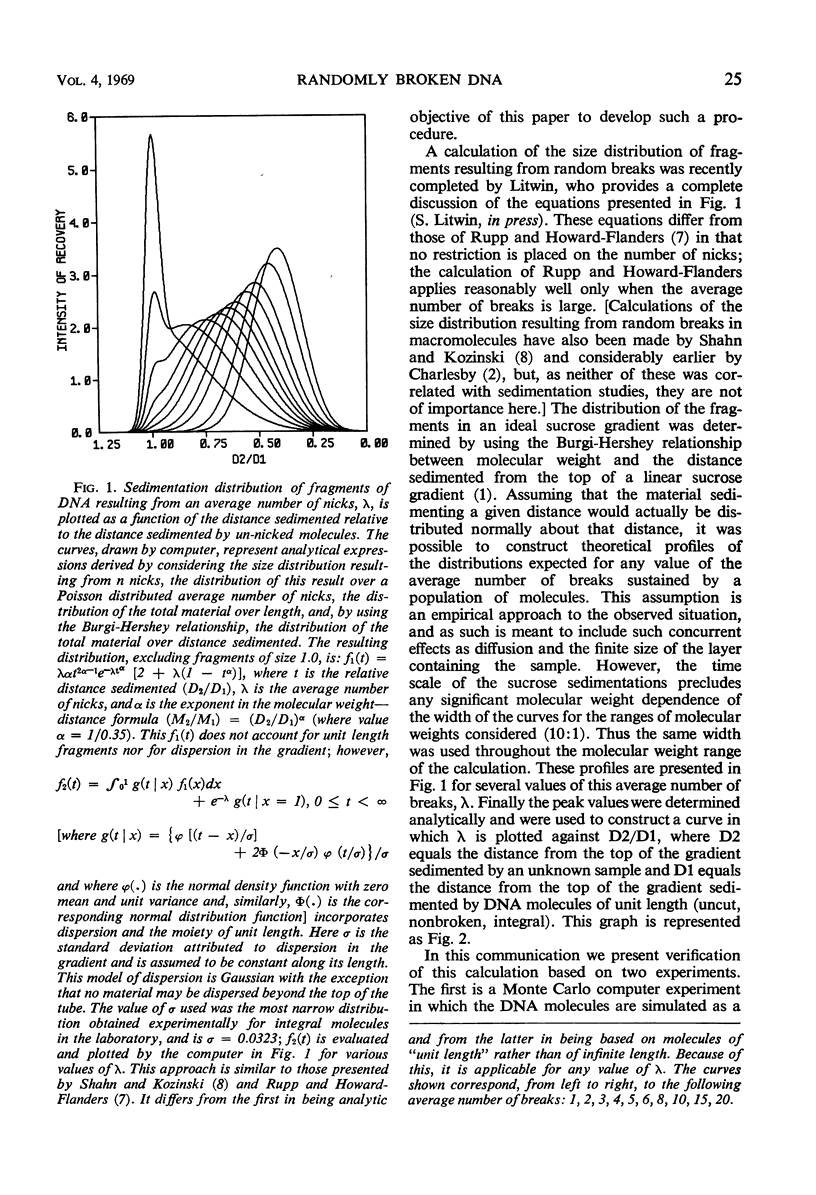
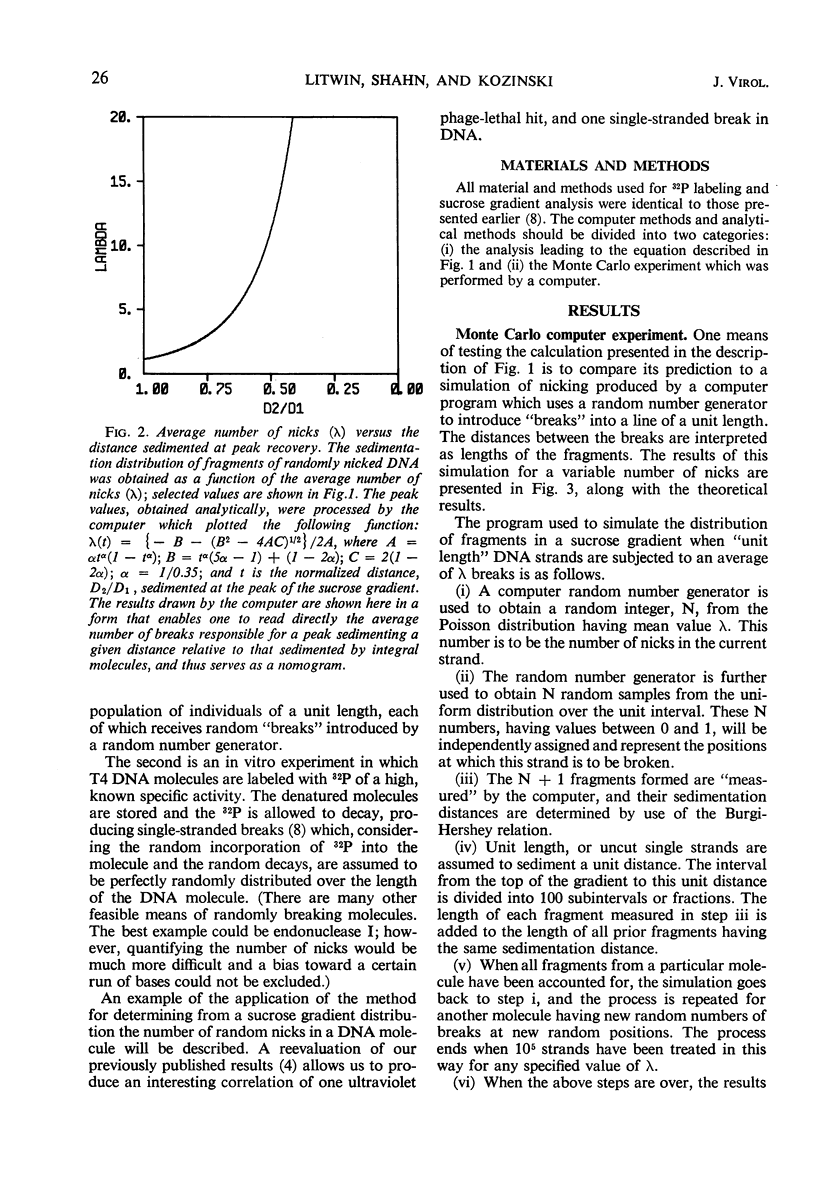
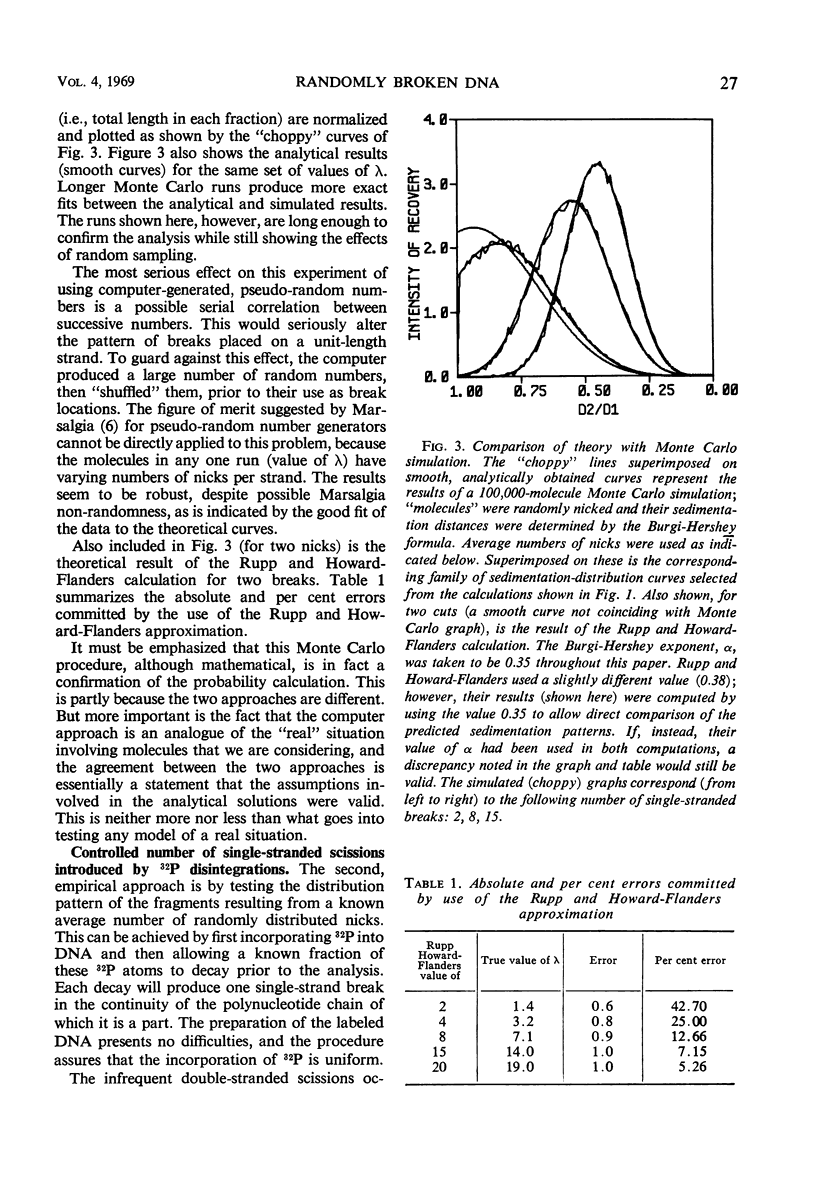
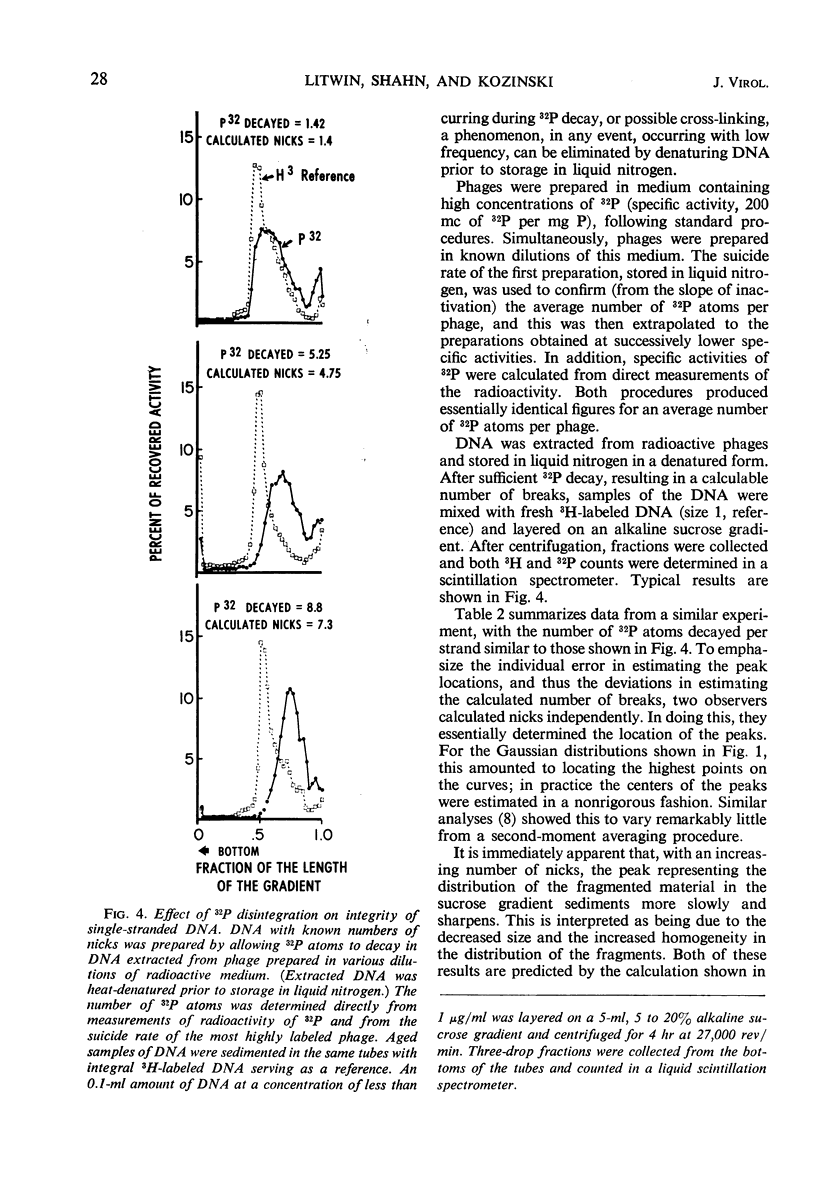
Abstract
Mass distribution in a sucrose gradient of deoxyribonucleic acid (DNA) fragments arising as a result of random breaks is predicted by analytical means from which computer evaluations are plotted. The analytical results are compared with the results of verifying experiments: (i) a Monte Carlo computer experiment in which simulated molecules of DNA were individuals of unit length subjected to random “breaks” applied by a random number generator, and (ii) an in vitro experiment in which molecules of T4 DNA, highly labeled with 32P, were stored in liquid nitrogen for variable periods of time during which a precisely known number of 32P atoms decayed, causing single-stranded breaks. The distribution of sizes of the resulting fragments was measured in an alkaline sucrose gradient. The profiles obtained in this fashion were compared with the mathematical predictions. Both experiments agree with the analytical approach and thus permit the use of the graphs obtained from the latter as a means of determining the average number of random breaks in DNA from distributions obtained experimentally in a sucrose gradient. An example of the application of this procedure to a previously unresolved problem is provided in the case of DNA from ultraviolet-irradiated phage which undergoes a dose-dependent intracellular breakdown. The relationship between the number of lethal hits and the number of single-stranded breaks was not previously established. A comparison of the calculated number of nicks per strand of DNA with the known dose in phage-lethal hits reveals a relationship closely approximating one lethal hit to one single-stranded break.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozinski A. W., Felgenhauer Z. Z. Molecular recombination in T4 bacteriophage deoxyribonucleic acid. II. Single-strand breaks and exposure of uncomplemented areas as a prerequisite for recombination. J Virol. 1967 Dec;1(6):1193–1202. doi: 10.1128/jvi.1.6.1193-1202.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozinski A. W., Kozinski P. B., James R. Molecular recombination in T4 bacteriophage deoxyribonucleic acid. I. Tertiary structure of early replicative and recombining deoxyribonucleic acid. J Virol. 1967 Aug;1(4):758–770. doi: 10.1128/jvi.1.4.758-770.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozinski A., Lorkiewicz Z. K. Early intracellular events in the replication of T4 phage DNA, IV. Host-mediated single-stranded breaks and repair in ultraviolet-damaged T4 DNA. Proc Natl Acad Sci U S A. 1967 Nov;58(5):2109–2116. doi: 10.1073/pnas.58.5.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsaglia G. Random numbers fall mainly in the planes. Proc Natl Acad Sci U S A. 1968 Sep;61(1):25–28. doi: 10.1073/pnas.61.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahn E., Kozinski A. Fragmentary transfer of P32 labeled parental DNA to progeny phage. 3. Incorporation of a single parental fragment to the progeny molecule. Virology. 1966 Nov;30(3):455–470. doi: 10.1016/0042-6822(66)90122-x. [DOI] [PubMed] [Google Scholar]


