Abstract
Brain tumors have frequently been associated with a neural stem cell (NSC) origin and contain stem-like tumor cells, so-called brain tumor stem cells (BTSCs) that share many features with normal NSCs. A stem cell state of BTSCs confers resistance to radiotherapy and treatment with alkylating agents. It is also a hallmark of aggressive brain tumors and is maintained by transcriptional networks that are also active in embryonic stem cells. Advances in reprogramming of somatic cells into induced pluripotent stem (iPS) cells have further identified genes that drive stemness. In this review, we will highlight the possible drivers of stemness in medulloblastoma and glioma, the most frequent types of primary malignant brain cancer in children and adults, respectively. Signals that drive expansion of developmentally defined neural precursor cells are also active in corresponding brain tumors. Transcriptomal subgroups of human medulloblastoma and glioma match features of NSCs but also more restricted progenitors. Lessons from genetically-engineered mouse (GEM) models show that temporally and regionally defined NSCs can give rise to distinct subgroups of medulloblastoma and glioma. We will further discuss how acquisition of stem cell features may drive brain tumorigenesis from a non-NSC origin. Genetic alterations, signaling pathways, and therapy-induced changes in the tumor microenvironment can drive reprogramming networks and induce stemness in brain tumors. Finally, we propose a model where dysregulation of microRNAs (miRNAs) that normally provide barriers against reprogramming plays an integral role in promoting stemness in brain tumors.
Keywords: brain tumor, progenitor, neural stem cell, glioma, medulloblastoma, reprogramming, miRNA
Introduction
Defined gradients of signaling factors coordinate self-renewal and differentiation in NSC populations during neural development. Genetic alterations or epigenetic regulation of genes that disturb this delicate balance in NSCs and restricted progenitors may lead to development of brain tumors.
The incidence of histologically and genetically distinct brain tumors peaks in defined time windows during childhood and in adults. In this review, we will focus on medulloblastoma and glioma, the most common primary malignant brain tumors in childhood and adults, respectively. Current therapies for medulloblastomas and gliomas include surgical resection, radiation and chemotherapy (Huse and Holland, 2010). Traditional histological classification defines three classes of human medulloblastoma that are associated with specific outcomes. Nodular/desmoplastic tumors (17%) with nodular accentuation of reticulin-free pale nodules/stromal reticulin have a more favorable outcome. Classic tumors (72%) have small and relatively uniform cells with nuclear molding, which tend to be associated with an intermediate outcome. Large cell/anaplastic tumors (LC/A) (11 %) with features of anaplasia; including large pleomorphic tumor cells with nuclear atypia, are often associated with poor prognosis (Ellison et al., 2011). Gliomas include ependymomas, astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas. The World Health Organization (WHO) classification divides glioma into four grades (I–IV) after malignancy. Grade I glioma, like pilocytic astrocytoma, is considered least malignant and is more prevalent in children or young adults. Within infiltrating gliomas the grading (II–IV) is based on histopathologic features of anaplasia including nuclear atypia, mitotic activity, microvascular proliferation and/or necrosis. The most malignant glioma, GBM (Table 1), can either present de-novo or arise from a lower-grade glioma (Louis et al., 2007). Advances in gene expression profiling have identified subgroups of human brain tumors that can be indistinguishable on histology but show distinct trancriptomal and/or genetic signatures (Figure 1). The transcriptomal signatures in tumors are associated with gene expression profiles reminiscent of NSCs or more differentiated progeny. Studies of GEM models support the notion that established brain tumors can be traced back to a defined precursor cell based on their gene expression profile (Chen et al., 2012; Gibson et al., 2010; Johnson et al., 2010; Schuller et al., 2008; Swartling et al., 2012).
Table 1.
Abbreviations.
| ABBREVIATION | WORD |
|---|---|
| ATRX | alpha thalassemia/mental retardation syndrome X-linked |
| bHLH | Basic helix-loop-helix |
| BMI1 | BMI1 polycomb ring finger oncogene |
| BTSC | Brain tumor stem cell |
| C/EBPβ | CCAAT-enhancer-binding protein β |
| CIC | Homolog of the Drosophila gene capicua |
| CNS | Central nervous system |
| CTNNB1 | β-catenin |
| DAXX | Death domain associated protein |
| DIPG | Diffuse intrinsic pontine glioma |
| EGF | Epidermal growth factor |
| EZH2 | Enhancer of zeste homologue 2 |
| FGF | Fibroblast growth factor |
| GBM | Glioblastoma |
| GEM | Genetically-engineered mouse |
| GNP | Granule neuron precursor |
| HES | Hairy and enhancer of split |
| HGF | Hepatocyte growth factor |
| HIF2α | Hypoxia inducible factor 2α |
| ID4 | Inhibitor of differentiation 4 |
| IDH1/2 | Isocitrate dehydrogenase 1/2 |
| iPS | Induced pluripotent stem |
| MAD | Mitotic arrest deficient |
| MADM | Mosaic analysis with double marker |
| MAPK | Mitogen-activated protein kinase |
| MAX | Myc-associated factor X |
| MGMT | Methylated-DNA-protein-cysteine methyltransferase |
| MLL | Myeloid/lymphoid or mixed-lineage leukemia |
| NF1 | Neurofibromatosis type 1 |
| NG2 | Neuron-glial antigen 2 |
| NSC | Neural stem cell |
| OLIG | Oligodendrocyte transcription factor |
| OPC | Oligodendrocyte progenitor cell |
| OTX2 | Orthodenticle homeobox 2 |
| PA | Pilocytic astrocytoma |
| PDGFRA | Platelet-derived growth factor α |
| PI3K | Phosphatidylinositide 3-kinase |
| PTCH1 | Patched homolog 1 or rather: Patched 1 |
| PTEN | Phosphatase and tensin homolog |
| REST/NRSF | Repressor element-1 silencing transcription factor/neuron-restrictive silencer factor |
| SHH | Sonic Hedgehog |
| SMO | Smoothened homolog or rather: Smoothened, frizzled family receptor |
| SOX | SRY-related HMG box |
| STAT3 | Signal transducer and activator of transcription 3 |
| SVZ | Subventricular zone |
| TAM | Tumor-associated macrophage |
| TAPs | Transit-amplifying progenitors |
| TAZ | Transcriptional coactivator with PDZ-binding motif |
| TLX | Transcription of nuclear receptor tailess |
| WNT | Wingless or rather: Wingless-type MMTV integration site |
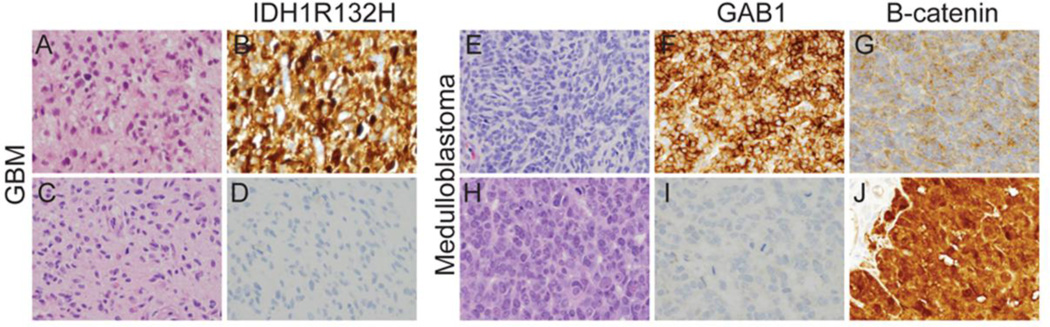
Figure 1. Molecular heterogeneity in GBM and medulloblastoma.
Representative H&E stained sections of GBM from two different patients demonstrate similar histologic features (A,C) despite distinct molecular profiles. Tumor in (A) is IDH1R132H mutant (B) while tumor in (C) is negative for the mutation (D) based on immunoreactivity with an antibody directed against the R132H epitope (H09). In addition, tumor in (A) is negative and tumor in (B) is positive for amplification of EGFR by FISH (data not shown). H&E stained sections of medulloblastoma can also appear similar on H&E (E,H) despite prominent molecular differences. Tumor in (E) has upregulation of GAB1 (F) suggesting activity of the SHH signaling pathway and the tumor in (H) exhibits prominent nuclear immunoreactivity for B-catenin (J) suggesting activation of the WNT signaling pathway. Magnification 400×.
Brain tumors that harbor stem cell-like tumor cells and display stemness signatures are found in highly malignant childhood and adult brain tumors of patients characterized by a poor prognosis (Ben-Porath et al., 2008; Clement et al., 2007; Hemmati et al., 2003; Laks et al., 2009; Singh et al., 2003). Subpopulations of these so-called BTSCs survive current therapies which is why considerable efforts aim to identify therapeutic approaches that also target these cells. Studies by Yamanaka et al. have elegantly demonstrated a small set of reprogramming genes that generate iPS cells from terminally differentiated somatic cells (Takahashi and Yamanaka, 2006). Such reprogrammed iPS cells resemble embryonic stem cells and have implications for how we think about brain tumor heterogeneity. In fact iPS cells like embryonic stem cells are similar to cancer cells and form teratoma or sometimes even malignant teratocarcinoma (Okita et al., 2007; Shih et al., 2007) when injected in immunodeficient mice (Knoepfler, 2009).
In this review, we will discuss the signals and reprogramming networks that drive stemness in brain tumors. The clonal evolution model suggests that all tumor cells to some extent can sustain tumor growth. In contrast, the cancer stem cell model proposes that a stable hierarchy exists, where cancer stem cells undergo self-renewal and promote long-term tumor growth. We describe stemness as a fluid state in brain tumors that can be influenced by the tumor microenviroment or emerge from genetic alterations over time. Finally, we suggest that microRNAs (miRNAs), small non-coding RNAs that block translation or induce degradation of target mRNAs, function as switches that can modulate stemness in brain tumors.
Signals that drive cellular expansion in forebrain and hindbrain regions
Gradients of secreted molecules balance self-renewal and differentiation of embryonic NSCs and progenitors in a coordinated manner along rostrocaudal and dorsoventral axes during central nervous system (CNS) development. Radial glia and embryonic NSCs generate neurons, glial cells, and ependymal cells in temporal waves during neural development (Rakic, 1990). In the hindbrain, primary and secondary germinal zones give rise to defined neuronal populations in the cerebellum (Hatten and Heintz, 1995; Hoshino et al., 2005; Miale and Sidman, 1961). Postnatal NSC-derived neurogenesis was first restricted to the dentate gyrus of the subgranular zone and the subventricular zone (SVZ) lining the lateral ventricles (Curtis et al., 2007; Sanai et al., 2011). Recent studies suggest that NSCs may also be found lining the third and fourth ventricles (Weiss et al., 1996; Xu et al., 2005). Stemness has also been found in postnatal Bergmann glia in the cerebellum, another possible NSC population that can give rise to brain tumors (Koirala and Corfas, 2010; Sottile et al., 2006). While NSCs are rare, neuron-glial antigen 2 (NG2)-expressing oligodendrocyte progenitor cells (OPCs), also denoted polydendrocytes and synantocytes, constitute an abundant and widespread population of cycling cells in the adult rodent brain (Dawson et al., 2003; Nishiyama et al., 2009). In contrast to reduced numbers of NSCs, the population of OPCs may actually increase during life (Dawson et al., 2003; Shook et al., 2012). Coordinated activation of transcription factors drives networks that give regional patterning of NSCs and their progeny. This regional specification of NSCs and differentiated progeny continues into adulthood (Merkle et al., 2007; Robinson et al., 2012). Thus positional identity is an organizing principle underlying cellular subtype diversification in the brain and is controlled by a homeodomain transcriptional code (Hochstim et al., 2008). Below, we will review signals that expand neural precursors and brain tumors.
Interactions between growth factors and receptor tyrosine kinases lead to expansion of neural precursors during embryonic and adult phases. Fibroblast growth factor (FGF) signaling regulates patterning of both embryonic forebrain and hindbrain regions (Hebert and Fishell, 2008). In the telencephalic germinal zone, FGF2 promotes self-renewal of NSCs in the anterior neural plate followed by emergence of epidermal growth factor (EGF)-responsive NSCs (Kilpatrick and Bartlett, 1995; Tropepe et al., 1999). Multipotent NSCs continue to respond to mitogenic EGF and FGF2 stimulation throughout adulthood (Gritti et al., 1999). The platelet-derived growth factor receptor α (PDGFRA) is expressed in the neural plate and allows NSCs to respond to platelet-derived growth factor-AA (PDGF-AA) (Forsberg-Nilsson et al., 1998). Later in development, PDGFA acts as a potent mitogen of OPCs expressing PDGFRA (Hall et al., 1996). It is controversial if postnatal SVZ NSCs express PDGFRA (Chojnacki et al., 2011; Jackson et al., 2006). Similar to embryonic development, SVZ NSC generate EGF-responsive NSCs and transitamplifying progenitors (TAPs) (Gonzalez-Perez et al., 2009). Infusion of FGF, PDGF-AA, or EGF into the ventricles and SVZ induce massive expansion of OLIG2+ cells (Gonzalez-Perez et al., 2009; Jackson et al., 2006; Kuhn et al., 1997), implicating a possible involvement of growth-factor signaling and NSCs in the development of gliomas.
A wealth of literature has demonstrated the roles of sonic hedgehog (SHH), wingless (WNT), and NOTCH signaling during expansive stages of brain development. A gradient of SHH is formed in a dorsal-ventral manner along the neural tube (Lee et al., 1997). SHH ligand binds to its receptors, patched homolog 1 (PTCH1) and smoothened homolog (SMO), and leads to the activation of GLI transcription factors that control normal brain growth and stem cell behavior during both embryonic and adult stages (Lai et al., 2003; Palma and Ruiz i Altaba, 2004). In the adult forebrain, SHH activation of SVZ NSCs produces specific neuronal progeny from dorsal and ventral regions (Ihrie et al., 2011). In the cerebellum, SHH is released by Purkinje neurons that act as a mitogen, stimulating granule neuron precursor (GNP) proliferation (Dahmane and Ruiz i Altaba, 1999; Wechsler-Reya and Scott, 1999). WNT proteins antagonize SHH and thereby regulate the dorso-ventral patterning of the neural tube (Alvarez-Medina et al., 2008). The WNT pathway maintains pluripotency in murine embryonic stem cells via paracrine and autocrine signaling (ten Berge et al., 2011) as well as controlling neuronal precursor cell fates (Chenn and Walsh, 2002). The WNT signals are divided into two different pathways: the canonical, or WNT/β-catenin, pathway is involved in cell-fate determination, whereas the non-canonical pathways are involved in the control of cell movement and tissue polarity (Katoh, 2007). Activation of β-catenin leads to amplification of the neural progenitor pool (Chenn and Walsh, 2002; Megason and McMahon, 2002). If stabilised β-catenin is overexpressed in transgenic mice, the brain is enlarged, and the neural precursor population is expanded (Chenn and Walsh, 2002). In adult forebrain, inhibition of WNT signaling abolished hippocampal neurogenesis (Lie et al., 2005). In the developing cerebellum, aberrant WNT signaling expands the stem cell pool and impairs differentiation (Pei et al., 2012a). The third pathway, NOTCH signaling, is known as a master regulator of NSCs and neural development (Louvi and Artavanis-Tsakonas, 2006; Yoon and Gaiano, 2005). NOTCH receptors are commonly activated by ligands expressed on neighboring cells. Activation of NOTCH receptors leads to cleavage of the NOTCH intracellular domain (NICD) by γ-secretase followed by induced expression of target genes such as (hairy and enhancer of split) Hes and Hes-related (HESR/HEY) family of basic helix-loop-helix (bHLH) transcription factors. NICD can also signal through a non-canonical pathway through protein-protein interactions and recombining binding protein suppressor of hairless (RBPJ)-independent gene activation (Sanalkumar et al., 2010). NOTCH signaling has been suggested as an integral component that regulates maintenance of NSCs (Ables et al., 2011). The RNA-binding protein Musashi1 with expression restricted to NSCs and progenitor cells positively regulate NOTCH signaling through translational repression of numb mRNA (Imai et al., 2001; Kaneko et al., 2000). In the postnatal cerebellum, NOTCH2 expression prevents differentiation of GNPs and support continued proliferation (Fan et al., 2004; Solecki et al., 2001) while NOTCH1 is mainly expressed in postmitotic cells (Fan et al., 2004). In adult SVZ, activated, but not dormant, NSCs are regulated by NOTCH1 signaling (Basak et al., 2012; Chapouton et al., 2010; Mizutani et al., 2007).
Growth factors and SHH, NOTCH and WNT can all regulate MYC expression. There are three members of the MYC gene family (MYC, MYCN and MYCL). The role of MYC proteins, in particular MYCN, is essential for normal brain development (Knoepfler et al., 2002; Swartling, 2012). Using the Cre/Lox system, Knoepfler et al. (2002) demonstrated that loss of Mycn in Nestin+ embryonic neural precursors lead to a marked reduction in growth of the brain. The reduced size of the cerebellum was especially pronounced and is likely explained by the normally elevated MYCN expression in the rapidly proliferating cerebellar primordium. Strikingly, a further reduction of cerebellar granule neurons was found in Myc and Mycn double knockout mice (Wey et al., 2010). Together with myc-associated factor X (MAX), MYC can promote cell proliferation and maintain an undifferentiated state. However, this is normally disrupted by mitotic arrest deficient (MAD) which displaces MYC in the complex allowing formation of post-mitotic differentiated states. MYC belongs to a small family of reprogramming genes that cooperate to turn terminally differentiated cells into iPS cells (Takahashi and Yamanaka, 2006). In fact MYCN is able to replace MYC in inducing pluripotency (Park et al., 2008). In conclusion, multiple developmental pathways converge on transcriptional nodes that drive stemness and expansion of progeny.
Matching origin with transcriptomal profiles in childhood brain tumors
Genetic alterations in pathways that drive embryonic and perinatal expansion of neural precursors are found in human pediatric brain tumors, including medulloblastoma, pilocytic astrocytoma, and ependymoma. Design of GEM models based on these occurrences has advanced our current understanding of the developmental origins of brain tumors. The studies suggest that cooperation of genetic alterations and developmental programs determine the phenotype of the resulting brain tumor. In this section, we will review the cell of origin for human pediatric brain tumors based on data from current GEM models (Table 2). Furthermore, to understand the differential response of residual malignant ependymomas and medulloblastomas to therapy comparisons based on cross-species approaches of GEM models and profiling of human tumors have been highly informative.
Table 2.
Examples of selected Murine brain tumor models with defined cells of tumor origin.
| Brain Tumor Type | GEM Model | Cell of Origin | Reference |
|---|---|---|---|
| Astrocytoma/GBM | K-RASG12D:AKT1 | Astrocyte/Neuron/NSC | (Friedmann-Morvinski, et al., 2012) |
| Astrocytoma/GBM | Nestin-ΔTK | Stem Cell | (Chen, et al., 2012) |
| Astrocytoma/GBM | Ink4a/Arf−/−, EGFR+ | Astrocyte | (Bachoo, et al., 2002) |
| Astrocytoma/GBM | Nf1; Trp53+/− | Astrocyte (GFAP+) | (Reilly, et al., 2000, Zhu, et al., 2005) |
| Oligodendroglioma | S100b-verbB | OPC | Weiss et al. (2003), Persson et al. (2010) |
| Oligodendroglioma | RCAS-PDGFB/CNP-TVA | OPC | (Lindberg, et al., 2009) |
| Oligodendroglioma | MADM-Nf1;Trp53 mut | OPC | (Liu, et al., 2011) |
| PNET | Rb;Trp53LoxP/LoxP | Stem Cell (GFAP+) | (Jacques, et al., 2010) |
| Ependymoma | Ink4a/Arf−/−, EphB2+ | Radial Glia (BLBP+) | (Johnson, et al., 2010) |
| DIPG | Olig2-SmoM2 | Brain Stem Progenitor | (Monje, et al., 2011) |
| Medulloblastoma (WNT) | Ctnnb1lox(ex3), Trp53flx/flx | Brain Stem Precursor (BLBP+) | (Gibson, et al., 2010) |
| Medulloblastoma (SHH) | Ptch loss | EGL Progenitor / Cerebellar Stem Cell | (Goodrich, et al., 1997, Yang, et al., 2008) |
| Medulloblastoma (SHH) | ND2-SmoA or SmoM2 | EGL Progenitor / Rhombic Lip Progenitor | (Hallahan, et al., 2004, Mao, et al., 2006, Schuller, et al., 2008) |
| Medulloblastoma (SHH) | Retroviral Shh in utero or RCAS-Shh/Nestin-TVA | Embryonic Cerebellar Cell or Nestin+ Cerebellar Cell | (Rao, et al., 2003, Weiner, et al., 2002) |
| Medulloblastoma (mostly non-SHH) | Glt1-MYCN | Non-EGL Cell (GLT1+) | (Swartling, et al., 2010) |
| Medulloblastoma (Group 3 or 4) | MycT58A/Trp53+/− or MycnT58A | Cerebellar Stem Cell | Kawauchi et al. (2012), Pei et al. (2012b), Swartling et al. (2012) |
The pathogenesis of the childhood tumor medulloblastoma implies an early embryonic initiating aberration in a number of important developmental genes. Gene expression profiling of human medulloblastoma has recently divided medulloblastoma into four molecularly distinct subgroups: WNT, SHH, Group 3 and Group 4 (Cho et al., 2011; Kool et al., 2008; Northcott et al., 2011; Northcott et al., 2012a; Taylor et al., 2012a; Thompson et al., 2006). Reliable signature pathway markers have been identified for both WNT and SHH medulloblastoma (Kool et al., 2008; Northcott et al., 2012b). The profile of Group 3 is of a photoreceptor/gamma-aminobutyric acid-ergic nature, showing moderate to high expression of retina-specific transcription factors, while Group 4 has neuronal/glutamatergic characteristics with overexpression of neuronal differentiation markers as well as glutamate receptor members (Kool et al., 2008).
Medulloblastoma of the SHH subgroup most likely develop from committed GNPs of the cerebellum (Yang et al., 2008) while the origin of the WNT subgroup has instead been located to progenitor cells in the embryonic dorsal brainstem/lower rhombic lip (Gibson et al., 2010) (Figure 2). MYC and MYCN amplifications are common (10–15%) in medulloblastoma (Pfister et al., 2009). In a cohort of 292 pediatric medulloblastoma patients, amplification of MYC and MYCN were significantly associated with a poor prognosis (Ryan et al., 2012). Interestingly, while MYC protein amplifications are common in Group 3 medulloblastoma, MYCN is often found amplified in either SHH or Group 4 medulloblastoma (Taylor et al., 2012a). To elucidate the origin of Group 3 medulloblastoma, Pei et al. soreted cerebellar cells based on expression of the stem cell marker with dominant negative TP53 (Pei et al. 2012a). Similarly, Kawauchi et al. introduced MYC to TP53 null GNPs sorted using neuronal lineage marker MATH1 (Kawauchi et al., 2012). Both murine models succeeded in recapitulating the LC/A phenotype of Group 3 medulloblastoma. The MYC-driven medulloblastoma demonstrated expression profiles overlapping with embryonic NSCs as well as iPS cells, suggesting a NSC origin or possibly de-differentiation as a result of MYC transformation (Eberhart, 2012). The definite origin of MYCN-amplified Group 4 is unclear, but MYCN-driven medulloblastoma models like the GTML model (Swartling et al., 2010) might aid in this purpose. Here GLT1-positive cerebellar cells drive an equal amount of either classic or LC/A medulloblastoma that are SHH-independent and orthodenticle homeobox 2 (OTX2)-positive (Swartling et al., 2010; Swartling et al., 2012). When GFAP-positive postnatal NSCs from the cerebellum were transduced with stabilized MYCN, Group 4 medulloblastoma formed after orthotopic transplantation. Similarly, MYCN drives SHH medulloblastoma from GFAP+ embryonic cerebellar NSCs suggesting the timing of transformation with MYCN determines medulloblastoma subtype (Swartling et al., 2012).
Figure 2. Different origins for stemness in brain tumors.
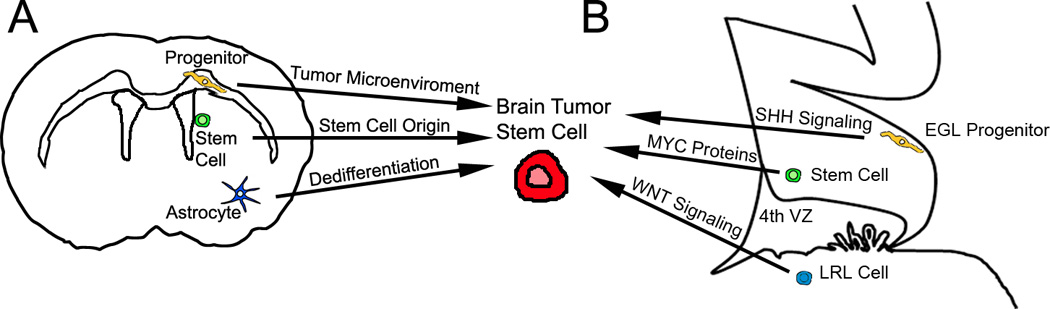
(A) Gliomas originating from a non-stem cell (progenitor or more differentiated cell) may acquire stemness from exposure to the surrounding microenviroment during tumor progression or after relapse. Secondly, stemness in gliomas can also be derived from a NSC origin. Finally, stemness in gliomas can result from genetic alterations that can reprogram more differentiated neural cells into stem-like tumor cells. (B) Recent studies suggest that SHH signaling in an EGL precursor, amplification or overexpression of MYC proteins (like MYC and MYCN) from cerebellar stem cells, or aberrant WNT signaling in precursors located in the LRL, are possible origins for stem cell-like medulloblastoma cells. The schematic figure depicts views of cerebrum and cerebellum of an adult and embryonic (E16) mouse brain, respectively. EGL: external granule layer. LRL: lower rhombic lip.
Pilocytic astrocytomas (PAs) can occur supra- and infratentorially and each is associated with a unique gene expression signature (Sharma et al., 2007). Activating BRAF alterations/gene fusions are common in PAs (Pfister et al., 2008) resulting in an activated mitogen-activated protein kinase (MAPK) pathway (Jones et al., 2008). Such PAs have been modeled in mice where a mutated active form (V600E) of BRAF is introduced into Nestin-expressing cells using the RCAS/tv-a system (Gronych et al., 2011). Children with the neurofibromatosis type 1 (NF1) tumor predisposition syndrome are prone to the development of NF1-associated optic or hypothalamic PAs (Listernick et al., 1995). The transcriptomal profile of NF1-associated tumors can be distinguished from other PAs (Sharma et al., 2007). Lee da et al., (Lee da et al., 2012) demonstrated that PAs arise from third ventricle NSCs, but not from lateral ventricle NSCs, after NF1 loss in a GEM model. Lee et al also found that activating BRAF mutation specifically increased proliferation of third ventricle NSCs (Lee da et al., 2012).
Infiltrating gliomas localized to the brainstem account for 10–15% of childhood brain tumors. Interestingly, tumors centered in the dorsal pons, midbrain, or medulla show a more favorable prognosis than those located in the ventral pons which are highly aggressive and termed diffuse intrinsic pontine gliomas (DIPGs). A wave of nestin expressing cells in the medulla and ventral pons peaks at 6 years of age in humans, a time that coincides with the incidence for DIPGs (Monje et al., 2011). Approximately half of the Nestin positive cells also express oligodendrocyte transcription factor 2 (OLIG2), a bHLH factor expressed in progenitor cells. Using GEM models, Monje et al. showed that SHH-responding OLIG2+ cells in the ventral pons expand at a time that corresponds to the peak of nestin-expressing cells in human pons and medulla. In human DIPG cultures, a subpopulation of NSC-like tumor cells displays a marker profile that corresponds to the suggested cell of origin. Improved GEM models of DIPG should help to discern if NSCs or OLIG2 expressing progenitors can give rise to this disease.
Recent studies suggest that mutations in the histone H3- alpha-thalassemia X-linked mental retardation protein (ATRX)- death domain associated protein (DAXX) chromatin remodeling pathway frequently occur in both pediatric and adult gliomas (Schwartzentruber et al., 2012; Sturm et al., 2012). Almost half of the studied pediatric GBMs displayed mutations in this chromatin remodeling pathway (Schwartzentruber et al., 2012). Histone H3 mutations was found in 78% of DIPGs and 22% of non-brainstem pediatric GBMs (Wu et al., 2012). Modeling of histone H3 mutations in murine GEM models will help to define the origin for these pediatric gliomas. Interestingly, IDH1 and H3F3A-ATRX-DAXX mutations seem to be mutually exclusive in both pediatric and adult gliomas, which can be further divided into several subgroups using IDH1 and H3F3A mutation status and global DNA methylation patterns (Schwartzentruber et al., 2012; Sturm et al., 2012). In medulloblastoma, distinct subclasses show amplification and inactivating mutations in regulators of histone H3 methylation (Northcott et al., 2009b; Robinson et al., 2012). Inactivating mutations of the histone-lysine N-methyltransferase genes MLL2 and MLL3 in 16% of medulloblastoma underscore the important role of histone methylation in human brain tumorigenesis (Parsons et al., 2011). Future studies will be required to dissect how alterations in the chromatin machinery contribute to malignant transformation.
Ependymomas are gliomas with evidence of ependymal differentiation, associated with the ventricular wall along the cerebrospinal axis (Louis et al., 2007). Both the layer of ependymal cells lining the ventricle wall and the intermixed SVZ NSCs are derived from radial glial cells (Merkle et al., 2004). Radial glia-like cancer stem cells were shown to propagate tumor growth of intracranial and spinal ependymomas (Taylor et al., 2005). Using a genomic cross-species approach, Johnson et al. matched the transcriptomal profile of human ependymoma samples with temporally and regionally defined NSCs (Johnson et al., 2010). Subsequent experiments demonstrated that radial glia isolated from the cerebrum of embryonic day 14.5 represents a likely source for supratentorial ependymomas.
Stemness reflects the origin or is acquired by the surrounding niche in adult gliomas
Molecular profiling of human high-grade astrocytoms lead to definition of proneural, proliferative, and mesenchymal astrocytomas that were associated with genetic alterations, pathway activation, and profiles analogous to NSCs and progenitors (Phillips et al., 2006). A more recent gene expression study classified GBMs into proneural, neural, classical, and mesenchymal subgroups that were highly associated with somatic mutations and copy number changes (Verhaak et al., 2010). Proneural gliomas express progenitor-associated genes whereas classical and mesenchymal tumors display stem cell signatures. GEM models have been an invaluable tool to study how genetic alterations drive gliomagenesis in a cell-context manner to shape the resulting gliomas.
Loss of TP53 alone is not sufficient to transform SVZ NSCs but will induce pleiotropic accumulation of cooperative oncogenic alterations that promote gliomagenesis (Gil-Perotin et al., 2006; Wang et al., 2009). Instead, combinations of TP53, phosphatase and tensin homolog (PTEN), and NF1 loss in NSCs generate gliomas whereas initial Rb loss leads to production of a primitive neuroectodermal neoplasm (PNET) phenotype (Alcantara Llaguno et al., 2009; Jacques et al., 2010; Zheng et al., 2008; Zhu et al., 2005). In patients, PNETs occur more frequently in hereditary retinoblastoma patients (Pacal and Bremner, 2006). Furthermore, patients diagnosed with supratentorial PNETs commonly show amplifications of cell cycle regulatory genes suggesting that the retinoblastoma-cyclin axis may be particularly important in this group of tumors. Introduction of activated MYCN into SVZ NSCs produce glioma-like tumors with PNET-like features (Swartling et al., 2012). Similarly, PNETs have been induced using amplification of MYC in combination with β-catenin or together with TP53 loss (Momota et al., 2008). Interestingly, amplification of MYC is common in human CNS PNETs (Behdad and Perry, 2010). In conclusion, these studies suggest different combinations of transforming events in NSCs produce PNETs and gliomas, respectively.
A lentiviral approach using a combination of oncogenic K-RASG12D and AKT1 showed that SVZ NSCs are more easily transformed into glioma cells than differentiated astrocytes (Jacques et al., 2010; Marumoto et al., 2009) (Figure 2). Another study found that activated K-RASG12D and loss of TP53 resulted in transformation of both SVZ NSCs and cortical astrocytes (Ghazi et al., 2012). Interestingly, cortical astrocytes gave rise to more aggressive GBM-like tumors displaying a mesenchymal phenotype. Surprisingly, NSC-derived tumors displayed progenitor-like markers while astrocyted-derived tumors displayed proteins commonly expressed in NSCs. In another model, combined overexpression of BMI1 polycomb ring finger oncogene (BMI1) and Ink4/Arf loss in primary astrocytes produced more malignant tumors compared to NSC counterparts. These studies suggest that TP53 loss (in contrast to AKT1) dedifferentiate mature astrocytes and allow oncogenic K-RASG12D to generate gliomas displaying a high degree of stemness. In addition to NSCs and astrocytes, a recent study suggests that even neurons can give rise to malignant gliomas when transduced by oncogenic lentiviral vectors (including K RASG12D) (Friedmann-Morvinski et al., 2012). As will be discussed below, it exemplifies how reprogramming of a non-stem cell origin can produce stem-like brain tumors.
As the largest population of cycling cells in the postnatal brain, OPCs represent a likely source of brain tumors. A transgenic mouse model, an RCAS/TVA model, and a mosaic analysis with double markers (MADM)-system based on different oncogenic drivers demonstrated that OPCs give rise to the proneural oligodendrogliomas (Lindberg et al., 2009; Liu et al., 2011; Persson et al., 2010). Proneural human GBMs commonly display TP53 mutations, IDH1R132H mutations, PDGFRA amplifications, and H3F3A (Histone H3) mutations (Sturm et al., 2012; Verhaak et al., 2010). Interestingly, GBMs displaying H3F3A mutations were mutually exclusive with IDH1R132H mutations, showed distinct global methylation patterns, and were found in separate anatomic compartments (Sturm et al., 2012). In contrast, classical GBM are characterized by amplification of EGFR and EGFRvIII mutations, and many mesenchymal GBMs are associated with mutations and copy number loss of NF1. Profiling of grade II–III infiltrating gliomas demonstrated that most oligodendrogliomas and the majority of astrocytomas display a proneural signature (Cooper et al., 2010). Considering that the majority of infiltrating gliomas and commonly secondary GBMs display IDH1R132H mutations, the molecular evolution of human GBMs displaying wild-type or mutant IDH1R132H may result from largely nonoverlapping sets of molecular events in separate cell types of origin (Lai et al., 2011). Proneural grade II–IV gliomas that display a CpG island methylator phenotype are linked to IDH1R132H somatic mutations (Noushmehr et al., 2010; Turcan et al., 2012). As for the IDH1R132H mutant GBMs, methylation status can influence the tumor phenotype. Hypermethylation of the CpG island for the transcriptional coactivator with PDZ-binding motif (TAZ) ensure low levels in proneural GBMs (Bhat et al., 2011). Modulation of TAZ expression allowed GBM stem cells to toggle between a proneural and mesenchymal phenotype. Is it then possible that therapy-induced changes in the tumor microenviroment can promote epigenetic changes that influence the phenotype? Of interest, proneural GBMs tend to shift to a mesenchymal subclass upon recurrence (Phillips et al., 2006). Recurrent GBMs that received radiotherapy display approximately 10-fold increased levels of tumor-associated macrophages (TAMs) (Kioi et al., 2010). Radio-resistant GBM stem cells produce cytokines known to polarize TAMs to a tumor-promoting phenotype (Bao et al., 2006; Wu et al., 2010). Is it possible that TAM-produced cytokines can drive a mesenchymal phenotype considering that microglia/macrophage number and microglia/macrophage-related gene expression are highest in the mesenchymal GBM subtype (Engler et al., 2012). A mesenchymal GBM phenotype can also be promoted by hepatocyte growth factor (HGF)-dependent MET activity (De Bacco et al., 2012; Li et al., 2011). An elegant study by Lu et al. demonstrates that anti-angiogenic therapy lead to highly invasive tumors through increased phosphorylation of C-MET in a hypoxia-independent manner (Lu et al., 2012). Finally, necrosis in GBMs was found to promote a mesenchymal phenotype, including upregulation of signal transducer and activator of transcription 3 (STAT3) and CCAAT-enhancer-binding protein β (C/EBPβ), in a hypoxia-dependent manner (Cooper et al., 2012). Ectopic co-expression of these two factors reprograms NSCs towards a mesenchymal phenotype, while elimination leads to collapse of the mesenchymal signature in GBMs (Carro et al., 2010). These studies exemplify how a brain tumor phenotype can be regulated by factors in the tumor microenvironment and not solely by genetic alterations.
Reprogramming networks drive stem cell-ness in brain tumors
Stemness reflects a state rather than a physical entity. The epigenetic and gene expression profiles of many cancers show significant overlap with embryonic stem cells, suggesting that similar transcriptional networks are active in both stem cells and cancer cells (Easwaran et al., 2012). MYC has been defined as one major player that account for the similar transcription programs in embryonic stem and cancer cells (Ben-Porath et al., 2008; Kim et al., 2010; Widschwendter et al., 2007). To reprogram differentiated somatic cells iPS cells, Yamanaka and colleagues identified combinations of a small set of genes (OCT4, SOX2, NANOG, C-MYC, KLF4, Lin28), previously known to maintain pluripotency and self-renewal in both human and murine embryonic stem cells (Takahashi and Yamanaka, 2006). These factors are highly expressed in stem cell-like cancer cells (e.g. cancer stem cells), suggesting that cancer stem cells derive from normal stem cells or alternatively more differentiated cells reprogrammed by genetic aberrations or environmental cues.
Stemness in brain tumors is correlated with poor prognosis. The tumor suppressor genes TP53 and Ink4/Arf, frequently deleted or mutated in gliomas, function as barriers for somatic cell reprogramming (Choi et al., 2011; Marion et al., 2009). In murine and human fibroblasts, Arf and Ink4a, respectively, play dominant roles as the main barriers to reprogramming by activation of TP53 and p21 / WAF1 (Li et al., 2009a). Such species-specific differences may be evident for other reprogramming factors. PTEN is a tumor suppressor that negative regulates phosphatidylinositide 3-kinase (PI3K)-Akt signaling in high-grade gliomas. PTEN knockdown induces the levels of reprogramming genes OCT4 and NANOG (Alva et al., 2011). Amplification of RTKs (MET, EGFR, PDGFRA) drive self-renewal, stemness, and tumor growth in human GBMs. For mesenchymal GBMs, c-MET activation was found to induce the expression of reprogramming genes and promote stemness in human GBMs (Li et al., 2011), an effect that could be reversed by NANOG blockade. Unfortunately, the authors did not investigate if c-MET-induced activation of MEK-ERK, PI3K-AKT, and STAT3 signaling pathways alone, or in combination, contributes to stemness and upregulation of reprogramming genes. STAT3 has been shown to mediate stemness and tumorigenicity down-stream of the nonreceptor tyrosine kinase BMX, PDGFRB, and the erythropoietin receptor (Guryanova et al., 2011; Kim et al., 2012). Less is known about the link between stemness and MEK-ERK/PI3K-AKT signaling. Blockade of EGFR reduced tumorigenesis and induced robust differentiation of human GBMs (Mazzoleni et al., 2010). On the other hand, PDGFRA amplification is characteristic in a subset of proneural GBMs that display an OPC rather than NSC signature. Intra-tumoral variation of EGFR, MET, and PDGFRA amplifications in GBM makes it more difficult to discern the contribution of individual RTKs to stemness (Little et al., 2012; Snuderl et al., 2011; Szerlip et al., 2012). These studies illustrate that genetic changes in glioma may contribute to stemness and reprogramming genes in stem cell-like tumor cells.
The SHH-Gli signaling pathway promotes an embryonic stem cell-like signature in both GBM and medulloblastoma (Wang et al., 2012; Zbinden et al., 2010). GLI1 was shown to bind to the BMI1 promoter, a member of the Polycomb group (PcG) genes that partly regulate stemness by repressing the levels of Ink4a/Arf (Valk-Lingbeek et al., 2004). SHH-GLI signaling also regulates stemness in cerebellar NSCs through expression of NANOG in cerebellar NSCs (Po et al., 2010). A MYC network was suggested to account for similarities between embryonic stem and cancer cell transcription programs (Kim et al., 2010). Amplified MYC and MYCN in Group 3 and Group 4 medulloblastoma can directly contribute to stemness (Taylor et al., 2012a). MYC amplification can initiate medulloblastoma formation in combination with TP53 loss and stabilizing mutations of MYCT58A or aberrant expression of the repressor element-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) (Pei et al., 2012b; Su et al., 2006). These studies suggest that induction of multiple reprogramming genes is necessary for medulloblastoma formation. In human GBMs, MYC was identified as a major down-stream target gene of the enhancer of zeste homologue 2 (EZH2), shown to be essential for stemness and tumorigenicity (Suva et al., 2009). Nucleostemin, a transcriptional target of MYC, is expressed in BTSCs and can substitute for MYC as a reprogramming factor in embryonic stem cells (Qu and Bishop, 2012). MYC can also be activated through WNT/β-catenin signaling (Hoffmeyer et al., 2012). Hoffmeyer et al., found that WNT/β-catenin signaling can also regulate telomerase activity in stem cells and cancer cells through the interaction with KLF4, a core component of the pluripotency transcriptional network. In the developing cerebellum, WNT signaling increased self-renewal and impaired differentiation through regulation of MYC, BMPs and the CDK inhibitor p21 (Pei et al., 2012a). In conclusion, many pathways converge on MYC that is contributing, but not sufficient, to drive stemness in brain tumors.
Stemness in brain tumor cells can also be induced by changes in the tumor microenviroment (Figure 2). A hypoxic environment was found to promote stem cell-ness in cultured human primary GBM cells (Li et al., 2009c). Hypoxia inducible factor 2 alpha (HIF2-alpha) was found to increase the levels of reprogramming genes (OCT4, NANOG, and C-MYC) and induced a stem cell phenotype in human GBM cells (Heddleston et al., 2009). In GBM patients, CD133+ cells were enriched in the inner core of the tumor mass compared to peripheral and neovascularised areas (Pistollato et al., 2010). Importantly, tumor cells expressing the stem cell marker CD133 also displayed higher levels of MGMT genes, implicating increased chemoresistance to the alkylating agent temozolomide. An acidic environment has been found in brain tumors (Vaupel et al., 1989). A recent study showed that HIF2α also mediates acidic stress-induced regulation of the reprogramming genes OCT4 and NANOG (Hjelmeland et al., 2011). In summary, genetic changes, activation of SHH and WNT signaling, or altered tumor microenviroment can all drive reprogramming networks and promote stemness in human brain tumors.
MiRNAs as regulators of stemness in brain tumors
Given that stemness correlates with aggressive behavior of brain tumors, it is important to identify signaling effectors that mediate stemness from intrinsic and microenviromental cues (Li et al., 2009c). Recent studies demonstrate an important role for miRNAs in regulating stem cell self-renewal and differentiation by repressing the translation of selected mRNAs in stem cells and differentiated daughter cells (Fineberg et al., 2009; Gangaraju and Lin, 2009). REST/NRSF together with the corepressor for REST (coREST), have been shown to coordinate neural induction and neuronal differentiation programs during brain development (Abrajano et al., 2010; Ballas et al., 2005; Soldati et al., 2012). In brain tumors, overexpression of REST in medulloblastoma and glioblastoma promotes stemness and is associated with poor prognosis in patients (Conti et al., 2012; Fuller et al., 2005; Taylor et al., 2012b). REST can directly interact with cis elements upstream of the miR-21 gene, a repressor of NANOG, OCT4, and SOX2 (Singh et al., 2008). MiR-21 is highly expressed in human GBMs and recognized as an oncogene in glioblastoma (Chan et al., 2005) and cooperates with growth factors like PDGF to drive SOX2 expression in normal developing brain and in human glioblastoma (Polajeva et al., 2012). Enforced expression of the bHLH factor inhibitor of differentiation 4 (ID4) in Ink4a/Arf null NSCs drives gliomagenesis in a murine glioma model (Jeon et al., 2008). ID4 was found to repress miR-9 expression that results in reduced chemoresistance and stemness in human glioblastoma cells (Jeon et al., 2011). Jeon et al. found that reduced miR-9 levels promoted SOX2 expression in human glioblastoma cells. Interestingly, SOX2 expression increases transcription of nuclear receptor tailess (TLX, also known as NR2E1) in adult NSCs (Shimozaki et al., 2012). As a direct target of miR-9, enforced expression of TLX in SVZ NSCs resulted in glioma formation (Liu et al., 2010; Zhao et al., 2009). TLX is also a target of miR-137, a miRNA that is depleted in human glioblastoma, and when expressed, induced differentiation of GBM cells into a neuronal phenotype (Silber et al., 2008). In this publication, we found that introduction of miR-124a also promotes neuronal differentiation of human GBM cells. In the normal brain, the levels of miR-124a increase as adult SVZ NSCs differentiate into mature neurons (Cheng et al., 2009). Cheng et al. suggest that miR-124a directly targets SOX9, a gene that is required for maintenance of stemness and blocks neurogenesis in adult SVZ NSCs (Scott et al., 2010).
For induction of iPS cells, cooperative actions of miR-34a and the cyclin-dependent kinase inhibitor p21 were found to regulate somatic reprogramming downstream of TP53 (Choi et al., 2011). MiR-34a cooperated with miR-34b and c to repress Nanog, SOX2, and MYCN. In human ES cells, low expression of deacetylated and inactive TP53 activates miR-34a and miR-145, which in turn repress reprogramming factors (Jain et al., 2012). In murine E14 NSCs, miR-34a expression induces neuronal differentiation and neurite elongation (Aranha et al., 2011). In another study, miR-34a levels were further down-regulated in TP53 mutant GBMs, and when introduced, down-regulated NOTCH1/2 signaling and MET activity, leading to suppressed stemness in human GBM (Guessous et al., 2010). In addition to GBMs, miR-34a down-regulated MET activities in human medulloblastoma cells (Li et al., 2009b). MicroRNA-34a induces apoptosis, G2 arrest, and senescence in medulloblastoma and renders these cells more sensitive to chemotherapeutic agents in part via the oncogenic MAGE-A gene family and TP53 (Weeraratne et al., 2011). In a GEM model (Ptch1+/−:TP53−/−) of medulloblastoma, transduction of miR-34a into tumor spheres reduced expression of the NOTCH ligand Delta-like 1 (Dll1) and other targets, leading to a reduced stem cell-like phenotype and induction of neuronal differentiation (de Antonellis et al., 2011). MicroRNA-199b-5p also impairs NOTCH pathway through negative regulation of HES1 in medulloblastoma initiating cells (Garzia et al., 2009). MiR-199b-5p over-expression blocks expression of several stem-cell genes and decreases the population of CD133-positive medulloblastoma cells.
Through different mechanisms, all four transcriptional subgroups of medulloblastoma display aberrant expression of the miR-17/92 cluster (Fernandez et al., 2009). Aberrant expression of miR-17-92 in primary cerebellar granule precursors is not sufficient to induce medulloblastoma formation, but rather cooperate with SHH signaling to drive their expansion (Northcott et al., 2009a; Uziel et al., 2009). Other miRs important for the SHH pathway are miR-125b and miR-326 that suppresses SMO and miR-324-5p, which targets the downstream transcription factor GLI1 (Ferretti et al., 2008). Finally, miRNAs can also be regulated indirectly by drug treatment. For example, a lovastatin-regulated miRNA, miR-33, represses human MYC expression in miR-33 positive medulloblastoma cells (Takwi et al., 2012).
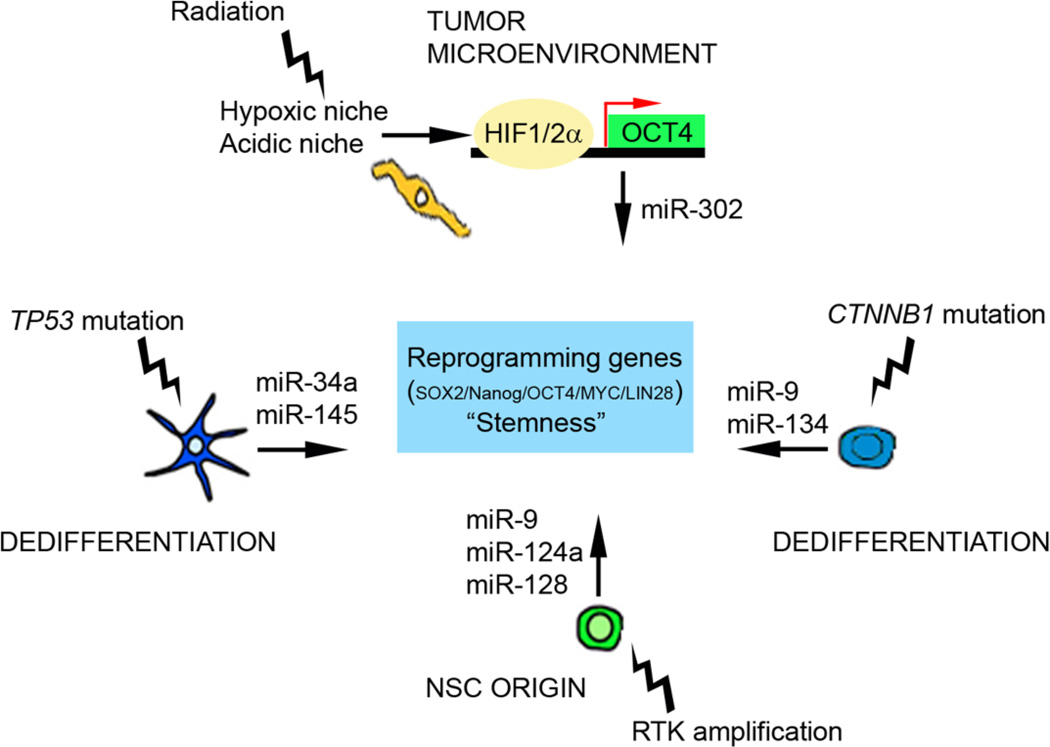
So-called “onco-miRs” have been found to regulate expression of tumor suppressor genes and oncogenes in cancers (for a review of “onco-miRs” and glioma see (Gonzalez-Gomez et al., 2011)). Future studies are needed that reveal how genetic alterations in turn regulate miRNAs that modulate self-renewal and differentiation in brain tumors. In conclusion, changes in the tumor microenvironment and genetic alterations regulate miRNA levels in a cell-context dependent manner that promotes expression of reprogramming genes and stemness in malignant brain tumors (Figure 3).
Figure 3. MiRNAs as mediators of reprogramming and stemness in brain tumors.
P53 mutations in brain tumors can drive stemness in tumor cells by down-regulating miR-34a and miR-145. Tumor microenviroment can drive stemness in heavily treated brain tumors displaying an hypoxic niche, by induction of reprogramming genes such as OCT4 reduced levels of miR-302. We propose that activating genetic alterations in the RAS pathway of NSCs produce reduced levels of miRNAs that promote self-renewal and stemness. To exemplify how a dedifferentiated cell might promote a stem cell-like tumor phenotype, CTNNB1 mutations (β-catenin) reduce levels of miR-9 and miR-134.
Conclusions
To effectively understand the susceptibility of neural precursor populations to generate brain tumors, important driver mutations needs to be defined and passenger mutations needs to be sorted out. Large scale whole genome sequencing and gene expression analyses of glioma samples have offered better genetic details; including frequencies of amplifications of known cancer genes like the EGFR, PDGFRA or PIK3CA/PIK3R1 but also identified novel cancer gene alterations in the IDH1/2, in CIC (homolog of the Drosophila gene capicua) as well as mutations in the far-upstream element (FUSE) binding protein, FUBP1 (Bettegowda et al., 2011; Parsons et al., 2008; Verhaak et al., 2010). Similar recent large-scale efforts in medulloblastoma found for instance inactivating mutations of the histone-lysine N-methyltransferase genes MLL2 or MLL3, regulators of H3K27 and H3K4 trimethylation like KDM6A and ZMYM3 and mutations in the RNA helicase gene DDX3X (Jones et al., 2012; Parsons et al., 2011; Pugh et al., 2012; Robinson et al., 2012). Such novel candidate genes should be carefully studied in sophisticated GEM models to understand if they are drivers or passengers in brain tumors.
Even though GEM models have been extremely valuable to understand the etiology of brain tumors, cell transformation differences between humans and rodents suggest that more studies should probe the ability to transform isolated human brain precursor cells (Rangarajan et al., 2004; Rangarajan and Weinberg, 2003). Studies by Kriegstein et al. highlight differences between mouse and human cortical development, including species-specific precursor populations that can serve as the cell of origin for human brain tumors (Hansen et al., 2010; Lamonica et al., 2012). Comparisons of the transcriptional profiles of murine and human OPCs reveal significant differences between fetal and human counterparts, as well as between rodent and human OPCs (Sim et al., 2009). These cellular differences and the disparity in developmental timing between human and mouse brains needs to be clarified in order to fully understand brain tumor development mechanisms.
In this review, we have discussed the signals and reprogramming networks that drive stemness in brain tumors. We argue that BTSCs and stemness in brain tumors can reflect a NSC origin but also be induced in restricted progenitors and differentiated glial cells. The traditional clonal evolution model suggests that all tumor cells are equivalent and can sustain tumor growth. In contrast, the cancer stem cell model proposes that a stable hierarchy exists, where only cancer stem cells undergo self-renewal and promote long-term tumor growth. We envision stemness as a fluid state in brain tumors that can be influenced by the tumor microenviroment, and emerge from genetic alterations over time. Is stemness then a driver or a passenger state in brain tumorigenesis and progression? If brain tumor stemness is a driver and needs to be controlled we suggest that modulation of miRNAs and blockade of HIF induction could function as excellent switches to reverse stemness and obstruct brain tumor progression.
Acknowledgements
Supported by research grants to AIP from the TDC Foundation, the American Cancer Society, and NIH/U54CA163155-01, to FJS from the Swedish Childhood Cancer Foundation, the Swedish Cancer Society, the Swedish Research Council, the Swedish Society of Medicine, Hjärnfonden, Åke Wibergs stiftelse, Lions Cancerforskningsfond, Stiftelsen Lars Hiertas Minne and the Association for International Cancer Research, and to JJP from the NIH (K08NS063456 and 1R01 NS081117-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We apologize to authors whose work we did not cite, due to space restrictions in this review. Authors further declare no conflict of interest.
References
- Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano JJ, Qureshi IA, Gokhan S, Molero AE, Zheng D, Bergman A, Mehler MF. Corepressor for element-1-silencing transcription factor preferentially mediates gene networks underlying neural stem cell fate decisions. Proc Natl Acad Sci U S A. 2010;107:16685–16690. doi: 10.1073/pnas.0906917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva JA, Lee GE, Escobar EE, Pyle AD. Phosphatase and tensin homolog regulates the pluripotent state and lineage fate choice in human embryonic stem cells. Stem Cells. 2011;29:1952–1962. doi: 10.1002/stem.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–247. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- Aranha MM, Santos DM, Sola S, Steer CJ, Rodrigues CM. miR-34a regulates mouse neural stem cell differentiation. PLoS One. 2011;6:e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Basak O, Giachino C, Fiorini E, Macdonald HR, Taylor V. Neurogenic subventricular zone stem/progenitor cells are Notch1-dependent in their active but not quiescent state. J Neurosci. 2012;32:5654–5666. doi: 10.1523/JNEUROSCI.0455-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behdad A, Perry A. Central nervous system primitive neuroectodermal tumors: a clinicopathologic and genetic study of 33 cases. Brain Pathol. 2010;20:441–450. doi: 10.1111/j.1750-3639.2009.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, Velculescu VE, Oba-Shinjo SM, Marie SK, Vogelstein B, Bigner D, Yan H, Papadopoulos N, Kinzler KW. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, Turski A, Azodi Y, Yang Y, Doucette T, Colman H, Sulman EP, Lang FF, Rao G, Copray S, Vaillant BD, Aldape KD. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chapouton P, Skupien P, Hesl B, Coolen M, Moore JC, Madelaine R, Kremmer E, Faus-Kessler T, Blader P, Lawson ND, Bally-Cuif L. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, Lau CC, Olson JM, Gilbertson RJ, Gajjar A, Delattre O, Kool M, Ligon K, Meyerson M, Mesirov JP, Pomeroy SL. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacki A, Mak G, Weiss S. PDGFRalpha expression distinguishes GFAP-expressing neural stem cells from PDGF-responsive neural precursors in the adult periventricular area. J Neurosci. 2011;31:9503–9512. doi: 10.1523/JNEUROSCI.1531-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Crisafulli L, Caldera V, Tortoreto M, Brilli E, Conforti P, Zunino F, Magrassi L, Schiffer D, Cattaneo E. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS One. 2012;7:e38486. doi: 10.1371/journal.pone.0038486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y, Kurc T, Van Meir EG, Saltz JH, Moreno CS, Brat DJ. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180:2108–2119. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, Saltz JH, Brat DJ, Moreno CS. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Kam M, Nannmark U, Anderson MF, Axell MZ, Wikkelso C, Holtas S, van Roon-Mom WM, Bjork-Eriksson T, Nordborg C, Frisen J, Dragunow M, Faull RL, Eriksson PS. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science. 2007;315:1243–1249. doi: 10.1126/science.1136281. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- de Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, De Vita G, Carotenuto M, Bello A, Formiggini F, Galeone A, De Rosa G, Virgilio A, Scognamiglio I, Sciro M, Basso G, Schulte JH, Cinalli G, Iolascon A, Zollo M. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One. 2011;6:e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bacco F, Casanova E, Medico E, Pellegatta S, Orzan F, Albano R, Luraghi P, Reato G, D'Ambrosio A, Porrati P, Patane M, Maderna E, Pollo B, Comoglio PM, Finocchiaro G, Boccaccio C. The MET Oncogene Is a Functional Marker of a Glioblastoma Stem Cell Subtype. Cancer Res. 2012;72:4537–4550. doi: 10.1158/0008-5472.CAN-11-3490. [DOI] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, Collisson E, Zhu J, Yegnasubramanian S, Matsui W, Baylin SB. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart CG. Three down and one to go: modeling medulloblastoma subgroups. Cancer Cell. 2012;21:137–138. doi: 10.1016/j.ccr.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, Taylor RE, Bailey S, Clifford SC, Gilbertson RJ. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, James CD, Molinaro A, Phillips JJ. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS One. 2012;7:e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Northcott PA, Taylor MD, Kenney AM. Normal and oncogenic roles for microRNAs in the developing brain. Cell Cycle. 2009;8:4049–4054. doi: 10.4161/cc.8.24.10243. [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, Bozzoni I, Gulino A. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Forsberg-Nilsson K, Behar TN, Afrakhte M, Barker JL, McKay RD. Platelet-derived growth factor induces chemotaxis of neuroepithelial stem cells. J Neurosci Res. 1998;53:521–530. doi: 10.1002/(SICI)1097-4547(19980901)53:5<521::AID-JNR2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of Neurons and Astrocytes by Oncogenes Can Induce Gliomas in Mice. Science. 2012;338(6110):1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, Majumder S. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol Cancer Ther. 2005;4:343–349. doi: 10.1158/1535-7163.MCT-04-0228. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De Martino D, Esposito V, Galeone A, Navas L, Esposito S, Gargiulo S, Fattet S, Donofrio V, Cinalli G, Brunetti A, Vecchio LD, Northcott PA, Delattre O, Taylor MD, Iolascon A, Zollo M. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi SO, Stark M, Zhao Z, Mobley BC, Munden A, Hover L, Abel TW. Cell of origin determines tumor phenotype in an oncogenic Ras/p53 knockout transgenic model of high-grade glioma. J Neuropathol Exp Neurol. 2012;71:729–740. doi: 10.1097/NEN.0b013e3182625c02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, Finkelstein D, Pounds S, Weiss A, Patay Z, Scoggins M, Ogg R, Pei Y, Yang ZJ, Brun S, Lee Y, Zindy F, Lindsey JC, Taketo MM, Boop FA, Sanford RA, Gajjar A, Clifford SC, Roussel MF, McKinnon PJ, Gutmann DH, Ellison DW, Wechsler-Reya R, Gilbertson RJ. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia-Bonnefil P. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J Neurosci. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gomez P, Sanchez P, Mira H. MicroRNAs as regulators of neural stem cell-related pathways in glioblastoma multiforme. Mol Neurobiol. 2011;44:235–249. doi: 10.1007/s12035-011-8196-y. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez O, Romero-Rodriguez R, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Epidermal growth factor induces the progeny of subventricular zone type B cells to migrate and differentiate into oligodendrocytes. Stem Cells. 2009;27:2032–2043. doi: 10.1002/stem.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Gritti A, Frolichsthal-Schoeller P, Galli R, Parati EA, Cova L, Pagano SF, Bjornson CR, Vescovi AL. Epidermal and fibroblast growth factors behave as mitogenic regulators for a single multipotent stem cell-like population from the subventricular region of the adult mouse forebrain. J Neurosci. 1999;19:3287–3297. doi: 10.1523/JNEUROSCI.19-09-03287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronych J, Korshunov A, Bageritz J, Milde T, Jugold M, Hambardzumyan D, Remke M, Hartmann C, Witt H, Jones DT, Witt O, Heiland S, Bendszus M, Holland EC, Pfister S, Lichter P. An activated mutant BRAF kinase domain is sufficient to induce pilocytic astrocytoma in mice. J Clin Invest. 2011;121:1344–1348. doi: 10.1172/JCI44656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, Purow B, Abounader R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM, Shou W, Hambardzumyan D, Lee J, Hjelmeland AB, Sloan AE, Bredel M, Stark GR, Rich JN, Bao S. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Giese NA, Richardson WD. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development. 1996;122:4085–4094. doi: 10.1242/dev.122.12.4085. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, Olson JM. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan A, Rich JN. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kriegstein AR, Alvarez-Buylla A. Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron. 2011;71:250–262. doi: 10.1016/j.neuron.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Jacques TS, Swales A, Brzozowski MJ, Henriquez NV, Linehan JM, Mirzadeh Z, O'Malley C, Naumann H, Alvarez-Buylla A, Brandner S. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29:222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001268. e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HM, Jin X, Lee JS, Oh SY, Sohn YW, Park HJ, Joo KM, Park WY, Nam DH, DePinho RA, Chin L, Kim H. Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev. 2008;22:2028–2033. doi: 10.1101/gad.1668708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, Rand V, Leary SE, White E, Eden C, Hogg T, Northcott P, Mack S, Neale G, Wang YD, Coyle B, Atkinson J, DeWire M, Kranenburg TA, Gillespie Y, Allen JC, Merchant T, Boop FA, Sanford RA, Gajjar A, Ellison DW, Taylor MD, Grundy RG, Gilbertson RJ. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, McCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison DW, Gilbertson RJ, Roussel MF. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick TJ, Bartlett PF. Cloned multipotential precursors from the mouse cerebrum require FGF-2, whereas glial restricted precursors are stimulated with either FGF-2 or EGF. J Neurosci. 1995;15:3653–3661. doi: 10.1523/JNEUROSCI.15-05-03653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim E, Wu Q, Guryanova O, Hitomi M, Lathia JD, Serwanski D, Sloan AE, Weil RJ, Lee J, Nishiyama A, Bao S, Hjelmeland AB, Rich JN. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26:1247–1262. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioi M, Vogel H, Schultz G, Hoffman RM, Harsh GR, Brown JM. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J Clin Invest. 2010;120:694–705. doi: 10.1172/JCI40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Corfas G. Identification of novel glial genes by single-cell transcriptional profiling of Bergmann glial cells from mouse cerebellum. PLoS One. 2010;5:e9198. doi: 10.1371/journal.pone.0009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, Mrsic A, Ylstra B, Grajkowska W, Hartmann W, Pietsch T, Ellison D, Clifford SC, Versteeg R. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH. Epidermal growth factor and fibroblast growth factor-2 have different effects on neural progenitors in the adult rat brain. J Neurosci. 1997;17:5820–5829. doi: 10.1523/JNEUROSCI.17-15-05820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, Ellingson BM, Bowers CW, Soriano RH, Schmidt NO, Mohan S, Yong WH, Seshagiri S, Modrusan Z, Jiang Z, Aldape KD, Mischel PS, Liau LM, Escovedo CJ, Chen W, Nghiemphu PL, James CD, Prados MD, Westphal M, Lamszus K, Cloughesy T, Phillips HS. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, Yong WH, Vinters HV, Liau LM, Lazareff JA, Mischel PS, Cloughesy TF, Horvath S, Kornblum HI. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr Opin Neurobiol. 2012;22(5):747–753. doi: 10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee da Y, Gianino SM, Gutmann DH. Innate neural stem cell heterogeneity determines the patterning of glioma formation in children. Cancer Cell. 2012;22:131–138. doi: 10.1016/j.ccr.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009a;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009b;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, Xia S, Trageser D, Guerrero-Cazares H, Eberhart CG, Quinones-Hinojosa A, Scheffler B, Laterra J. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108:9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009c;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Desire L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Lindberg N, Kastemar M, Olofsson T, Smits A, Uhrbom L. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28:2266–2275. doi: 10.1038/onc.2009.76. [DOI] [PubMed] [Google Scholar]
- Listernick R, Darling C, Greenwald M, Strauss L, Charrow J. Optic pathway tumors in children: the effect of neurofibromatosis type 1 on clinical manifestations and natural history. J Pediatr. 1995;127:718–722. doi: 10.1016/s0022-3476(95)70159-1. [DOI] [PubMed] [Google Scholar]
- Little SE, Popov S, Jury A, Bax DA, Doey L, Al-Sarraj S, Jurgensmeier JM, Jones C. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72:1614–1620. doi: 10.1158/0008-5472.CAN-11-4069. [DOI] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Wang Y, Belz T, Bock D, Takacs A, Radlwimmer B, Barbus S, Reifenberger G, Lichter P, Schutz G. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev. 2010;24:683–695. doi: 10.1101/gad.560310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, Isachenko N, Fouse SD, Phillips JJ, Cheresh DA, Park M, Bergers G. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, Verma IM. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, Falini A, De Palma M, Bulfone A, Poliani PL, Galli R. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci U S A. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miale IL, Sidman RL. An autoradiographic analysis of histogenesis in the mouse cerebellum. Exp Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Momota H, Shih AH, Edgar MA, Holland EC. c-Myc and beta-catenin cooperate with loss of p53 to generate multiple members of the primitive neuroectodermal tumor family in mice. Oncogene. 2008;27:4392–4401. doi: 10.1038/onc.2008.81. [DOI] [PubMed] [Google Scholar]
- Monje M, Mitra SS, Freret ME, Raveh TB, Kim J, Masek M, Attema JL, Li G, Haddix T, Edwards MS, Fisher PG, Weissman IL, Rowitch DH, Vogel H, Wong AJ, Beachy PA. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci U S A. 2011;108:4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009a;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, Ra YS, Zilberberg K, McLeod J, Scherer SW, Sunil Rao J, Eberhart CG, Grajkowska W, Gillespie Y, Lach B, Grundy R, Pollack IF, Hamilton RL, Van Meter T, Carlotti CG, Boop F, Bigner D, Gilbertson RJ, Rutka JT, Taylor MD. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009b;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, Beroukhim R, Ellison DW, Marshall CR, Lionel AC, Mack S, Dubuc A, Yao Y, Ramaswamy V, Luu B, Rolider A, Cavalli FM, Wang X, Remke M, Wu X, Chiu RY, Chu A, Chuah E, Corbett RD, Hoad GR, Jackman SD, Li Y, Lo A, Mungall KL, Nip KM, Qian JQ, Raymond AG, Thiessen NT, Varhol RJ, Birol I, Moore RA, Mungall AJ, Holt R, Kawauchi D, Roussel MF, Kool M, Jones DT, Witt H, Fernandez LA, Kenney AM, Wechsler-Reya RJ, Dirks P, Aviv T, Grajkowska WA, Perek-Polnik M, Haberler CC, Delattre O, Reynaud SS, Doz FF, Pernet-Fattet SS, Cho BK, Kim SK, Wang KC, Scheurlen W, Eberhart CG, Fevre-Montange M, Jouvet A, Pollack IF, Fan X, Muraszko KM, Gillespie GY, Di Rocco C, Massimi L, Michiels EM, Kloosterhof NK, French PJ, Kros JM, Olson JM, Ellenbogen RG, Zitterbart K, Kren L, Thompson RC, Cooper MK, Lach B, McLendon RE, Bigner DD, Fontebasso A, Albrecht S, Jabado N, Lindsey JC, Bailey S, Gupta N, Weiss WA, Bognar L, Klekner A, Van Meter TE, Kumabe T, Tominaga T, Elbabaa SK, Leonard JR, Rubin JB, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012a;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C, Eberhart CG, Dubuc A, Guettouche T, Cardentey Y, Bouffet E, Pomeroy SL, Marra M, Malkin D, Rutka JT, Korshunov A, Pfister S, Taylor MD. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012b;123:615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pacal M, Bremner R. Insights from animal models on the origins and progression of retinoblastoma. Curr Mol Med. 2006;6:759–781. doi: 10.2174/1566524010606070759. [DOI] [PubMed] [Google Scholar]
- Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Brun SN, Markant SL, Lento W, Gibson P, Taketo MM, Giovannini M, Gilbertson RJ, Wechsler-Reya RJ. WNT signaling increases proliferation and impairs differentiation of stem cells in the developing cerebellum. Development. 2012a;139:1724–1733. doi: 10.1242/dev.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, Witt H, Korshunov A, Read TA, Sun JL, Schmitt EM, Miller CR, Buckley AF, McLendon RE, Westbrook TF, Northcott PA, Taylor MD, Pfister SM, Febbo PG, Wechsler-Reya RJ. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012b;21:155–167. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S, Ayers-Ringler J, Nishiyama A, Stallcup WB, Berger MS, Bergers G, McKnight TR, Goldman SA, Weiss WA. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, Toedt G, Wittmann A, Kratz C, Olbrich H, Ahmadi R, Thieme B, Joos S, Radlwimmer B, Kulozik A, Pietsch T, Herold-Mende C, Gnekow A, Reifenberger G, Korshunov A, Scheurlen W, Omran H, Lichter P. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J, Wittmann A, Devens F, Gerber NU, Joos S, Kulozik A, Reifenberger G, Rutkowski S, Wiestler OD, Radlwimmer B, Scheurlen W, Lichter P, Korshunov A. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pistollato F, Abbadi S, Rampazzo E, Persano L, Della Puppa A, Frasson C, Sarto E, Scienza R, D'Avella D, Basso G. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells. 2010;28:851–862. doi: 10.1002/stem.415. [DOI] [PubMed] [Google Scholar]
- Po A, Ferretti E, Miele E, De Smaele E, Paganelli A, Canettieri G, Coni S, Di Marcotullio L, Biffoni M, Massimi L, Di Rocco C, Screpanti I, Gulino A. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 2010;29:2646–2658. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polajeva J, Swartling FJ, Jiang Y, Singh U, Pietras K, Uhrbom L, Westermark B, Roswall P. miRNA-21 is developmentally regulated in mouse brain and is co-expressed with SOX2 in glioma. BMC Cancer. 2012;12:378. doi: 10.1186/1471-2407-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Bishop JM. Nucleostemin maintains self-renewal of embryonic stem cells and promotes reprogramming of somatic cells to pluripotency. J Cell Biol. 2012;197:731–745. doi: 10.1083/jcb.201103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Principles of neural cell migration. Experientia. 1990;46:882–891. doi: 10.1007/BF01939380. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- Rao G, Pedone CA, Coffin CM, Holland EC, Fults DW. c-Myc enhances sonic hedgehog-induced medulloblastoma formation from nestin-expressing neural progenitors in mice. Neoplasia. 2003;5:198–204. doi: 10.1016/S1476-5586(03)80052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SL, Schwalbe EC, Cole M, Lu Y, Lusher ME, Megahed H, O'Toole K, Nicholson SL, Bognar L, Garami M, Hauser P, Korshunov A, Pfister SM, Williamson D, Taylor RE, Ellison DW, Bailey S, Clifford SC. MYC family amplification and clinical risk-factors interact to predict an extremely poor prognosis in childhood medulloblastoma. Acta Neuropathol. 2012;123:501–513. doi: 10.1007/s00401-011-0923-y. [DOI] [PubMed] [Google Scholar]
- Sanai N, Nguyen T, Ihrie RA, Mirzadeh Z, Tsai HH, Wong M, Gupta N, Berger MS, Huang E, Garcia-Verdugo JM, Rowitch DH, Alvarez-Buylla A. Corridors of migrating neurons in the human brain and their decline during infancy. Nature. 2011;478:382–386. doi: 10.1038/nature10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanalkumar R, Dhanesh SB, James J. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell Mol Life Sci. 2010;67:2957–2968. doi: 10.1007/s00018-010-0391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Mansur DB, Reifenberger G, Perry A, Leonard JR, Aldape KD, Albin MG, Emnett RJ, Loeser S, Watson MA, Nagarajan R, Gutmann DH. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67:890–900. doi: 10.1158/0008-5472.CAN-06-0973. [DOI] [PubMed] [Google Scholar]
- Shih CC, Forman SJ, Chu P, Slovak M. Human embryonic stem cells are prone to generate primitive, undifferentiated tumors in engrafted human fetal tissues in severe combined immunodeficient mice. Stem Cells Dev. 2007;16:893–902. doi: 10.1089/scd.2007.0070. [DOI] [PubMed] [Google Scholar]
- Shimozaki K, Zhang CL, Suh H, Denli AM, Evans RM, Gage FH. SRY-box-containing gene 2 regulation of nuclear receptor tailless (Tlx) transcription in adult neural stem cells. J Biol Chem. 2012;287:5969–5978. doi: 10.1074/jbc.M111.290403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook BA, Manz DH, Peters JJ, Kang S, Conover JC. Spatiotemporal changes to the subventricular zone stem cell pool through aging. J Neurosci. 2012;32:6947–6956. doi: 10.1523/JNEUROSCI.5987-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Windrem MS, Goldman SA. Fate determination of adult human glial progenitor cells. Neuron Glia Biol. 2009;5:45–55. doi: 10.1017/S1740925X09990317. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, Louis DN, Iafrate AJ. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Soldati C, Bithell A, Johnston C, Wong KY, Teng SW, Beglopoulos V, Stanton LW, Buckley NJ. Repressor element 1 silencing transcription factor couples loss of pluripotency with neural induction and neural differentiation. Stem Cells. 2012;30:425–434. doi: 10.1002/stem.1004. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Sottile V, Li M, Scotting PJ. Stem cell marker expression in the Bergmann glia population of the adult mouse brain. Brain Res. 2006;1099:8–17. doi: 10.1016/j.brainres.2006.04.127. [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Fruhwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Durken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, Snyder E, Eberhart CG, Majumder S. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, Marquez VE, Clement V, Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- Swartling FJ. Myc proteins in brain tumor development and maintenance. Ups J Med Sci. 2012;117:122–131. doi: 10.3109/03009734.2012.658975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, Lau J, Masic S, Nguyen K, Yakovenko S, Zhe XN, Gilmer HC, Collins R, Nagaoka M, Phillips JJ, Jenkins RB, Tihan T, Vandenberg SR, James CD, Tanaka K, Taylor MD, Weiss WA, Chesler L. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010;24:1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A, Phillips JJ, Taylor MD, Weiss WA. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, Brennan CW. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takwi AA, Li Y, Becker Buscaglia LE, Zhang J, Choudhury S, Park AK, Liu M, Young KH, Park WY, Martin RC. A statin-regulated microRNA represses human c-Myc expression and function. EMBO Mol Med. 2012;4:896–909. doi: 10.1002/emmm.201101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012a;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, Magdaleno S, Dalton J, Calabrese C, Board J, Macdonald T, Rutka J, Guha A, Gajjar A, Curran T, Gilbertson RJ. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Taylor P, Fangusaro J, Rajaram V, Goldman S, Helenowski IB, Macdonald T, Hasselblatt M, Riedemann L, Laureano A, Cooper L, Gopalakrishnan V. REST Is a Novel Prognostic Factor and Therapeutic Target for Medulloblastoma. Mol Cancer Ther. 2012b;11:1713–1723. doi: 10.1158/1535-7163.MCT-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, Gajjar A, Gilbertson RJ. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Sibilia M, Ciruna BG, Rossant J, Wagner EF, van der Kooy D. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev Biol. 1999;208:166–188. doi: 10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Venugopal C, Manoranjan B, McFarlane N, O'Farrell E, Nolte S, Gunnarsson T, Hollenberg R, Kwiecien J, Northcott P, Taylor MD, Hawkins C, Singh SK. Sonic hedgehog regulates Bmi1 in human medulloblastoma brain tumor-initiating cells. Oncogene. 2012;31:187–199. doi: 10.1038/onc.2011.232. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Weeraratne SD, Amani V, Neiss A, Teider N, Scott DK, Pomeroy SL, Cho YJ. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13:165–175. doi: 10.1093/neuonc/noq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner HL, Bakst R, Hurlbert MS, Ruggiero J, Ahn E, Lee WS, Stephen D, Zagzag D, Joyner AL, Turnbull DH. Induction of medulloblastomas in mice by sonic hedgehog, independent of Gli1. Cancer Res. 2002;62:6385–6389. [PubMed] [Google Scholar]
- Weiss S, Dunne C, Hewson J, Wohl C, Wheatley M, Peterson AC, Reynolds BA. Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J Neurosci. 1996;16:7599–7609. doi: 10.1523/JNEUROSCI.16-23-07599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Burns MJ, Hackett C, Aldape K, Hill JR, Kuriyama H, Kuriyama N, Milshteyn N, Roberts T, Wendland MF, DePinho R, Israel MA. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–1595. [PubMed] [Google Scholar]
- Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum. 2010;9:537–547. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, Sawaya R, Heimberger AB. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Tamamaki N, Noda T, Kimura K, Itokazu Y, Matsumoto N, Dezawa M, Ide C. Neurogenesis in the ependymal layer of the adult rat 3rd ventricle. Exp Neurol. 2005;192:251–264. doi: 10.1016/j.expneurol.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]