Abstract
Among ~5,000,000 fungal species,1 Candida albicans is exceptional in its lifelong association with humans, either within the gastrointestinal microbiome or as an invasive pathogen.2 Opportunistic infections are generally ascribed to defective host immunity 3 but may require specific microbial programs. Here, we report that exposure of C. albicans to the mammalian gut triggers a developmental switch, driven by the Wor1 transcription factor, to a commensal cell type. Wor1 expression was previously observed only in rare genetic backgrounds,4–6 where it controls a white-opaque switch for mating.4–7 We show that passage of wild-type cells through the murine gastrointestinal tract triggers WOR1 expression and a novel phenotypic switch. The resulting GUT (Gastrointestinally-IndUced Transition) cells differ morphologically and functionally from previously defined cell types, including opaque, and express a transcriptome that is optimized for the digestive tract. The white-GUT switch illuminates how a microorganism utilizes distinct genetic programs to transition between commensalism and invasive pathogenesis.
The yeast Candida albicans is well-known as the most common agent of symptomatic fungal disease,8,9 but its more typical role is as a permanent resident of the healthy gastrointestinal microbiome.2 Longitudinal molecular typing studies indicate that disseminated C. albicans infections originate from patients’ own commensal strains,10 and the transition to virulence is generally thought to reflect impaired host immunity.3 Nevertheless, the ability of this commensal-pathogen to thrive in radically different host niches speaks to the existence of functional specializations for commensalism and disease. To investigate the C. albicans commensal lifestyle, we developed a murine model of stable gastrointestinal (GI) candidiasis in which the animals remain healthy despite persistent infection with high titers of yeasts.11 Using this model, we found that a C. albicans mutant lacking the Efg1 transcriptional regulator exhibits enhanced commensalism, such that it strongly outcompetes wild type in mixed infections (Figure 1A); similar findings were recently reported elsewhere.12
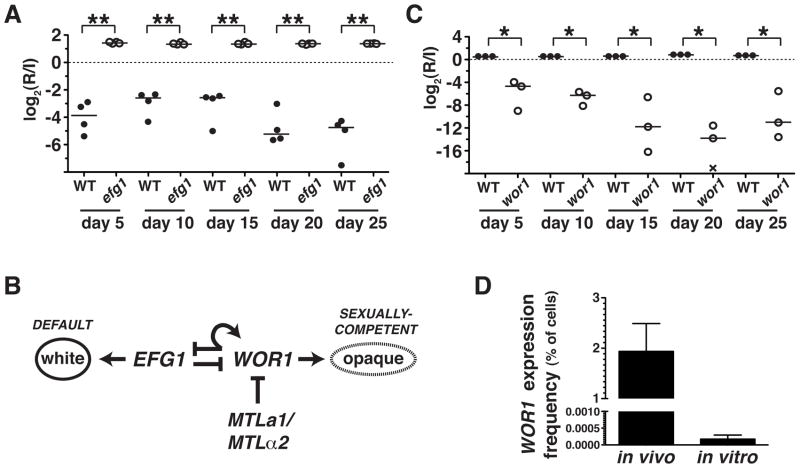
Figure 1. EFG1 inhibits and WOR1 promotes C. albicans fitness in the commensal milieu.
A) Competition experiment between wild-type (WT, SN250) and efg1ΔΔ mutant (SN1011) strains in 4 mice. Relative abundance was determined by qPCR, using strain-specific primers. Log2(R/I) represents the log ratio of the abundance of each strain after recovery from the feces of a given animal (R) compared to the level in the infecting inoculum (I). * indicates p<0.001 by the t-test (two-tailed, unpaired samples). Similar results were obtained when the efg1ΔΔ mutant was competed in a pool of 48 strains, as described in the text (Pande and Noble, unpublished). B) Transcriptional regulation of the white-to-opaque switch. C) Competition experiment between WT (SN250) and wor1ΔΔ (SN881) in 3 mice, as above. * p<0.05. Similar results were obtained in tests of two additional wor1ΔΔ strains, shown in Supplementary Figure 1, A–B. D) Comparison of WOR1 expression in MTLa/α cells grown in the commensal model or in vitro. The WOR1promoter-FLP strain (SN1020) was propagated for 3 days in the murine model or for 8 generations in liquid culture medium, followed by 5-FOA selection and PCR of 5-FOAR colonies to verify Flp-mediated deletion URA3. Mean percentages of cells meeting these criteria are plotted on the y-axis. Error bars reflect the standard deviation among 6 (in vivo) or 4 (in vitro) biological replicates.
Efg1 has diverse cellular functions, including inhibition of WOR1,13 the master regulator of a “white-opaque” epigenetic switch that controls C. albicans sexual competency 4–6 (Figure 1B). We asked whether WOR1 also regulates commensalism by testing a wor1ΔΔ deletion mutant in the same murine model. Indeed, as shown in Figure 1C, wor1ΔΔ was rapidly depleted from the GI tract, indicating that WOR1 is required for normal commensal fitness. Similar defects were observed with two additional wor1ΔΔ mutants (Supplementary Figure 1A–1B), confirming linkage between WOR1 and fitness. Moreover, the substantial commensal defect of a heterozygous knockout mutant (wor1Δ/WOR1, Supplementary Figure 1C) indicates that WOR1 dosage is also important. Note that a his1ΔΔ, leu2ΔΔ auxotroph exhibited wild-type fitness in the same assay (Supplementary Figure 1D), indicating that these gene disruption markers are neutral for commensalism, as they are for virulence.14
These in vivo results were remarkable in light of multiple reports in the C. albicans literature suggesting that only rare cell types are competent for WOR1 expression in vitro.4–6 Under laboratory conditions, WOR1 is repressed in most isolates of this diploid species by a potent transcriptional repressor, a1-α2, whose subunits are encoded by distinct alleles of the MTLa/α Mating Type-Like locus.4–6 Therefore, only strains that have undergone loss of either MTLa or MTLα were considered capable of expressing WOR1.4–6 However, our analysis was performed with MTLa/α strains.
We hypothesized that signals present in the mammalian GI tract might elicit WOR1 expression in MTLa/α cells. To test this idea, we utilized a technique for permanently marking cells when a promoter is activated in vivo 15,16 (Supplementary Figure 2A). We created a WOR1prom-FLP strain containing an endogenous WOR1 promoter fused to the gene for Flp recombinase, as well as a copy of URA3 (conferring sensitivity to 5-fluoroorotic acid, 5-FOA) flanked by FRT recombination sites. Activation of the WOR1 promoter results in expression of FLP, deletion of URA3, and resistance to 5-FOA. Using this strain, we determined the frequency of WOR1 expression in MTLa/α C. albicans propagated in the laboratory or in the murine commensal model. After 8 generations of mid-log growth at 37°C, only 0.0002% of cells propagated in vitro exhibited Flp-medicated excision of URA3 (Figure 1D, Supplementary Figure 2B–2C). In contrast, after 3 days of growth in the host, 1.9% of cells exhibited this event (Figure 1D, Supplementary Figure 2B–2C). These figures likely underestimate the true frequency of WOR1 expression, since Flp is reportedly unstable at mammalian body temperature.17 Nevertheless, comparison of the rates suggests that propagation of MTLa/α cells within the mammalian GI tract increases the probability of WOR1 expression by roughly 10,000-fold.
To determine whether overexpression of WOR1 in MTLa/α cells would confer a gain of function phenotype, we constructed a strain (WOR1OE) in which a single copy of WOR1 is driven by the strong, constitutively expressed TDH3 promoter (Supplementary Figure 3A–3C). As shown in Figure 2A, WOR1OE exhibited a transient competitive deficit in the murine commensal model (day 5), followed by the predicted competitive advantage over wild type (days 10 to 25).
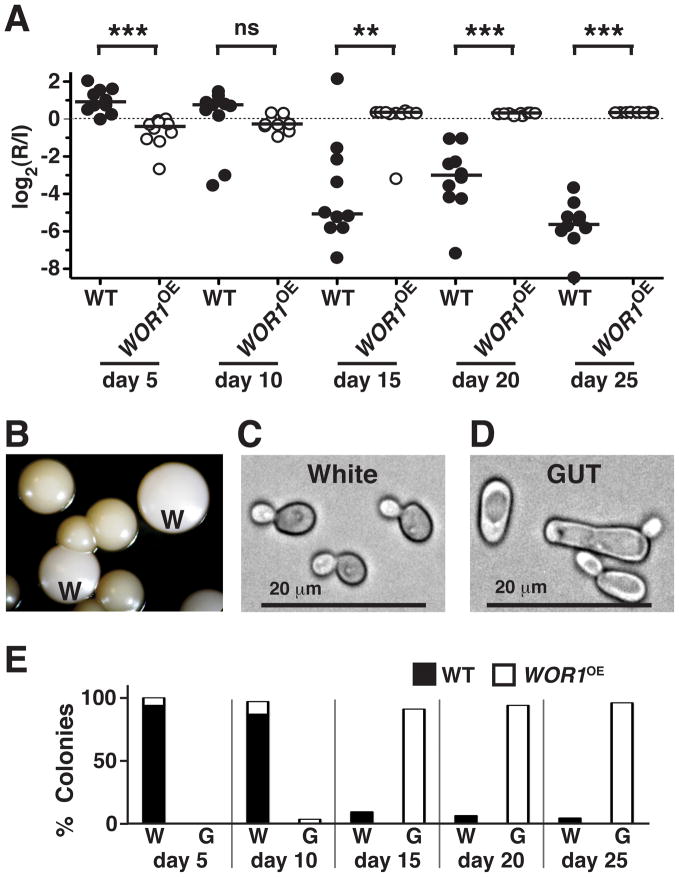
Figure 2. Wor1 promotes a white-to-GUT transition that confers enhanced fitness in the mammalian gastrointestinal tract.
A) Competition between wild-type (SN425) and WOR1OE (SN928) MTLa/α strains in the murine commensal model (n = 10 animals). The t-test (two-tailed, unpaired samples) was used to determine significance. *p<0.05, **p<0.001, ***p<0.0001, n.s. nonsignificant. A replicate of this experiment yielded similar results (Pande and Noble, unpublished). B) Appearance of distinct colony morphologies after recovery from the mouse. W signifies colonies with typical white morphology. Colonies with GUT morphology are unmarked. C) and D) Light microscopic appearance of cells recovered from white and GUT colonies, respectively. E) Analysis of colonies recovered from the WT vs. WOR1OE commensal competition experiment. White (W) vs. GUT (G) phenotypes were visually assessed, followed by determination of strain identity (WT vs. WOR1OE) by colony PCR.
Inspection of colonies recovered from animals revealed an unexpected consequence of WOR1 overexpression. C. albicans usually exists in the “white” phase, characterized by round to oval yeast morphology and white, domed colonies.17 Whereas white strains were used to infect the commensal model, yeasts recovered 10 days later produced two types of colonies (Figure 2B). One was white, like that of the infecting strains (W, Figure 2B), but the second was dark and flattened (unmarked, Figure 2B), resembling the appearance of sexually-competent “opaque” colonies.18 By light microscopy, cells from white colonies were round to oval (Figure 2C), but cells from dark colonies were elongated, with prominent vacuoles (Figure 2D), resembling the morphology of opaque cells. Unlike true opaques, however, cells from both colony morphotypes generally retained heterozygosity at MTL (Supplementary Table 1). An analysis of colony morphology and strain identity over the experimental time course (Figure 2E) revealed that dark colonies were formed exclusively by WOR1OE and that, once apparent, these colonies rapidly dominated the population.
These results established that WOR1 overexpression within the mammalian gut produces two profound changes in C. albicans biology. First, WOR1OE cells acquire enhanced commensal fitness, such that they outcompete wild type in mixed infections. Second, although the WOR1OE strain remains white indefinitely when propagated strictly in vitro, exposure to the mammalian digestive tract triggers a heritable switch to the “dark” phenotype. We hypothesized that this phenotype might signify a larger program of changes that are adaptive in the gut, and that fixed expression of WOR1 somehow stabilizes the program even after host cues are removed. We tested the first part of this hypothesis by performing competitive infections between wild-type C. albicans and WOR1OE yeasts recovered from dark colonies. As shown in Supplementary Figure 4A, dark phase WOR1OE was almost immediately hypercompetitive, unlike the original white strain (Figure 2A). The specificity of this fitness advantage was determined by competing dark and white cells in two other systems. Strikingly, dark cells were significantly attenuated in both liquid culture medium (Supplementary Figure 4B) and in a murine bloodstream model of virulence (Supplementary Figure 4C). These results support a specific role in commensalism for the dark developmental program, hereafter termed GUT for Gastrointestinally-IndUced Transition.
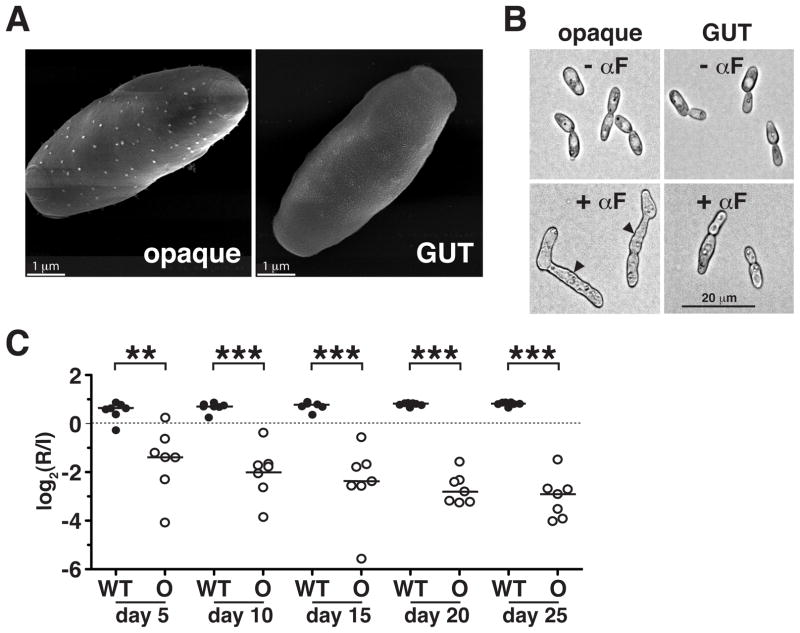
We explored the relationship between MTLa/α GUT cells and morphologically similar MTLa opaque cells by assaying each cell type for characteristics of the other. Apart from loss of heterozygosity at MTL, opaque cells are characterized by: 1) heat-sensitivity, such that they rapidly convert to white at temperatures >25°C; 2) cell surface “pimple” structures, detectable by scanning electron microscopy (SEM); 3) production of mating filaments in response to mating pheromone; 4) capacity for mating with a partner of the opposite sex; and 5) an “opaque-specific” transcriptome, defined by comparison with white cells.19 The first indication that WOR1OE GUT cells are not the same as opaque cells was their stability to incubation at 30°C (the temperature at which they were initially recovered) and 37°C (Supplementary Figure 5A). Visualization of GUT cells by SEM revealed few if any pimple structures (Figure 3A; see Supplementary Figure 5B for white controls), and GUT cells proved unresponsive to mating pheromone (Figure 3B; see Supplementary Figure 5C for white controls). Similarly, quantitative mating assays revealed that GUT cells mate with approximately 2 million-fold lower efficiency than opaque cells (<3×10−7 vs. 8.0×10−1, respectively, after 5 days; Supplementary Table 2). By these criteria, GUT cells lack the defining characteristics of opaque cells.
Figure 3. GUT cells are distinct from previously identified opaque cells.
A) Scanning electron micrographs showing “pimples” on the surface of opaque (SN967) but not GUT (SN1045) cells. B) Opaque but not GUT cells form mating filaments in response to mating pheromone (αF). Arrowheads indicate mating projections. All images were obtained at the same magnification. C) Opaque cells (SN967) are significantly outcompeted by wild type (SN425) in the murine commensal model (n = 7 animals). ** p<0.005, *** p<0.001 by the t-test. A replicate of this experiment yielded similar results (Pande and Noble, unpublished).
The major characteristic of GUT cells, identified in this study, is enhanced fitness within the mammalian gastrointestinal tract. We therefore tested opaque cells in the same commensal model, using the same white phase competitor as in previous tests of GUT cells (Figure 2A and Supplementary Figure 4A). The opaque and white strains were isogenic, apart from allelism at MTL. As shown in Figure 3C, opaque cells are severely attenuated for commensalism. Taken together with the failure of GUT cells to exhibit opaque cell characteristics, the failure of opaque cells to establish robust commensal infections indicates that the two cell types are distinct.
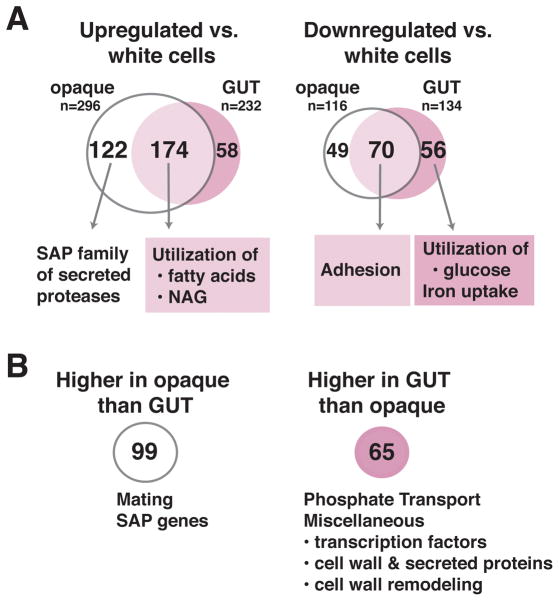
The mammalian GI tract differs from the bloodstream and from standard in vitro conditions in well-described nutritional and physical parameters.11,20 We hypothesized that an analysis of the GUT transcriptome in light of these differences might yield insights into its functional specializations. GUT cells, opaque cells, and isogenic white cells were propagated in glucose-containing medium, maintained at room temperature to preserve the phenotype of opaque cells, and profiled using custom C. albicans ORF microarrays. The results are schematized in Figure 4A, with the full dataset and results for genes of interest appearing in Supplementary Tables 3 and 4, respectively. Compared to white phase controls, both GUT and opaque cells demonstrated significant, 2-fold or greater upregulation of a common set of 174 genes. These were enriched for GO-terms associated with the catabolism of fatty acids (p=1.11×10−9) and N-acetylglucosamine (p=5.69×10−3) compared to the genome as a whole. Opaque cells but not GUT cells also upregulated six members of the SAP gene family, which encode secreted aspartyl proteases with previously demonstrated roles in nutrient acquisition 21 and virulence.22 GUT and opaque cells downregulated a common set of 70 genes, including six genes associated with “biological adhesion” (p=3.95×10−2). Notably, GUT cells but not opaque cells also downregulated genes associated with iron acquisition (p=4.96×10−5) and glucose catabolism (p=7.40×10−4).
Figure 4. GUT and opaque cells exhibit overlapping but distinct patterns of transcriptional expression.
RNAs from WOR1OE MTLa/α GUT cells (SN1045, n = 4 biological replicates), MTLa opaque cells (SN967, n = 2 biological replicates), and isogenic white controls (n = 2–4 biological replicates) were profiled using custom Agilent C. albicans ORF microarrays and a pooled reference. Significant changes in gene expression were defined as ones with BAGEL p-values <0.05, after implementation of a Bonferroni correction for comparison of multiple variables. A) Venn diagram of genes that are significantly upregulated or downregulated in both opaque and GUT cells, relative to white cells. B) Genes that are differentially expressed by GUT vs. opaque cells.
Consistent with our prediction, GUT cells exhibited a striking reorientation of cellular metabolism towards nutrients available in the distal mammalian GI tract. Because mammals digest and absorb dietary starch and oligosaccharides in the proximal small bowel,23 glucose is relatively depleted more distally. The large bowel is instead replete with short chain fatty acids, which is produced by microbial fermentation of indigestible carbohydrates,23 and N-acetylglucosamine, which is a component of host mucin 24 and bacterial peptidoglycan.25 Iron is abundant throughout the GI tract 26 but is predicted to be more bioavailable in the anaerobic atmosphere of the large bowel, where microbial limitation of iron uptake may defend against iron-related toxicity.11,27 This optimization of GUT cell metabolism towards conditions encountered in the mammalian digestive tract may contribute to its success as a commensal, although additional factors are likely important given that opaque cells, which are attenuated for commensalism (Figure 3C), share several of these metabolic features (Figure 4A and 28).
Direct comparison of the GUT and opaque transcriptomes highlights important differences between the two cell types. In addition to secreted aspartyl proteases, opaque cells express significantly higher levels of genes required for mating (STE2, STE4, STE18, CAG1; Figure 4B and Supplementary Table 4). Conversely, GUT cells express higher levels of genes for phosphate uptake (p=3.00×10−2), as well as certain predicted transcription factors, cell surface or secreted proteins, and cell wall remodeling enzymes (Supplementary Table 4) that could potentially modulate cell identity.
Our results indicate that the ability of a commensal organism to produce disease is not merely a consequence of impaired host immunity. In the case of C. albicans, we propose that a wholesale change of cell identity underlies its transition from commensal to pathogen. Pathogenic C. albicans consists of white phase budding yeasts, pseudohyphae, and hyphae.29 Based on results described here, we propose that cues from the mammalian GI tract trigger WOR1 expression and the white-GUT switch in at least a fraction of host-associated yeasts (Supplementary Figure 6). GUT phase MTLa/α cells thrive within the GI tract because of metabolic adaptations to the locally available nutrients, as well as other, undefined specializations. Continuous exposure to GI-specific signals is probably important to maintain the GUT state, since wild-type cells are recovered in the white phase after exit from the host. Such signals may include CO2 and N-acetylglucosamine, which have very recently been reported to trigger morphological elongation and WOR1 RNA accumulation in a subset of MTLa/α clinical isolates.30 Yet, our recovery from animals of WOR1OE isolates that retain the GUT phenotype over generations of growth in the laboratory suggests that WOR1 expression is sufficient to maintain the phenotype. It is also possible that, under particular conditions, GUT cells naturally give rise to sexually competent opaque cells via loss of one allele of MTL; however, the observation that 95% of clinical isolates remain heterozygous at MTL 31,32 argues that this may be rare in humans. In conclusion, we describe a new cell type, programmed by Wor1, that drives the commensal lifestyle of Candida albicans. The identification of specialized states for C. albicans commensalism and virulence offers opportunities for prevention as well as treatment of clinical diseases produced by this important human pathogen.
Online Methods
Strain Construction
All strains used in this study are derivatives of the clinical isolate, SC5314.33 Strains are listed in Supplementary Table 5, plasmids in Supplementary Table 6, and primers in Supplementary Table 7. The wor1ΔΔ mutant (SN881) was constructed by fusion PCR as previously described.14
The WOR1OE/wor1Δ strain (SN928) used for animal experiments was created in two steps. PCR and homologous recombination in S. cerevisiae 34 were used to engineer a plasmid (pSN209) containing: PmeI restriction site, WOR1 promoter fragment ending ~300 bp upstream of the ORF, SAT1 (nourseothricin resistance gene), C. albicans TDH3 promoter, first ~300 bp of WOR1 ORF, PmeI site. PmeI-digested pSN209 was transformed into Arg− reference strain SN250 to generate WOR1OE/WOR1. The wild-type allele of WOR1 was next disrupted using C. dubliniensis ARG4 as in 14. In these and subsequent strains, colony PCR was used to verify the 5′ and 3′ junctions of DNA integration events.
The MTLa/a opaque strain (SN967) used for transcriptome analysis was kindly provided by the Johnson laboratory.35 The white version of this strain (SN966) was isolated after incubation of freshly plated opaque cells at 37°C. The His− Arg+ MTLa opaque strain (SN1038) used for mating assays was created by replacing ~9.2 kb of MTLα in reference strain SN152 with C. dubliniensis ARG4, followed by visual inspection of colonies for opaque sectors. The His+ Arg− MTLα opaque strain (SN1008) was constructed by replacing ~10kb of the MTLa locus with C. dubliniensis HIS1.
The His+Arg− WOR1OE/wor1Δ white strain (SN1001) was constructed via transformation of PmeI-digested pSN209 into SN999 (His+Arg− WOR1/wor1Δ). The corresponding GUT phase isolate (SN1046) was obtained after passage of SN1001 through the murine commensal model and visual inspection of colonies for the GUT phenotype.
WOR1prom-FLP (SN1020) was created in two steps. To exchange FLP for the WOR1 ORF, plasmid pSN288 was engineered to contain: PmeI restriction site, terminal ~500 bp of WOR1 promoter, FLP ORF, SAT1, ~500 bp of WOR1 downstream sequence, PmeI site. The source of FLP and SAT1 was pSFS2A.36 PmeI-digested pSN288 was transformed into Ura− reference strain SN78 to create SN1013. To introduce a copy of URA3 flanked by FRT recombination sites into the LEU2 locus, plasmid pSN290 was engineered to contain: PmeI site, terminal ~440 bp of LEU2 promoter, FRT, URA3, FRT, ~550 bp of LEU2 downstream sequence, PmeI site. Finally, PmeI-digested pSN290 was transformed into SN1013.
Competitive Infections
All procedures involving animals were approved by the UCSF Institutional Animal Care and Use Committee, which enforces the ethical and humane use of animals. Each experiment used the (estimated) minimum number of animals required for detection of a significant biological effect. Blinding and randomization were not necessary because, for a given competition, pooled C. albicans strains were assessed in the same animals. Commensalism experiments: 6–10 week female BALB/c mice were infected by gavage with 1:1 mixtures of competing C. albicans strains as previously reported.11 Relative abundances of strains in the infecting inoculum and after recovery from murine fecal pellets were determined by qPCR,11 using strain-specific primers (Supplementary Table 7). Virulence experiments: 6–10 week female BALB/c mice were infected via the lateral tail vein as in 11. Relative abundances of strains in the infecting inoculum and after recovery from murine kidneys were determined by qPCR, using the same primers as above. Competition experiments are also plotted in “% abundance of the less fit strain” format in Supplementary Figure 7.
Estimation of WOR1 expression by MTLa/α cells
In vitro: Four independent cultures of the WOR1prom-FLP strain (SN1020) were diluted to OD600 0.1 in liquid SC medium 37 and incubated with rolling at 37°C. To maintain cells in mid-log phase, cultures were diluted 1:20 with prewarmed SC when they reached OD600 2–3. After ~8 doublings, aliquots of cells were plated on nonselective Sabouraud medium (Difco) and 5-FOA-containing medium 37 (with 25 μg/ml uridine), which selects for uracil auxotrophs. 5-FOAR colonies were further analyzed by colony PCR to determine their genotype at MTL and to verify Flp-mediated deletion of URA3. In the latter assay (depicted in Supplementary Figure 2), primers SNO509 and SNO840 were used to amplify a 346 bp PCR product from strains that had undergone Flp-mediated recombination. The frequency of cells expressing WOR1 was estimated as the ratio of (5-FOAR, PCR-positive colonies) / (total number of colonies). In vivo: SN1020 was used in commensal infections of six BALB/c mice. After 3 days, C. albicans was recovered from fecal pellets and analyzed as above.
RNA Extraction and RT-qPCR Analysis
Total RNA was prepared using a hot-phenol method 7 and treated with DNaseI using the Turbo DNA-free kit (Ambion). Ten micrograms of RNA was used in standard RT reactions using oligo [(dT)20-N] primers. cDNAs were quantified by qPCR with primers listed in Supplementary Table 7 and normalized against ACT1.
Protein Extraction and Immunoblotting
C. albicans protein extracts were prepared as described.11 Lysates corresponding to 1 OD600 cells were analyzed by SDS-PAGE and immunoblotted using polyclonal anti-Wor1 antibodies (kindly provided by the Johnson laboratory) and anti-α-tubulin antibody (Novus Biologicals, NB100-1639).
Expression profiling
Saturated overnight cultures of wild-type MTLa/α white cells (SN425), MTLa white cells (SN966), MTLa opaque cells (SN967), WOR1OE MTLa/α white cells (SN1044), and WOR1OE MTLa/α GUT cells (SN1045) were inoculated into SC medium 37 (with 100 μg/ml uridine) to OD600=0.1 and incubated with shaking at room temperature for 6–8 hours before harvesting at OD600=0.8–1.0. Two to four biological replicates were performed per strain. RNA isolation, cDNA labeling, and hybridization to custom Agilent C. albicans ORF microarrays were performed as described.11
Equal amounts of cDNA from each sample were pooled to prepare a mixed reference. Cy5-labeled cDNAs from each strain were directly hybridized against the pooled reference (Cy3). Arrays were scanned using a GenePix 4000A Axon scanner, and spots were filtered using GenePix Pro software. Data were normalized using Goulphar 38 (LOWESS normalization) and subjected to Bayesian Analysis of Gene Expression Levels (BAGEL)39. Significant changes in expression were defined as ones with BAGEL p-values <0.05, after implementing a correction for comparison of multiple variables (i.e. multiplication by the number of ORFs on the microarray [6168]).
Mating filament assay
Saturated overnight cultures of MTLa opaque (SN967), MTLa white (SN966), WOR1OE MTLa/α white (SN1044), and WOR1OE MTLa/α GUT (SN1045) cells were tested as described.40
Quantitative mating assay
SN1038 (His− Arg+ MTLa opaque) was used as a common partner for the following His+ Arg− strains: SN1008 (MTLα opaque), SN235 (MTLa/α white), SN1001 (WOR1OE MTLa/α white), and SN1046 (WOR1OE MTLa/α GUT). Mating assays were performed as described,7 except that mating mixtures were spotted onto sterile filter paper rather than using a vacuum manifold.
Scanning Electron Microscopy (SEM)
Wild-type MTLa/α white cells (SN425), MTLa opaque cells (SN967), WOR1OE MTLa/α white cells (SN928), and WOR1OE MTLa/α GUT cells (SN1045) were plated from frozen glycerol stocks onto SC medium and incubated for 48 hours at room temperature. Cells were applied to poly-L-lysine-coated silicon wafers and fixed overnight at 4°C with 2.5% gluteraldehyde in 0.1 M Na-cacodylate pH 7.4. Washed cells were post-fixed in the dark in 1% aqueous osmium tetroxide in 0.1 M Na-cacodylate pH 7.4 for 90 minutes, progressively dehydrated in ethanol (33%–66%–95%–100%), and dried in a Tousimis autoSamdri 815 critical point dryer. Samples were mounted onto stubs and coated with gold palladium in a Tousimis sputter coater prior to scanning using a Hitachi S-5000 scanning electron microscope.
Statistical analysis
The t-test (two-tailed, comparison of unpaired samples) was used for evaluation of competitive fitness, with significance defined as p<0.05. Bayesian Analysis of Gene Expression Levels (BAGEL) 39 was used for analysis of C. albicans transcriptomes. The Bonferroni Correction for Multiple Hypothesis Testing (multiplication by 6168 or the total number of ORFs) was applied, and significance was defined as p<0.05. The GO Term Finder tool on the Candida Genome Database website (http://www.candidagenome.org/) 41 was used to identify functional groups with altered gene expression. This algorithm utilizes a hypergeometric distribution with Bonferroni correction, and significance was defined as p<0.05.
Supplementary Material
Acknowledgments
We are grateful to Richard Bennett for provision of protocols for C. albicans SEM and response to mating pheromone and to Alexander Johnson for strains and α-Wor1 antibodies. We thank Sinem Beyhan and Mark Voorhies for guidance with BAGEL software. Martin Mwangi assisted with the preparation of images of GUT and opaque cells and with colony PCR. Jeffery Cox, Hiten Madhani, Quinn Mitrovich, and Anita Sil provided helpful comments on the manuscript. This work was supported by NIH R21AI099659-01, a UCSF Program for Breakthrough Biomedical Research award, a Burroughs Wellcome Fund CABS award, and a Pew Foundation scholarship.
Footnotes
Database Accession Numbers:
Transcriptional profiling data are available at the Gene Expression Omnibus website (GEO, http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE43972.
Author Contributions:
K.P. identified C. albicans mutants with altered commensal fitness, characterized the white-GUT switch, and analyzed mating and pheromone response. C.C. performed strain construction, expression profiling, and SEM. S.M.N. oversaw the work and wrote the manuscript.
References
- 1.Blackwell M. The fungi: 1, 2, 3 … 5.1 million species? Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Odds FC. Candida and Candidosis, a Review and Bibliography. W.B. Saunders; London: 1988. [Google Scholar]
- 3.Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6:2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang G, et al. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srikantha T, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 8.Edmond MB, et al. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 9.Zaoutis TE, et al. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–1239. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 10.Odds FC, et al. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol. 2006;44:3647–3658. doi: 10.1128/JCM.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Pande K, French SD, Tuch BB, Noble SM. An Iron Homeostasis Regulatory Circuit with Reciprocal Roles in Candida albicans Commensalism and Pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce JV, Kumamoto CA. Variation in Candida albicans EFG1 Expression Enables Host-Dependent Changes in Colonizing Fungal Populations. MBio. 2012;3:e00117–00112. doi: 10.1128/mBio.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camilli A, Beattie DT, Mekalanos JJ. Use of genetic recombination as a reporter of gene expression. Proc Natl Acad Sci U S A. 1994;91:2634–2638. doi: 10.1073/pnas.91.7.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staib P, et al. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol Microbiol. 1999;32:533–546. doi: 10.1046/j.1365-2958.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 17.Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slutsky B, et al. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 20.Wilson M. Microbial Inhabitants of Humans. University of Cambridge Press; Cambridge: 2005. The gastrointestinal tract and its indigenous microbiota; pp. 251–317. [Google Scholar]
- 21.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson BE, Wilhelmus KR, Hube B. The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci. 2007;48:3559–3565. doi: 10.1167/iovs.07-0114. [DOI] [PubMed] [Google Scholar]
- 23.Wong JM, Jenkins DJ. Carbohydrate digestibility and metabolic effects. J Nutr. 2007;137:2539S–2546S. doi: 10.1093/jn/137.11.2539S. [DOI] [PubMed] [Google Scholar]
- 24.Goodman MJ, Kent PW, Truelove SC. Glucosamine synthetase activity of the colonic mucosa in ulcerative colitis and Crohn’s disease. Gut. 1977;18:219–228. doi: 10.1136/gut.18.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miret S, Simpson RJ, McKie AT. Physiology and molecular biology of dietary iron absorption. Annu Rev Nutr. 2003;23:283–301. doi: 10.1146/annurev.nutr.23.011702.073139. [DOI] [PubMed] [Google Scholar]
- 27.Pierre JL, Fontecave M, Crichton RR. Chemistry for an essential biological process: the reduction of ferric iron. Biometals. 2002;15:341–346. doi: 10.1023/a:1020259021641. [DOI] [PubMed] [Google Scholar]
- 28.Lan CY, et al. Metabolic specialization associated with phenotypic switching in Candidaalbicans. Proc Natl Acad Sci U S A. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockhart SR, Wu W, Radke JB, Zhao R, Soll DR. Increased virulence and competitive advantage of a/alpha over a/a or alpha/alpha offspring conserves the mating system of Candida albicans. Genetics. 2005;169:1883–1890. doi: 10.1534/genetics.104.038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie J, et al. White-opaque switching in natural MTLa/alpha isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 2013;11:e1001525. doi: 10.1371/journal.pbio.1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legrand M, et al. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol Microbiol. 2004;52:1451–1462. doi: 10.1111/j.1365-2958.2004.04068.x. [DOI] [PubMed] [Google Scholar]
- 32.Lockhart SR, et al. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 34.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitrovich QM, Tuch BB, Guthrie C, Johnson AD. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 2007;17:492–502. doi: 10.1101/gr.6111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Guthrie C, Fink CG. Guide to Yeast Genetics and Molecular and Cell Biology. 2002 [Google Scholar]
- 38.Lemoine S, Combes F, Servant N, Le Crom S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics. 2006;7:467. doi: 10.1186/1471-2105-7-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:RESEARCH0071. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin CH, Choi A, Bennett RJ. Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Mol Biol Cell. 2011;22:4918–4930. doi: 10.1091/mbc.E11-09-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inglis DO, et al. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 2011;40 doi: 10.1093/nar/gkr945. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 44.Tuch BB, et al. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2011;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.