1 were a kind gift of Dr. W.-K. Chan (NU, Signapore) (Liu et al., 2000). Coding sequences for TRαA1, TRαA1-F, and TRβ1 and of corresponding N-terminally HA-tagged derivatives were amplified by PCR and cloned into the Eco RI/Xba I cloning sites of pXT7, a pGEM-4Z derivative that contains the 5′ and 3′ UTRs of the globin gene. The PCR primers used for the amplification were (sequences homologous to TRαA1 or TRβ1 are shown in italic, restriction sites are underlined): zTRαA1/fw: 5′ AAGAATTCATGGAAAACACAGAGCAGGAG 3′; HA-zTRαA1/fw: 5′ AAGAATCATGTACCCTTATGATGTGCCAGATTATGCCGAAAACACAGAGCAGGAGCAC 3′; zTRαA1/rev: 5′ GACTAGTTCTAGATCACCTTAAGCAGGAACCGTC 3′; zTRαA1-F/rev: 5′ GACTAGTTCTAGATCAATCCTCGAAGACCTCCAG 3′; zTRβ1/fw: 5′ AAGAATTCATGTCAGAGCAAGCAGACAAA 3′; HA-zTRβ1/fw: 5′ AAGAATTCATGTACCCTTATGATGTGCCAGATTATGCCTCAGAGCAAGCAGACAAA 3′; zTRβ1/rev: 5′ GACTAGTTCTAGATCAGTCTTCAAACACTTCCAG 3′. All PCR-amplified sequences were confirmed by sequence analysis. Eco RI/Xba I fragments of the pXT7 clones were subcloned into pcDNA3 (Invitrogen).
The zebrafish NCoA-2 NID (amino acids 558-758) was PCR amplified from a cDNA library of 1 dpf zebrafish embryos (kind gift of Dr. J. Postlethwait, UO) using the following primers: zNCoA-2/fw: 5′ AAAAAAGGATCCGCTGCACATTCGGTAGCAGTAAG 3′; zfNCoA-2/rev: 5′ ATATATCTCGAGTCCAGGCTCCATTTTAATACCTTTAC 3′. This fragment was cloned into the Bam HI and Xho I cloning sites of a pGEX4-T1 derivative that expresses proteins as fusion with an N-terminal GST-tag and a C-terminal His6-tag (Darimont et al., 1998).
2.3 Antisense morpholino oligonucleotide (MO) and peptide synthesis
MOs against TRαA1 (5′ CTCCTGCTCTGTGTTTTCCATTCAC 3′) and TRβ1 (5′ GTTGCATTTGTCTGCTTGCTCTGAC 3′) were purchased from GeneTools (Start codons are underlined). Sequence, purification and quantification of the NCoA2 (GRIP1) NR-box 2 and 3 peptides have been described (Darimont et al., 1998).
2.4 Trypsin proteolysis
TRαA1 and TRαA1-F were expressed and 35S-labeled using corresponding pXT7 clones and a coupled transcription/translation reticulocyte expression system (TNT, Promega). Typsin digestions were performed in 10 μl reactions containing 1.5 μl of the respective in vitro expression reaction, 20 mM TrisHCl pH 8.0, 0.1 M NaCl, 10% glycerol and 0-300 μg/ml trypsin (Roche) diluted in 1 mM HCl. Reactions were incubated at 25°C for 25 min and stopped by the addition of 2× sodium dodecyl sulfate (SDS) loading buffer. Proteolytic fragments were separated by SDS-polyacrylamide gelelectrophoresis (PAGE) and visualized by autoradiography.
2.5 Saturation hormone binding
Hormone binding reactions were performed in 100 μl containing 1μl (~1-5 ng) in vitro expressed TRαA1 (see above), 20 mM KPO4 pH 8.0, 0.4 M NaCl, 0.5 mM EDTA, 1.0 mM MgCl2, 10% glycerol, 1 mM monothioglycerol, 50 μg calf thymus histones (Calbiochem) and 0.1-15 nM L-3,5,3′-[125I]-T3 (NEN Life Science Products). Reactions were incubated overnight at 4°C. Receptor-bound [125I]-T3 was isolated by gravity flow through a 2 ml course Sephadex G25 column (Pharmacia) and quantified using a γ-counter. The results from three independent repeats were fit by nonlinear regression and the Kd calculated using a single site saturation binding model.
2.6 Zebrafish in vivo hormone accumulation
T3 and [125I]-T3 were mixed in a ratio of 100:1 and added in final concentrations of 0.01-1 μM to embryo media. Per hormone concentration, 50 embryos were incubated in batches of 10 embryos in 2 ml embryo medium for 16 h at 28°C. Embryos were dechorionated and washed 8 times in 5 ml hormone-free embryo medium. Washed embryos were lysed by adding scintillation fluid and the amount of radioactivity quantified using a scintillation counter.
2.7 Zebrafish reporter assay
To produce the TR mRNAs, 10 μg of the corresponding pXT7 plasmid were digested with Xba I, precipitated, resuspended in 20 μl RNase-free water and quantified by gelelectrophoresis using a mass ladder. 1.5 μg of this DNA was transcribed using the mMessage mMachine T7 Kit (Ambion) and the resulting purified RNA quantified spectroscopically. The concentration was adjusted to 0.4 μM and frozen in aliquots at −80°C. Zebrafish embryos (1-4 cell stage) were microinjected with approximately 1 nl of a mix containing Phenol red (5% Phenol Red and 0.2 M KCl in RNase-free water), the DR4 luciferase reporter (0.25 μg/μl), a β-galactosidase reporter (0.25 μg/μl), and, dependent on the experiment, TR mRNAs (0.06 μM) and/or TRα1/TRβ1 MOs (1.5 mM each). The DR4 luciferase reporter and the constitutively expressed β-galactosidase reporter (p6Rβ-gal) have been described (McKnight and Kingsbury, 1982; Pearce and Yamamoto, 1993). At about 3 hpf injected embryos were transferred in batches of 50 into 30 ml embryo medium that contained either vehicle (DMSO) or the appropriate concentration of T3 (0.1-10 μM) and incubated for 16-20 h at 28° C. Embryos were dechorionated, transferred into Eppendorf tubes (15 embryos/tube) and lysed in 1× BD luciferase lysis buffer (total volume 50 μl), followed by a freeze-thaw cycle in liquid nitrogen and vortexing. Lysates were cleared by centrifugation (20800 × g, 15 min, 4°C), the total protein concentration determined using a Biorad protein assay and adjusted to 4 μg/μl. 60 μg and 40 μg total protein was used to measure β-galactosidase and luciferase activity, respectively, as described by Iniguez-Lluhi et al. (1997). The β-galactosidase activities were used to correct for variability in injection volumes.
2.8 Mammalian cells reporter assay (transient transfection)
Monkey CV1 kidney fibroblasts (American Tissues and Clones Collection) were maintained in Dulbecco’s modified Eagle’s medium (4.5 g/l glucose) and 5% fetal bovine serum (FBS). For transient transfections, 70,000 CV1 cells/well were seeded in 24-well plates and 24 h later transfected with 800 ng total plasmid DNA containing 20 ng p6Rβ-gal (Pearce and Yamamoto, 1993), 200 ng DR4 luciferase reporter (McKnight and Kingsbury, 1982) and 0-20 ng pcDNA3 (HA-)zTRαA1 or pcDNA3 (HA-)zTRαA1-F using Lipofectamine (1.34 μl) and Lipofectamine Plus (2 μl) (Invitrogen). About 6 h after transfection, CV1 cells were washed with PBS and incubated for 20-24 h in DMEM containing 5% FBS and 0.1-50 nM T3 as indicated. Cells were harvested in 100 μl 1× lysis buffer (BD), lysed by freeze-thawing and assayed for luciferase activity (6 μl lysate) and β-galactosidase activity (20 μl lysate) as described above. The β-galactosidase activity was used to correct for differences in transfection efficiency.
2.9 Immunoblot analysis
For immunoblot analysis of ectopically expressed HA-tagged TRαA1 and TRαA1-F zebrafish embryos (1-4 cell stage) were microinjected as described in 2.7 (~ 80 embryos/construct). At 24 hpf zebrafish embryos were frozen, resuspended in three volumes of cold lysis buffer (1% (w/v) TritonX-100, 50 mM Hepes pH 7.5, 0.4 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 25 mM β-mercaptoethanol, 10% (w/v) glycerol, protease inhibitors (Complete-EDTA (Roche), aprotinin and leupeptin (2 μg/ml each)) and lysed by 6-8 freeze-thaw cycles. Lysates were cleared by centrifugation (20800 × g, 30 min, 4°C) and protein concentrations determined using a Biorad protein assay. Proteins (100 μg) were separated by PAGE, probed with a monoclonal HA-tag antibody (Covance) and a polyclonal actin antibody (A2066; Sigma), and analyzed using a LI-COR Odyssey imager.
For immunoblot analysis of transfected CV1 cells, ~350,000 transfected cells (see above) were harvested in PBS + 1 mM EDTA. Cell pellets were frozen in liquid nitrogen, resuspended in 100 μl lysis buffer (see above), and lysed using 3 freeze-thaw cycles. Lysates were cleared by centrifugation (20800 × g, 30 min, 4°C), the protein concentration determined using the Biorad protein assay and adjusted to 1.5 μg/μl. Proteins (30 μg) were separated by PAGE, transferred to nitrocellulose and the HA-tagged TRs visualized using a monoclonal HA antibody (Covance). Blots were developed with horseradish peroxidase-conjugated anti-mouse secondary antibodies and an enhanced chemiluminescence substrate (SuperSignal WestPico, Pierce).
For immunoblot analysis of in vitro expressed TRα variants and TRβ1, 2 μl of the coupled in vitro expression reactions (TNT, Promega) were separated by PAGE, transferred to nitrocellulose, and incubated with a polyclonal antibody that recognizes the conserved α-helix H12 in the TR LBD (Zhu et al., 1996). This antibody was a kind gift of Dr. S.-Y. Cheng (NIH). Blots were developed with horseradish peroxidase-conjugated anti-rabbit secondary antibodies and developed as described above.
For immunoblot analysis of endogenous zebrafish TRs, frozen zebrafish embryos or adult tissues were homogenized in an equal volume of cold harvest buffer (0.8 M KCl, 0.1 M KPO4 pH 8.0, 4 mM EDTA, 10 mM β-mercaptoethanol, 2% TritonX-100, 20% glycerol, 1 mM PMSF, aprotinin and leupeptin (4 μg/ml, each)) using a pellet pestle® motor (KONTES). Lysates were cleared by centrifugation (20800 × g, 30 min, 4°) and protein concentrations determined using a Biorad protein assay. Proteins (200 μg) were separated by PAGE, probed with the polyclonal TR antibody mentioned above and analyzed using a LI-COR Odyssey imager.
2.10 GST-pull down assays
The GST fusion of zfNCoA2 NID was expressed in BL21DE3 at 37°C. Cells were harvested 4 h after induction with 1 mM IPTG at OD600 0.7, resuspended in lysis buffer (20 mM TrisHCl pH 8.0, 0.1 M NaCl, 10% glycerol), and lysed using a French press. The cell lysate was cleared by centrifugation (20800 × g, 50 min, 4°C) and 20 ml lysate incubated with 2.5 ml Talon beads and 5 mM imidazole for 2-3 h at 4°C. GST-NCoA2 NID was eluted in 5 ml 500 mM imidazole in a concentration of ~ 7.5 mg/ml.
45 mg of Talon purified GST-NCoA2 NID were bound to 2 ml glutathione agarose as described in Darimont et al. (1998). The final concentration of glutathione agarose-bound GST-NCoA2 NID was about 80 μM. GST pull down experiments using in vitro expressed, 35S-labeled zTRαA1 or zTRαA1-F in the absence or presence of various concentrations of GRIP1 (mouse NCoA2) NR-box 2 and 3 peptides were performed as Darimont et al. (1998).
2.11 RNA and cDNA preparations
RNA was prepared from 50-100 μl of either unfertilized (squeezed) zebrafish eggs, zebrafish embryos at different stages (30-300 embryos dependent on stage and protocol) or tissues from various adult organs using either Trizole (Invitrogen) or RNeasy mini (Qiagen). For cDNA production, 1 μg of total RNA was reverse transcribed using the First Strand cDNA synthesis kit (New England Biolabs). Reactions were incubated for 1 h at 42°C followed by treatment with 0.5 ng RNase at 37°C for 30 min.
2.12 Non-quantitative reverse transcriptase PCR
Non-quantitative PCR reactions were performed in 50 μl containing 4.5 μl of the cDNA preparations (see above), 5 μl 10× reaction buffer (0.2 M Tris-HCl pH 8.4, 0.5 M KCl, 15 mM MgCl2), 0.25 μM of each a forward and a reverse primer (see below), 200 μM dNTP mix, and 2.5 units Taq polymerase. Primers used in these reactions: zTRα_E8/fw: 5′ TGCCCTGTGAAGACCAGATCATCTTGCTGAAAGGC 3′; zTRα_E9/rev: 5′ CCGCTGTGTCTCTGGGTCACACCTCCTGATCCTCG 3′; zTRα_I9/rev: 5′ GGTGGAGTTTGTTTTGCCGCTGTGTCTCTGGGTCA 3′; zTRα_E10/rev: 5′ AAGCAGGAACCGTCTTCCTGTGCTGCCACTCCAGT 3′. After denaturation at 94°C for 4 min, fragments were amplified using 30 cycles of the following sequence: 30 sec at 94°C, 45 sec at 55°C, 60 sec at 72°C. For sequence analysis, the PCR fragments were gel purified on 2% agarose using a Qiaquick Gel Extraction Kit (Qiagen) and eluted in 30 μl low TE. Samples were quantified spectroscopically and sequenced using the TRα_E8/forward primer.
2.12 Quantitative reverse transcriptase PCR
Quantitative reverse transcriptase PCR reactions were performed in 40 μl reactions containing either 10−12-10−16 M standard (see below) or 5 μl of the cDNA preparations (corresponding to 0.1 μg total RNA, see above), 4 μl 10× reaction buffer (0.2 M Tris-HCl pH 8.4, 0.5 M KCl), 1.5 mM MgCl2, 500 μM dNTP mix, 300 nM of each forward/reverse primer (listed below), 0.2× SYBR green I (Molecular probe) and 1 unit of Taq DNA polymerase (Invitrogen). The primers used in this analysis were: AE7/fw: 5′ GACAATGATAAAGTGGACCTG 3′; AE8/rev: 5′ GCACCGCTGCTCGCAATG 3′; AE9/fw: 5′ GTGTCCAACAGAACTGTTCC 3′; AE10/rev: 5′ GATGCACGTGAATGGATGTG 3′; AI9/rev: 5′ GAGGCGCACGAGCTTGTGAG 3′; BE7/fw: 5′ GATGGGGACAAAGTGGATTTG 3′; BE8/rev: 5′ GAACTGCAGCACGCAGAG 3′; BE9/fw: 5′ ATGCCCCACTGAACTCTTTC 3′; BE9*/rev: 5′ TGCAATTGCTGCTGGTGAGT 3′; zβ–actin1/fw: 5′ GAGAAGATCTGGCATCACAC 3′; zβ–actin1/rev: 5′ GTGTTGAAGGTCTCGAACATG 3′. Reactions were amplified in a MyiQ thermocycler (Bio-Rad) using the following conditions (95°C, 5 min followed by 35 cycles of 94°C, 30 sec; 57°C, 30 sec; 72°C, 2 min, 78°C, 10 sec, and a final incubation at 72°C for 10 min). Amplification products were analyzed by raising the temperature between 72°C and 94°C in 0.2°C increments. For each primer pair standard curves were produced by using as templates various concentrations of either AE7/AE10, AE7/AI9 or BE7/BE9 PCR fragments that have been gel purified and quantified spectroscopically. These standard curves were used to convert the threshold cycles calculated from Base Line Substracted PCR Amplification Cycle plots of reactions with unknown template concentration. The correlation coefficients for these conversions were typically between 0.998-1.0. β-Actin was used to normalize independent repeats of each sample.
3. Results and discussion
The C-terminal extension of zebrafish TRαA1 is not related to other “F-domains”
According to available sequence information, zebrafish TRαA1 is the only known thyroid hormone receptor whose ligand binding domain is extended by more than three amino acids (Marchand et al., 2001). Consistent with the poor sequence conservation of F-domains in general, the 17 amino acids at the C-terminus of zebrafish TRαA1 display no significant sequence homology to other F-domains other than a high content of glycine and serine residues that is typical for F-domains (Fig. 1B).
Deletion of the F-domain does not alter the structure of TRαA1
To investigate whether the zebrafish TRαA1 F-domain plays a structural role, we deleted the F-domain by PCR mutagenesis (TRαA1-F) and expressed TRαA1 and TRαA1-F in vitro. For both, TRαA1 and TRαA1-F, digestion with trypsin generated a proteolytic fragment pattern that is consistent with cleavage at two known conserved trypsin cleavage sites in the amino (N)-terminus of the TR LBD (Fig. 1C; note that TRαA1 is 1.9 kDa bigger than TRαA1-F). Moreover, in the presence of T3 (3,5,3′-L-triiodothyronine) both receptors display a trypsin resistant fragment (TRαA1 24.4 kDa; TRαA1-F 22.5 kDa) that is characteristic for hormone-bound TR ligand binding domains (B. Darimont, unpublished observation). These results indicate that deleting the F-domain has no significant effect on the overall structure of TRαA1 and does not prevent TRαA1 from binding hormone. Similar observations were obtained for F-domains from other nuclear receptors (Farboud and Privalsky, 2004).
Transcriptional activity of TRs in zebrafish embryos
To monitor the transcriptional activity of TRαA1 and TRαA1-F, we established a transcriptional reporter assay using microinjected zebrafish embryos. In this assay, 1-4 cell stage zebrafish embryos were microinjected with a constitutively expressed β-galactosidase reporter, a luciferase reporter that is regulated by a DR4-type TRE, mRNAs for zebrafish TRαA1 and TRβ1 and/or corresponding TRαA1/β1 antisense morpholino oligonucleotides (MOs) (Fig. 2A). The β-galactosidase activity was used to correct for variability in injection volumes. Following injection, embryos were treated with either DMSO (vehicle) or T3 for several hours, then dechorionated, and lysed. β-Galactosidase and luciferase activities in these lysates were determined spectroscopically.
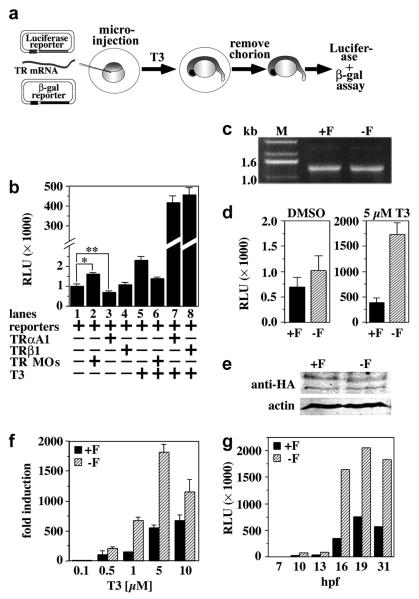
Fig. 2. Deletion of the F-domain modulates transcriptional activity of TRαA in zebrafish embryos.
A, Schematic representation of the zebrafish reporter assay. Zebrafish embryos were injected at the 1-4-cell stage with a DR4 luciferase reporter, a CMV-driven β-galactosidase reporter and, dependent on the experiment, TRαA1, TRαA1-F or TRβ1 mRNA and/or TRαA1/β1 MOs. Injected embryos were treated with vehicle (DMSO) or T3 and luciferase and β-galactosidase activities determined within 24 hpf. B, Activity of endogenous and exogenous TRs in zebrafish embryos in the absence or presence of exogenous T3 and/or TRαA1/β1 MOs. Transcriptional activity is given in relative luciferase units (RLU). Lane 1: activity of endogenous TRs; lane 2: activity of endogenous TRs in the presence of TRαA1/β1 MOs; lanes 3, 4: activity of microinjected TRαA1 and TRβ1, respectively; lanes 5-8: same as lanes 1-4 but instead with vehicle (DMSO), embryos were treated with 5 μM T3. Each lane represents the averages and standard deviations of the luciferase activity/β-galactosidase activity ratio of 2 independent experiments performed with 30-45 injected embryos per condition. A student’s t test shows that the differences in the activities shown in lane 1/lane 2 and lane 1/lane 3 are statistically significant (*, p=0.008; **, p= 0.028). C, Agarose gel analysis of the TRαA1 (+F) and TRαA1-F (−F) injection mixes. D, Activity of injected TRαA1 or TRαA1-F in the presence of either vehicle (DMSO) or T3 (5 μM). Shown are the averages and standard deviations of four injection groups (60 embryos total) per condition. E, Immunoblot analysis monitoring the expression of injected HA-tagged TRαA1 or TRαA1-F (100 μg total protein) in 1 dpf zebrafish embryos treated with 5 μM T3 using a HA-tag specific antibody. The lower molecular band in the anti-HA plot either represents an unspecific band or a C-terminally truncated deletion product of TRαA1 and TRαA1-F. To control for identical loading, the membrane was simultaneously probed with an antibody specific to actin. F, Dose response analysis of injected TRαA1 or TRαA1-F in the absence or presence of various concentrations of T3 (0.1-10 μM). Fold induction is defined as RLU(+H)/RLU(−H). Shown are the averages and standard deviations of 4-6 injection groups (60-90 embryos total) per condition. G, Kinetics of reporter gene activation. After microinjection, embryos were treated with 5 μM T3 (starting at 4 hpf). Luciferase and β-galactosidase activities were determined at various times as indicated. Shown is one of two experiments that gave similar results. Each time point represents at least 30 injected embryos.
Injection of zebrafish embryos with the DR4 luciferase reporter and β-galactosidase reporters alone gave low luciferase activity (Fig. 2B, lane 1), which increased by approximately 70% (p=0.008) upon co-injection of TRαA1/β1 MO (Fig. 2B, lane 2). Co-injection of in vitro transcribed TRαA1 mRNA enhanced the repression of the DR4 luciferase reporter by 30% (p=0.028) (Fig. 2B, lane 3), while co-injection of TRβ1 mRNA had very little effect on the expression of the DR4 luciferase reporter (Fig. 2B, lane 4). These results indicate that both endogenous and exogenous zebrafish TRα1 regulate expression of the DR4 luciferase reporter and confirm the results of others suggesting that TRαA1 can repress transcription in zebrafish embryos (Essner et al., 1997).
To determine whether THs can activate TRs in zebrafish embryos, we next repeated these reporter studies in the presence of T3 supplied to the embryo medium. In these experiments 0.5-5 μM T3 increased the expression of the DR4 luciferase reporter by endogenous or ectopically-expressed zebrafish TRs 2.5-fold or ~450-fold, respectively (Fig. 2B, lanes 5, 7-8). However, lower hormone concentrations were ineffective (Fig. 2F). Co-injection of TRαA1/β1 MOs suppressed the hormone-dependent activation of endogenous TRs (Fig. 2B, lanes 6), indicating that the increase in luciferase activity is TR-dependent.
Using an in vitro binding assay we found that zebrafish TRαA1 binds T3 with a dissociation constant of 1.3 ± 0.4 nM (data not shown). Therefore, in contrast to the results shown in figure 2F, TRαA1 should be fully activated in the presence of 0.1 μM T3. A possible explanation for the low hormone responsiveness of TRs in zebrafish embryos is that the uptake of the exogenous hormone supplied with the embryo medium is slow. To accommodate the relatively short lifetime of injected RNAs, injected embryos were harvested within 24 hours (h) post injection; therefore, the embryos were exposed to exogenous hormone for only 16-20 hours. To investigate this possibility, we quantified the amount of exogenous 125I-T3 that accumulates in the embryos during a 16 h incubation period. The results indicated that in our experiments on average the T3 concentration in the embryo is about 500 times lower than in the embryo medium (Table I), which correlates well with the hormone response of TRαA1 in figure 2F.
Table 1. In vivo accumulation of 125I-T3 in zebrafish embryos.
125I-T3 accumulation in zebrafish embryos after 16 h incubation with various concentrations of 125I-T3 added to the embryo medium. Shown are the averages and standard deviations of four experiments each performed with 50 embryos per hormone concentration.
|
125I-T3 in medium [μM] |
125I-T3 in embryos [nM] |
|---|---|
| 0.01 | 0.08±0.02 |
| 0.1 | 0.48±0.20 |
| 1 | 3.25±1.50 |
| 10 | 44.75±20.90 |
The ability of TR to activate transcription in the presence of exogenous T3 illustrates that zebrafish embryos contain the necessary cofactors to enable TR-mediated transcriptional activation. The more than 100-fold difference in the activity of endogenous and exogenous TRs indicates that this reporter assay can be used to monitor the activity of TRα mutants even though zebrafish embryos express endogenous TRs. Moreover, these results indicate that despite their relative high concentration maternally provided THs do not fully activate endogenous TRs in zebrafish embryos.
The F-domain modulates transcriptional activity of TRαA in zebrafish embryos
Using this reporter system, we compared the activity of TRαA1 and TRαA1-F by injecting zebrafish embryos with equal amounts of mRNA for either receptor in addition to the β-galactosidase and luciferase reporters (Fig. 2C). Whereas in the absence of exogenous hormone these receptors showed no statistically significant difference in their activities (p=0.35), in the presence of T3 the activity of TRαA1-F was at least 3-fold higher than the activity of TRαA1 (p<0.01) (Fig. 2D). Fusion of the N-termini of TRαA1 and TRαA1-F to an influenza hemagglutin (HA) tag had no effect on the activity of these receptors (data not shown). Immunoblot analysis of zebrafish embryos injected with HA-tagged TRαA1 or TRαA1-F using a HA-tag specific antibody revealed that these proteins are expressed in similar levels (Fig. 2E). Dose-response analysis [defined as activity(+H)/activity(−H)] indicated that the higher activity of TRαA1-F is not caused by changes in the affinity for hormone but rather reflects an increase in transcriptional activity (Fig. 2F). Since in these experiments activation of the luciferase reporter depends on the translation of the injected mRNAs, it is possible that differences in the stability of the TRαA1 and TRαA1-F mRNAs bias the luciferase expression. However, the similar kinetics of luciferase activation by TRαA1 and TRαA1-F argues against this possibility (Fig. 2G). We therefore conclude that the F-domain alters the transcriptional activity of TRαA.
Zebrafish TRαA1 is the only known TR that has an F-domain. Therefore the effect of this domain on TRα activity may be zebrafish-specific. To address this possibility, we compared the activity of TRαA1 and TRαA1-F in zebrafish and transiently transfected monkey CV1 kidney carcinoma cells. In CV1 cells, deletion of the F-domain slightly but significantly (p<0.01) reduced repression by unliganded TRαA1 and augmented hormone-induced transcriptional activation of the reporter (p<0.01) albeit to a lesser degree than in injected zebrafish embryos (Fig. 3A). Contrary to the situation in zebrafish embryos, in CV1 cells deleting the F-domain did not alter the responsiveness of TRαA1 to hormone [activity(+H)/activity(−H)] (Fig. 3B). A N-terminal HA-tag did not alter the activity of these receptors (data not shown). Immunoblot analysis of CV1 cells transfected with HA-tagged TRαA1 and TRαA1-F revealed that deletion of the F-domain does not affect the expression or stability of TRαA1 (Fig. 3C).
Fig. 3. The F-domain does not regulate the hormone responsiveness of TRαA in mammalian cells.
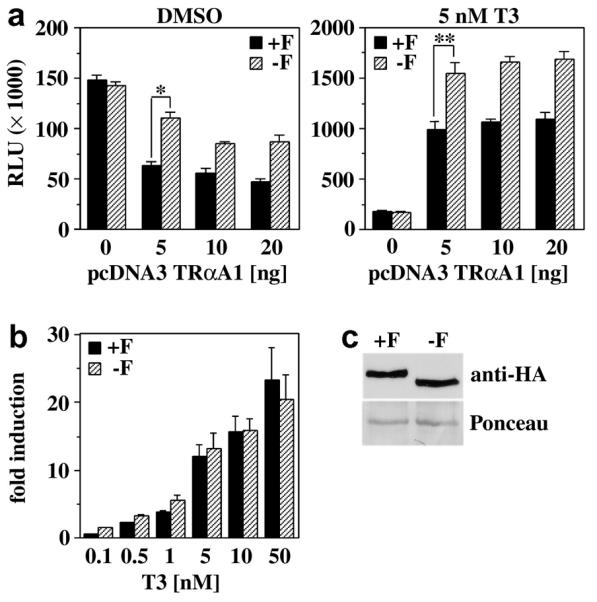
A, Luciferase activity (RLU) of CV1 cells transiently transfected with DR4 luciferase and CMV β-galactosidase reporters and various amounts of either pcDNA3 TRαA1 (+F) or pcDNA3 TRαA1-F (−F). Cells were treated with either DMSO (vehicle) or 5 nM T3. Shown are the averages and standard deviation of 3 independent experiments performed in triplicate. * A student’s t test shows that the difference in the activities of TRαA1 and TRαA1-F is statistically significant (p<0.001). B, Dose response analysis of the activity of TRαA1 and TRαA1-F (5 ng each) in the presence of various T3 concentrations (0.1 - 50 nM). Fold induction is defined as RLU(+H)/RLU(−H). Shown are the averages and standard deviations of three independent experiments performed in triplicate. C, Expression of TRαA1 and TRαA1-F in transiently transfected CV1 cells (30 μg total protein) monitored with a HA-tag specific antibody. Equal loading and transfer of CV1 expressed TRs were controlled by Panceau Red-staining of the blot. A selected Panceau Red-stained protein band is shown as loading control.
The F-domain regulates the interaction of TRαA1 with the zebrafish coactivator NCoA2
The observed differences in the activity of TRαA1 and TRαA1-F in zebrafish and CV1 cells suggest that the F-domain might be involved in species-specific interactions with coactivators. NCoA1-3 are well-studied TR coactivators for which sequences of (putative) zebrafish orthologs are available (Collingwood et al., 1999). These coactivators have a similar overall organization that includes an N-terminal basic helix-loop-helix (bHLH)/PAS domain, a nuclear receptor interaction domain (NID) and two activation domains (AD1, AD2) (Fig. 4A). AD1 and AD2 interact with the histone acetyltransferase CBP/p300, the protein methyltransferase CARM1 and various other enzymes involved in chromatin remodeling (Stallcup et al., 2003). Although the overall domain structure of mammalian and zebrafish NCoA1-3s appears to be conserved, their AD2 regions are quite distinct, suggesting that the activities of these coactivators may differ in mammals and zebrafish.
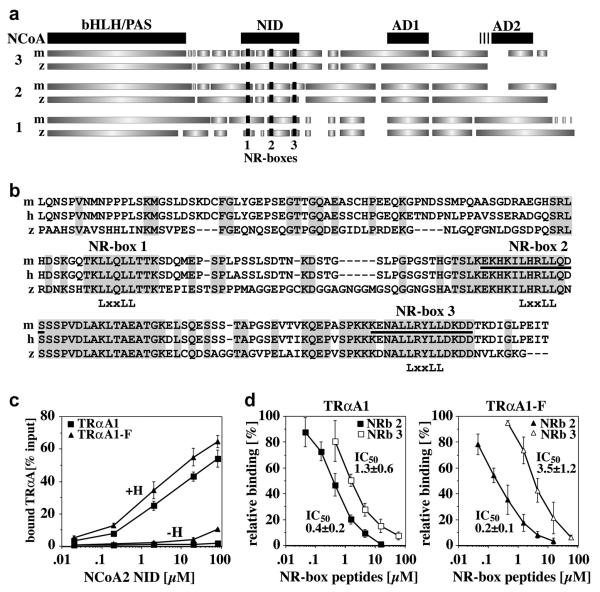
Fig. 4. The F-domain regulates the interaction of TRαA with the zebrafish coactivator NCoA2.
A, Domain structure of mouse and zebrafish NCoA1-3 based on a ClustalW sequence alignment (pHLH/PAS, helix-loop-helix/Per-Arnt-Sim domain; NID, nuclear receptor interaction domain; AD1 and AD2, activation domains 1 and 2. GeneBank accession numbers are: mNCoA3, NP_032705; zNCoA3, XP_692938 (predicted); mNCoA2, AAC53151; zNCoA2, AAK11608; mNCoA1, NM_010881; zNCoA1, CAI21172 (predicted). The nuclear receptor binding sites (NR-boxes 1-3) are shown in black. B, ClustalW alignment of the cloned zNCoA2 NID (aa 559-758) and the NIDs of mouse (m) and human (h) NCoA2 (both aa 563-764). The conserved “LxxLL” motifs of NR-boxes 1-3 are identified. Identical residues are grey shaded. The sequences of the NRb2 and NRb3 peptides used in (D) are underlined. C, Interaction of GST-bound zfNCoA2 NID (0.002-80 μM) with in vitro expressed, 35S-labeled TRαA1 and TRαA1-F in the presence of either DMSO (−H) or 10 μM T3 (+H). Shown are the averages and standard deviations of four binding experiments. The differences in the binding of TRαA1 and TRαA1-F at NCoA2 NID concentrations higher than 0.1 μM (+H) or higher than 10 μM (−H) are statistically significant (student’s t test p≤0.06). D, Competition of GST-bound zfNCoA2 NID (12 μM) and mNCoA2 NR-box 2 and 3 peptides (NRb 2, NRb 3) for binding to zebrafish TRαA1 and TRαA1-F. Peptides sequences are identified in (B) (underlined). Shown are the averages and standard deviations of four binding experiments. IC50 values are given in [μM].
Using a cDNA library of zebrafish embryos 1 day post fertilization (dpf), we cloned the NID of zebrafish NCoA2 (synonyms GRIP1, TIF2, SRC2). GST-pulldown experiments with a recombinantly expressed and affinity purified glutathione-S-transferase (GST) fusion of zebrafish NCoA2 NID revealed that deleting the F-domain increases the affinity of hormone-bound zebrafish TRαA1 and NCoA2 about 3-fold (p≤0.06) (Fig. 4C). This result suggested that the F-domain alters the interaction of TRαA1 with coactivators. The NID of p160 coactivators contains three conserved “LxxLL” binding motifs, called NR-boxes 1-3, which differ in their selectivity for nuclear receptors (Darimont et al., 1998) (Fig. 4B). The specificity of human TRβ for NCoA2 NR-box 2 depends on sequences N- and C-terminal of the conserved “LxxLL” motif. Based on the structures of a human TRβ:NR-box 2 complex (Darimont et al., 1998) and of steroid hormone receptor F-domains (Bledsoe et al., 2002; Fig. 1A), we hypothesized that interactions of the F-domain with the residues N-terminal of coactivator “LxxLL” motifs might alter the NR-box selectivity of zebrafish TRα. In agreement with this hypothesis, peptides representing NCoA2 NR-box 2 competed with GST-NCoA2 NID for binding to TRαA1-F with a 2-fold higher affinity than for binding to TRαA1 (0.01 ≤ p ≤ 0.06 for 0.15 μM ≤ [NR-box 2] ≤ 1.5 μM, student’s t test). In contrast, deletion of the F-domain decreased the affinity of TRαA1 for NR-box 3 about 3-fold (0.001 ≤ p ≤ 0.06 for 1.5 μM ≤ [NR-box 3] ≤ 15 μM), resulting in a more than 6-fold increase in the selectivity of TRαA1 for NR-box 2 (Fig. 4D). NCoA2 NR-box 3 binds human TRβ with a 4-fold lower affinity than NR-box 2, and TRβ interacts slightly better with a NCoA2 mutant that contains an inactive NR-box 3 (NCoA2 NR-box 3−) than with wild type NCoA2 (Darimont et al., 1998). In addition, in in vitro transcription assays performed with saturating NCoA2 concentrations a TRβ:NCoA2 NR-box3− complex displayed a 1.5-2-fold higher activity than TRβ:NCoA2 (Darimont, unpublished observation). These results indicate NR-boxes 2 and 3 compete for binding to TRβ and that TRβ: NCoA2 NR-box 2 interactions are not only more stable but also more active. In our interaction studies, zebrafish TRαA1 showed a similar NR-box preference as hTRβ. Hence, the observed decrease in affinity for NR-box 3 may explain the observed higher transcriptional activity of zebrafish TRαA1-F. If this hypothesis is true, we would expect the activity of TRαA1 in zebrafish embryos to increase more efficiently upon co-injection of the NCoA2 NR-box3− mutant than upon co-injection of wild type NCoA2. In addition to affecting the levels of target gene activation, the observed change in coactivator interaction may affect which genes are activated by TRα allowing cells to reprogram their response to THs.
Expression of the TRαA1 F-domain is regulated by alternative splicing
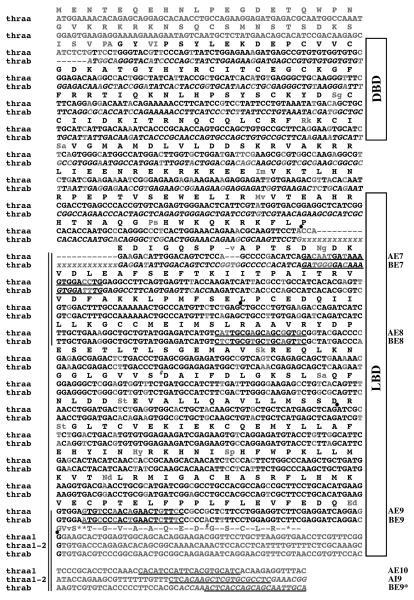
Information provided by the human and zebrafish genome analysis initiatives showed that the exon/intron structure of the zebrafish thraa gene (GeneBank access number NP_571471) is very similar to that of human THRA (GeneBank access number NP_955366) (Fig. 5A). Comparison of the predicted zebrafish thrab exon-intron structure (GeneBank access number XM_702123) with that of human THRA gave similar results (data not shown).
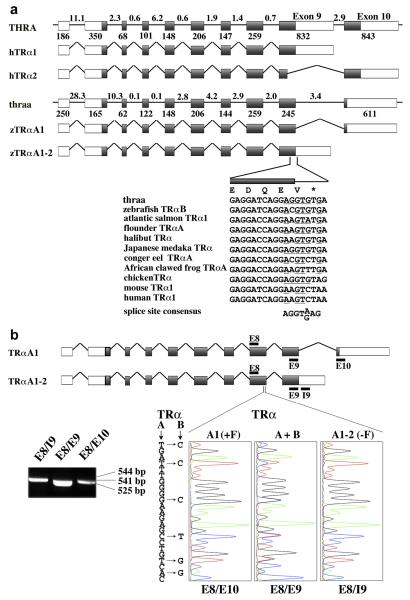
Fig. 5. Alternative splicing of Zebrafish TRαA exon 9 yields TRαA1 and TRαA1-2.
A, Comparison of exon-intron structure of the human THRA and zebrafish thraa genes and composition of THRA and thraa transcripts. Exons are identified by boxes and introns as lines; coding sequences are grey shaded. The lengths of introns (in kbp) are indicated above the introns; the length of exons (in bp) are shown below the exons. The sequences at the exon 9/intron 9-10 boundary of thraa is shown and compared with corresponding coding sequences for zebrafish TRαB (DQ017632), atlantic salmon TRα1 (AF146775), flounder TRαA (D16461), halibut TRα (AF143296), Japanese medaka TRα (AB114860), conger eel TRαA (AB183396), African clawed frog TRαA (M35343), chicken TRα (Y00987), mouse TRα1 (X5193), and human TRα1 (X55005). Nucleotides that are similar to the splice consensus sequence are underlined. B, Identification of TRαA1 (E8/E10), TRαA1-2 (E8/I9) and all TRα (E8/E9) transcripts by RT PCR using cDNA derived from 6 dpf embryos. Location of the PCR primers (black bars) and expected sizes of PCR fragments are indicated. Sequence analysis of these PCR products confirmed that the E8/E10 and E8/I9 products originate from TRαA1 and TRαA1-2 transcripts, respectively. The E8/E9 PCR product contained a mixture of TRαA and TRαB sequences (“C”- blue, “G”-black, “T”-red, “A”-green).
Two of the various isoforms (TRα1, TRα2) that are expressed from the human THR gene have different C-termini (Fig. 5A). Whereas expression of TRα1 terminates within exon 9, TRα2 is generated by an alternative splicing event in exon 9 that links part of exon 9 to exon 10. This splicing event replaces a significant part of the TRα1 LBD resulting in a receptor that is unable to bind hormone (Lazar and Chin, 1988). Like the mammalian TRα2 transcript, expression of the zebrafish TRαA1 transcript is generated by a splicing event in exon 9 that includes exon 10, which encodes the F-domain. However, contrary to the mammalian TRα2, the splicing event in exon 9 of zebrafish TRαA1 maintains a hormone-binding competent TRα LBD.
Closer inspection of the exon 9/intron 9-10 splice boundary of zebrafish thraa revealed that skipping of this splice site should enable the expression of a TRαA isoform (TRαA1-2) whose C-terminus corresponds to that of human TRα1 (Fig. 5A). This observation suggested that TRαA transcripts might be alternatively spliced allowing the F-domain to play a role in regulating the activity of TRα in vivo. Reverse transcriptase (RT)-PCR experiments using cDNA from 6 dpf zebrafish embryos and specific primers within exon 8 (E8), intron 9-10 (I9), exon 9 (E9) and exon 10 (E10) of the thraa gene yielded amplification products whose sizes and sequences are consistent with the presence of TRαA1 and TRαA1-2 transcripts (Fig. 5B). Moreover, sequence analyses of the E8/E9 PCR product confirmed that both the thraa and thrab genes are transcribed. Since the primers used in this analysis are located in different exons, PCR products resulting from genomic DNA would be at least 2 kbp longer than the obtained PCR product, which enables us to conclude that our PCR products are indeed derived from cDNA.
The exon 9/intron 9-10 junction of zebrafish thraa contains a classical splice site that is conserved in the TRα genes of Japanese medaka (Oryzias latipes) and chicken (Gallus gallus) (Fig. 5A). However, corresponding exon 9 sequences of other species are less similar to the splice site consensus sequence, with mammalian TRα genes displaying the lowest similarity. Based on these results it is unlikely that mammals express a TRα variant that contains an F-domain. Consistent with the identification of zebrafish TRαA1 as the only known TR transcript that contains a F-domain, blast searches did not identify any significant homologies to zebrafish thraa exon 10 in any species, nor were we able to find homologous sequences to thraa exon 10 downstream of zebrafish thrab. However, a definite answer to the question whether the F-domain is a unique feature of zebrafish TRαA1 will require experimental analysis of TRα transcripts from other species.
Comparison of the available TRαA1-2 and TRαB sequences suggested that the LBDs of these receptors are very similar (Fig. 6). With exception of a conservative glutamate to aspartate change at the C-terminus, sequence differences in the LBD of TRαA1-2 and TRαB mainly affect solvent-exposed loop regions, which are unlikely to influence the activity of these receptors. However, the predicted shorter N-terminus and splice variations at the junction between exon 6 and 7 of TRαB might have functional consequences. A very recently identified alternative spliceform of human TRα exchanges exon 6 for a microexon (6A) located in intron 6/7 (Casas et al., 2006). This isoform decreases the ability of TRα to inhibit MyoD transcriptional activity during myoblast proliferation.
Fig. 6. Sequence comparison of zebrafish TRαA1, TRαA1-2 and TRαB.
ClustalW alignment of the coding sequences and 3′ untranslated regions of TRαA (TRαA1, TRαA1-2) and TRαB. Sequence fragments that were amplified and sequenced in this study are identified by vertical bars. TRα-specific primers (AE7, AE8, AE9, AE10, AI9) and TRβ-specific primers (BE7, BE8, BE9, BE9*) are underlined. The cDNA sequence of TRαA1 has been previously described (Essner et al., 1997). Sequences that are based on predictions are shown in italic. The “xxxx” stretch in TRαB labels a region that contains several predicted splice variations of various lengths. In contrast to a previously published partial TRαB cDNA sequence (GeneBank accession number DQ017632), the TRαB sequence identified in this study corresponds well to the predicted TRαB sequence (GeneBank accession number XM_702123). Our sequence analysis of TRαB identified two positions that appear to be polymorphous (#). The positions of splice sites are labeled with dots. Sequences that are different in TRαA and TRαB are shown in grey. In case of deviations in the amino acid sequence, TRαA sequences are shown in capital letters (in the order of TRαA1 followed by TRαA1-2) and TRαB in small letters.
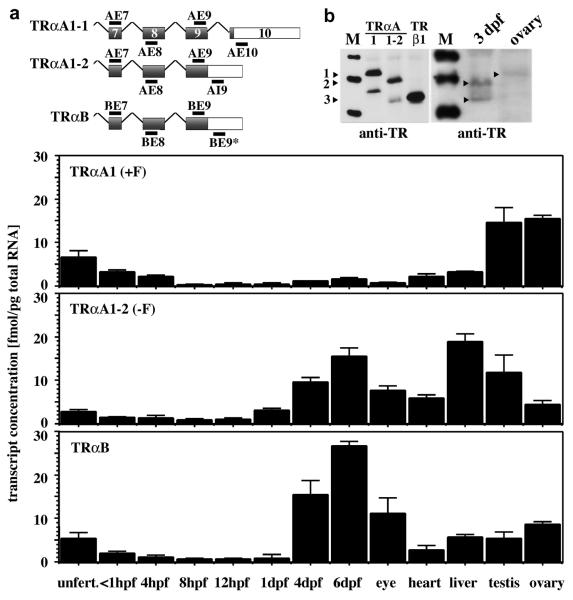
Expression of TRαA1, TRαA1-2 and TRαB is regulated in a stage- and tissue-specific manner
To investigate the expression patterns of TRαA1, TRαA1-2 and TRαB, we performed quantitative reverse transcriptase PCR experiments using primers that are specific for TRαA1, TRαA1-2 and TRαB (Fig. 6, 7A). By comparing the results generated with primers located within the same exon and primers located in different exons of thraa and thrab, we were able to control for genomic DNA contaminations in the cDNA samples. These experiments revealed that significant amounts of TRαA1-encoding transcripts are only present in testes, ovaries, unfertilized eggs and embryos up to 4 hpf (Fig. 7A). In embryos <1 hpf the concentration of TRαA1- and TRαA1-2-encoding transcripts were about 0.1 and 0.05 pg/embryo, respectively, which corresponds very well to the concentration of TRαA-encoding transcripts measured by RNase protection (Essner et al., 1999). In contrast to the expression of TRαA1, between 1 and 4 dpf the expression levels of TRαA1-2 increases 5-fold and that of TRαB 28-fold (Fig. 7A). The rise in the expression of these TR isoforms coincides with the onset of zygotic TH production (Elsalini and Rohr, 2003). In adult tissues the ratio of TRαA1, TRαA1-2 and TRαB varies in a tissue-specific manner with the eye and liver showing the highest level of TRαB and TRαA1-2, respectively, and testes and ovaries being particularly rich in TRαA1.
Fig. 7. Expression of TRαA1, TRαA1-2 and TRαB is regulated in a tissue-specific manner.
A, Quantitative PCR analysis of the expression of TRαA1, TRαA1-2 and TRαB in unfertilized zebrafish oocytes, zebrafish embryos of various stages and various adult zebrafish tissues. The sequences of the primers used in this study are shown in figure 6. While cDNA oocytes and zebrafish embryos did not contain detectable concentrations of genomic DNA, some of the tissue samples did (< 10%). In case of the primer pairs AE9/AI9 and BE9/BE9*, we cannot distinguish between amplification products resulting from cDNA or genomic DNA. The concentration of genomic DNA in these samples was quantified according to [AE9/AI9]genomic = [AE7/AE8] - [AE9/AE10] - [AE9/AE9*]cDNA. We observed that [BE9/BE9*] = [BE7/BE8] + [AE9/AI9]genomic, which indicates that TRαB is not spliced within exon 9. Shown are the averages and standard deviations of three amplification experiments of at least two independent samples performed in duplicate. Determination of template concentration followed the method outlined in Materials and Methods. B, Immunoblot analysis of the expression of TRs in lysates (200 μg total protein) from 3 dpf embryos and adult ovaries (right panel) and of in vitro expressed TRαA1 (47.0 kDa), TRαA1-2 (45.1 kDa) and TRβ1 (42.5 kDa) (left panel) using an antibody that recognizes the conserved sequence of α-helix 12 (Zhu et al., 1996). The predicted size of TRαB is 42.4 kDa, which is similar to that of TRβ1. Immunoreactive zebrafish bands corresponding to TRαA1 (1), TRαA1-2 (2), and TRβ1 or TRαB (3) are labeled.
Immunoblot analysis using an antibody that was generated against the conserved α-helix H12 in the TR LBD confirmed the expression of TRαA1 in zebrafish ovaries (Fig. 7B). A similar analysis of 3 dpf embryos showed two immunoreactive bands whose sizes are consistent with TRαA1 and TRαB/TRβ1 (Fig. 7B). Although this antibody readily detected in vitro expressed TRαA1, TRαA1-2 and TRβ, detection of endogenous TRs required high protein concentrations (>200 μg total protein) and a hypersensitive infrared detection system, indicating that not only the RNA but also the protein levels of endogenous TRs are very low. Although it is probably feasible to obtain antibodies that specifically recognize TRαA1, TRαA1-2 or TRαB, the low expression levels of these receptors will likely prevent their detection within cells.
Potential functional roles of the zebrafish TRαA1 F-domain
The high levels of TRαA1 in testes and ovaries suggest a role of this particular TRα isoform in reproduction. The ratio of TRαA1, TRαA1-2 and TRαB in unfertilized eggs corresponds well to the ratio of these transcripts in ovaries. Whether the elevated expression of TRαA1 in unfertilized eggs and early zebrafish embryos indicates a role of this TRα isoform in zebrafish development remains to be identified. The ability of the F-domain to attenuate hormone-dependent activation of TRα might render TRα less responsive to maternal THs, which in fish oocytes are present in high concentrations (Power et al., 2001). However, while the levels of maternal THs decrease gradually over the course of several days, TRαA1 transcript levels are only elevated during the first 8 hpf. Another possibility is that the F-domain modulates the ability of TRα to cross-talk with other pathways. Overexpression of TRαA1 in zebrafish embryos disrupted hindbrain patterning and resulted in growth retardation, which was interpreted in terms of functional interactions between TRαA1 and retinoid acid signaling (Essner et al., 1999). In contrast, in our experiments injection of zebrafish embryos with TRαA1 or TRαA1-F mRNAs had no significant effect on the viability, growth, morphology and cartilage/bone structure between 1 and 6 dpf.
The stage- and tissue-specific regulation of TR expression is key to understanding the mechanisms that underlie the cellular responses to THs. Dissection of these mechanisms will require the separate manipulation of the identified TRα isoforms in zebrafish embryos. Considering the possibility that in addition to RNA transcripts zebrafish oocytes might contain maternally deposited TRα protein, identifying the individual functions of these TRα isoforms will be technically difficult and might have to await the establishment of targeted gene replacements in zebrafish.
Acknowledgement
We are very grateful to the University of Oregon Zebrafish Community for fish husbandry, embryo production and access to equipment, to Adriana Rodriguez-Mari (UO) for help with zebrafish dissections, to Dr. Y. Honjo and members of the Eisen and Kimmel laboratories (UO) for help with microinjections and materials, to J. Talbot and C. Walker (UO) for help with cartilage/bone stains and analysis, and to Dr. C. Kentros (UO) for access to the MyiQ thermocycler. We acknowledge Dr. G. Kouzmitscheva for cloning of zNCoA2, and Drs. G. Fahid, N. Robinson (both LI-COR Biosciences, Inc) and J. Giebultowicz (Oregon State University) for assistance with the LI-COR Odyssey Scans. We would like to thank Drs. W.-K. Chan (NU, Signapore), S.- Y. Cheng (NIH), J. Postlethwait (UO), T. Scanlan (UCSF), and K. Yamamoto (UCSF) for providing materials and D. Hawley (UO) for advice and critical comments on the manuscript.
This work was supported by Grants from the Medical Research Foundation of Oregon to B.D. and National Institute of Health to J.E. (HD22786). M.H. was funded by a postdoctoral fellowship from the National Institute of Health (NS11170).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Due to the identification of the thrab gene (Genebank accession number XM_702123), the original zebrafish TRα gene (Genebank accession number NP_571471) is now called thraa. As shown in this study, thraa gives rise to at least two products: the F-domain encoding thraa1 (TRαA1, previously called TRα1 (Essner et al., 1997); Genebank accession number U54796) and thraa1-2 (TRαA1-2; Genebank accession number DQ991961). Evidence for the expression of thrab is given by Genebank accession numbers DQ017632 and DQ991962 (this study).
References
- Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- Bledsoe RK, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- Brown DD. The role of thyroid hormone in zebrafish and axolotl development. Proc Natl Acad Sci U S A. 1997;94:13011–6. doi: 10.1073/pnas.94.24.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DR, et al. Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol. 2006;145:1–19. doi: 10.1016/j.ygcen.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Bugge TH, et al. RXR alpha, a promiscuous partner of retinoic acid and thyroid hormone receptors. Embo J. 1992;11:1409–18. doi: 10.1002/j.1460-2075.1992.tb05186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas F, et al. Characterization of a novel thyroid hormone receptor alpha variant involved in the regulation of myoblast differentiation. Mol Endocrinol. 2006;20:749–63. doi: 10.1210/me.2005-0074. [DOI] [PubMed] [Google Scholar]
- Chan S, Kilby MD. Thyroid hormone and central nervous system development. J Endocrinol. 2000;165:1–8. doi: 10.1677/joe.0.1650001. [DOI] [PubMed] [Google Scholar]
- Chassande O, et al. Identification of transcripts initiated from an internal promoter in the c-erbA alpha locus that encode inhibitors of retinoic acid receptor-alpha and triiodothyronine receptor activities. Mol Endocrinol. 1997;11:1278–90. doi: 10.1210/mend.11.9.9972. [DOI] [PubMed] [Google Scholar]
- Collingwood TN, et al. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;23:255–75. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Escobar GM, et al. Maternal thyroid hormones early in pregnancy and fetal brain development. Best Pract Res Clin Endocrinol Metab. 2004;18:225–48. doi: 10.1016/j.beem.2004.03.012. [DOI] [PubMed] [Google Scholar]
- DeLong GR. Iodine and brain development. Dev Med Child Neurol. 1996;38:279–82. [PubMed] [Google Scholar]
- Elsalini OA, Rohr KB. Phenylthiourea disrupts thyroid function in developing zebrafish. Dev Genes Evol. 2003;212:593–8. doi: 10.1007/s00427-002-0279-3. [DOI] [PubMed] [Google Scholar]
- Essner JJ, et al. The zebrafish thyroid hormone receptor alpha 1 is expressed during early embryogenesis and can function in transcriptional repression. Differentiation. 1997;62:107–17. doi: 10.1046/j.1432-0436.1997.6230107.x. [DOI] [PubMed] [Google Scholar]
- Essner JJ, et al. Overexpression of thyroid hormone receptor alpha 1 during zebrafish embryogenesis disrupts hindbrain patterning and implicates retinoic acid receptors in the control of hox gene expression. Differentiation. 1999;65:1–11. doi: 10.1046/j.1432-0436.1999.6510001.x. [DOI] [PubMed] [Google Scholar]
- Farboud B, Privalsky ML. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–53. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- Flamant F, Samarut J. Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol Metab. 2003;14:85–90. doi: 10.1016/s1043-2760(02)00043-7. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Iniguez-Lluhi JA, et al. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–56. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- Kauppi B, et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem. 2003;278:22748–54. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- Lam SH, et al. Effects of thyroid hormone on the development of immune system in zebrafish. Gen Comp Endocrinol. 2005;142:325–35. doi: 10.1016/j.ygcen.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–93. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- Lazar MA, et al. Differential DNA binding by monomeric, homodimeric, and potentially heteromeric forms of the thyroid hormone receptor. Mol Cell Biol. 1991;11:5005–15. doi: 10.1128/mcb.11.10.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA, Chin WW. Regulation of two c-erbA messenger ribonucleic acids in rat GH3 cells by thyroid hormone. Mol Endocrinol. 1988;2:479–84. doi: 10.1210/mend-2-6-479. [DOI] [PubMed] [Google Scholar]
- Liu YW, Chan WK. Thyroid hormones are important for embryonic to larval transitory phase in zebrafish. Differentiation. 2002;70:36–45. doi: 10.1046/j.1432-0436.2002.700104.x. [DOI] [PubMed] [Google Scholar]
- Liu YW, et al. Temporal expression and T3 induction of thyroid hormone receptors alpha1 and beta1 during early embryonic and larval development in zebrafish, Danio rerio. Mol Cell Endocrinol. 2000;159:187–95. doi: 10.1016/s0303-7207(99)00193-8. [DOI] [PubMed] [Google Scholar]
- Mai W, et al. Thyroid hormone receptor alpha is a molecular switch of cardiac function between fetal and postnatal life. Proc Natl Acad Sci U S A. 2004;101:10332–7. doi: 10.1073/pnas.0401843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand O, et al. Molecular cloning and characterization of thyroid hormone receptors in teleost fish. J Mol Endocrinol. 2001;26:51–65. doi: 10.1677/jme.0.0260051. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Kingsbury R. Transcriptional control signals of a eukaryotic proteincoding gene. Science. 1982;217:316–24. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Montano MM, et al. The carboxy-terminal F domain of the human estrogen receptor: role in the transcriptional activity of the receptor and the effectiveness of antiestrogens as estrogen antagonists. Mol Endocrinol. 1995;9:814–25. doi: 10.1210/mend.9.7.7476965. [DOI] [PubMed] [Google Scholar]
- Pearce D, Yamamoto KR. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993;259:1161–5. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- Peters GA, Khan SA. Estrogen receptor domains E and F: role in dimerization and interaction with coactivator RIP-140. Mol Endocrinol. 1999;13:286–96. doi: 10.1210/mend.13.2.0244. [DOI] [PubMed] [Google Scholar]
- Pitt-Rivers R, Tata JR. The Thyroid Hormones. Pergamon Press; London: 1959. [Google Scholar]
- Power DM, et al. Thyroid hormones in growth and development of fish. Comp Biochem Physiol C Toxicol Pharmacol. 2001;130:447–59. doi: 10.1016/s1532-0456(01)00271-x. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–60. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Rastinejad F, et al. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–11. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- Renaud JP, Moras D. Structural studies on nuclear receptors. Cell Mol Life Sci. 2000;57:1748–69. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruse MD, Jr., et al. Competitive cofactor recruitment by orphan receptor hepatocyte nuclear factor 4alpha1: modulation by the F domain. Mol Cell Biol. 2002;22:1626–38. doi: 10.1128/MCB.22.6.1626-1638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JA, et al. Mutations targeted to a predicted helix in the extreme carboxylterminal region of the human estrogen receptor-alpha alter its response to estradiol and 4-hydroxytamoxifen. J Biol Chem. 2002;277:13202–9. doi: 10.1074/jbc.M112215200. [DOI] [PubMed] [Google Scholar]
- Sladek FM, et al. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol. 1999;19:6509–22. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup MR, et al. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85:139–45. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- Suaud L, et al. The activity of the activation function 2 of the human hepatocyte nuclear factor 4 (HNF-4alpha) is differently modulated by F domains from various origins. Biochem J. 1999;340(Pt 1):161–9. [PMC free article] [PubMed] [Google Scholar]
- Tata JR. Amphibian metamorphosis as a model for studying the developmental actions of thyroid hormone. Biochimie. 1999;81:359–66. doi: 10.1016/s0300-9084(99)80082-0. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A guide for the labratory use of zebrafish (Danio rerio) Univ. of Oregon Press; Eugene: 2000. The zebrafish book. [Google Scholar]
- Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–6. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- Yamano K, et al. Cloning of thyroid hormone receptor genes expressed in metamorphosing flounder. Dev Genet. 1994;15:378–82. doi: 10.1002/dvg.1020150409. [DOI] [PubMed] [Google Scholar]
- Yamano K, Inui Y. cDNA cloning of thyroid hormone receptor beta for the Japanese flounder. Gen Comp Endocrinol. 1995;99:197–203. doi: 10.1006/gcen.1995.1102. [DOI] [PubMed] [Google Scholar]
- Yamano K, Miwa S. Differential gene expression of thyroid hormone receptor alpha and beta in fish development. Gen Comp Endocrinol. 1998;109:75–85. doi: 10.1006/gcen.1997.7011. [DOI] [PubMed] [Google Scholar]
- Yaoita Y, Brown DD. A correlation of thyroid hormone receptor gene expression with amphibian metamorphosis. Genes Dev. 1990;4:1917–24. doi: 10.1101/gad.4.11.1917. [DOI] [PubMed] [Google Scholar]
- Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–66. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- Zhu XG, et al. Understanding the molecular mechanism of dominant negative action of mutant thyroid hormone beta 1-receptors: the important role of the wild-type/mutant receptor heterodimer. Endocrinology. 1996;137:712–21. doi: 10.1210/endo.137.2.8593822. [DOI] [PubMed] [Google Scholar]