Abstract
Itch, a HECT family E3 ligase, affects numerous cell functions by regulating ubiquitination and proteasomal degradation of target proteins. However, the role of Itch in osteoblasts has not been investigated. We report that Itch−/− mice have significantly increased bone volume, osteoblast numbers and bone formation rate. Using bone marrow stromal cells from Itch−/− mice and WT littermates as bone marrow mesenchymal precursor cells (BM-MPCs), we found BM-MPCs from Itch−/− mice have compatible numbers of cells expressing mesenchymal stem cell markers. However, Itch−/− BM-MPCs grew faster in an in vitro culture, formed more CFU-F mesenchymal colonies, and exhibited increased osteoblast differentiation and decreased adipogenesis. Importantly, Itch−/− mesenchymal colony cells formed significantly more new bone in a tibial defect of recipient mice compared with WT cells. The expression levels of JunB, an AP-1 transcription factor that positively regulate osteoblast differentiation, were significantly increased in Itch−/− BM-MPCs when proteasome function is intact. In contrast, the amount of ubiquitinated JunB protein was markedly decreased in Itch−/− cells when proteasome function was blocked. Over-expression of WT Itch, but not an Itch ligase-inactive mutant, rescued differentiation defects of Itch−/− BM-MPCs. Itch−/− BM-MPCs had a similar role in immune modulation as WT cells. Thus, Itch negatively controls osteoblast differentiation from BM-MPCs through the regulation of proteasomal degradation of positive osteoblast regulator JunB protein. Itch is a potential new target for bone anabolic drug development to treat patients with bone loss.
Keywords: Itch, E3 ligase, mesenchymal progenitor cells, osteoblasts, bone formation
INTRODUCTION
Ubiquitination is a post-translational modification that has many functional implications. Ubiquitination occurs through a well-defined three-step process. Ubiquitin is first transferred to an ubiquitin-activating enzyme, E1. The activated ubiquitin is then transferred to an ubiquitin-conjugating enzyme, E2. Finally, the ubiquitin–E2 complex is recruited to a third enzyme, an ubiquitin ligase, E3, that specifically binds a protein substrate and facilitates the transfer of ubiquitin from E2 to the substrate 1. Ubiquitinated proteins undergo proteasomal or lysosomal degradation 2. There are several types of ubiquitin ligases, which is classified according to structures and mechanisms of action to mediate target protein ubiquitination. WW domain-containing ubiquitin ligases are a subgroup of the HECT family of ubiquitin E3 ligases, which promote protein ubiquitination by binding to a PPXY motif on target proteins. Up to date, the WW domain-containing ubiquitin ligases, Smurf1, Smurf2, Wwp1 and Wwp2, have been reported to be involved in bone cell regulation through modification of the stability of multiple proteins including BMP-Smad-Runx2 protein 3, 4, Smad3 and GSK3β 5, JunB 6, and Goosecoid 7. Itch is another member of the WW domain-containing ubiquitin ligases. Itch−/− mice on a C57BL/6J background develop a progressive autoimmune disease with multi-organ inflammation 8. Patients with Itch mutations have autoimmune inflammatory cell infiltration in various tissues 9. However, the role of Itch in bone disease has not been investigated.
The molecular mechanisms by which Itch deficiency leads to autoimmune disease and multi-organ inflammation have been linked to persistent activation of the Jun amino-terminal kinase (JNK) and NF-κB signal pathways in T cells and macrophages in response to inflammatory cytokines. Itch−/− T cells show an activated phenotype, enhanced proliferation and augmented production of the type 2 T helper (TH2) cell cytokines interleukin 4 (IL-4) and IL-5, which are associated with decreased JunB ubiquitination 10. Furthermore, TH2 cell cytokins trigger phosphorylation-dependent activation of Itch through JNK signal protein kinase cascade, leading to accelerated degradation of c-Jun and JunB 11. Itch limits TNF-induced NF-κB activation by facilitating A20-mediated ubiquitination and degradation of the adaptor protein, RIP, in the TNF receptor complex in T cells and macrophages 12, 13. Itch is also required for negative regulation of TNF- and lipopolysaccharide (LPS)-mediated TNF receptor-associated factor 6 (TRAF6) ubiquitination induced by RING finger protein 11 14. Whether these signaling molecules mediate the effect of Itch on bone cells have not been investigated.
Recently we examined bone phenotype of Itch−/− mice and found that young Itch−/− mice have increased bone mass despite that they have increased osteoclast numbers, suggesting that Itch must affect cells in the osteoblast lineage to overcome osteoclast-mediated effect. The aim of this study is to elucidate cellular and molecular mechanisms by which osteoblast differentiation and function is elevated in Itch−/− mice. We demonstrate that Itch−/− mice have increased osteoblast numbers and bone formation rate. Bone marrow mesenchymal precursor cells (BM-MPCs) from Itch−/− mice have increased osteoblast differentiation in vivo and in viro. At the molecular level, Itch−/− BM-MPCs express increased amount of JunB protein, an positive regulator of osteoblast differentiation. Over-expression of WT Itch, but not an Itch ligase-inactive mutant, rescued differentiation defects of Itch−/− BM-MPCs. Our findings indicate that Itch depletion has a strong positive effect on osteoblast differentiation in a cell autonomous fashion and Itch is a new ubiquitin E3 ligase that negatively regulates osteoblast differentiation.
MATERIALS AND METHODS
Animals
Itch−/− mice were generated on a C57BL/6J background 8 and were genotyped by PCR analysis. Itch+/− mice were bred to generate Itch−/− mice. Primers for genotyping: Itch WT:5′-ATCGTCTACTCACCCCACATAAGG-3′, Itch knockout: 5′-AAGAAGCAGCAGAGACAACGAGTG-3′ Common: 5′-TCTATGCTCTGTTGTCTCCCATGC-3′. All animal procedures were conducted using procedures approved by the University of Rochester Committee for Animal Resources. One-month-old Itch−/− mice and WT littermates were used. SCID mice were purchased from Jackson Laboratories (Strain Name: B6.CB17-Prkdcscid/SzJ, Stock Number: 001913).
Antibodies and retrovirus
Antibodies to JunB, β-actin and Runx2 were purchased from Santa Cruz. FITC-anti-CD45, PE-anti-CD105, APC-anti-CD31, PE-cy7-anti-Sca1 and PE-cy5-anti-CD11b used in FACS were purchased from eBioscience. To generate retroviral expression vector encoding Itch or Itch ligase-inactive mutant, in which cysteine residue at the 830 position was mutated to alanine (ItchC830A), the coding region for Itch and ItchC830A was amplified by PCR from the Flag-Itch or ItchC830A plasmid and cloned into the pMX-GFP retroviral vector at the BamHl and Notl site, resulting in to the pMX-Itch-GFP and the pMX-ItchC830A-GFP expression vector. The pMX-GFP vector was used as a control for infection efficiency. These retroviral vectors were transiently transfected into the Plat-E retroviral packaging cell line, and viral supernatant was collected 48 h later, as we described previously 15.
Fluorescence-activated cell sorting (FACS)
Cells were harvested and red blood cells were lysed. Cells were stained with FITC-anti-CD45, PE-anti-CD105, APC-anti-CD31, PE-cy7-anti-Sca1 and PE-cy5-anti-CD11b for 30 mins, and subjected to FACS analysis according to the manufacture’s instruction 16. Results were analyzed by Flowjo7 software.
Mesenchymal progenitor cell preparation
All cells used in this study were primary cells from bone marrow after red blood cells were lysed. Several types of cell preparations were prepared based on individual experimental needs. 1) For CFU colony formation assay, an assay to evaluate the cell’s capacity to give rise to CFU-F, mesenchymal colonies, and CFU-Alkaline phosphatase (ALP) assay, an assay to examine the cell’s capacity to differentiate to osteoblasts, bone marrow cells were cultured in 10cm dish at 106 cells/dish in 10ml of α-MEM culture medium containing 10% FCS with or without 50μg/mL ascorbic acid and 10mM β-glycerophosphate. Media were changed every 4 days, and cultures were maintained for 28 days. At the end of the culture period, cells were stained for H&E or ALP. 2) For bone nodule formation, bone marrow cells were cultured in α-MEM culture medium containing 10% FCS for 7-10 days to generate bone marrow mesenchymal precursor cells (BM-MPCs). BM-MPCs were then cultured in osteoblast-inducing medium containing 50μg/mL ascorbic acid and 10mM β-glycerophosphate for 21-28 days and mineralized bone nodules were examined by Von Kossa staining. 3) For adipogenesis, BM-MPCs were cultured with the adipocyte differentiation medium for 12–16 days as described previously 17. Adipocyte staining with oil red O was performed. 4) For isolating CD45− cells which contain enriched mesenchymal progenitor cells (MPC-enriched cells), bone marrow cells were incubated with anti-CD45 antibody conjugated microbeads (Miltenyi Biotec, Auburn, CA, USA, www.miltenyibiotec.com). The CD45 negative population (CD45−) was isolated by negative selection according to the manufacturer’s instructions as we previously described 18. 5) For bone-derived mesenchymal progenitor cells (B-MPCs) were isolated using a recent published protocol 19. Long bones were flushed several times with PBS, cut into small pieces, and cultured in a plastic dish for 3 days. The bone pieces were transferred into a clear dish as the passage 1 and continually cultured for another 7 days to allow cells growing to confluent. The 3rd passage cells were used for characterization and experiments.
MicroCT, histology and histomorphometry of bone sections
Bones were dissected free of soft tissue, fixed overnight in 70% ethanol, and scanned at high resolution (10.5 μm) on a VivaCT40 micro-CT scanner (Scanco Medical, Basserdorf, Switzerland) using an integration time of 300 ms, energy of 55kVp, and intensity of 145 uA. The three-dimensional images were generated using a constant threshold of 275 for all samples. The hind limbs were fixed in 10% buffered formalin, decalcified in 10% EDTA, and embedded in paraffin. Sections (4 μm thick) were then stained with H&E or for Tartrate-resistant acid phosphatase (TRAP) activity for osteoclast identification. Histomorphometric analysis of osteoblast and osteoclasts numbers, expressed as the number per millimeter of bone surface, was performed on 2-3 sections/bone using an Osteometrics image analysis software system (Osteometrics, Atlanta, GA, USA), as we previously described 15.
Double calcein labeling
Double calcein labeling was performed by intraperitoneal injection of 10 mg calcein per gram of body weight (C-0875, Sigma, St. Louis, MO, USA) at 6 days and 1 day prior to sacrifice, as described previously 17. Bones were harvested and embedded in LR White acrylic resin. Serial sections were cut, and the freshly cut surface of each section was viewed and imaged using fluorescence microscopy. The double calcein–labeled morphometric analysis in trabecular bone was measured using an Osteometrics image analysis software system (Osteometrics, Atlanta, GA, USA). Two sections were assessed per bone. The mineral apposition rate (MAR), bone formation rate (BFR), and double label surface/bone surface (dLS/BS) were calculated as we previously described 18.
Tibial bone defect model
Two-month-old SCID mice (n=5) were anaesthetized and bilateral 2×5mm cortical bone defects were made in the anterior proximal tibiae of SCID mice and filled with bovine bone matrix. 5×105 cells from CFU colonies were injected into the bone matrix in defects. The right tibiae received cells from WT mice and the left tibiae received cells from Itch−/− mice. The mice were sacrificed 6 weeks post-surgery, and the volume of new bone (BV/TV, expressed as a percentage of the total defect volume) formed in the defects was measured by μCT followed by histomorphometric analysis of the area of newly formed trabecular bone in decalcified H&E-stained sections. Three sections were assessed per bone.
Quantitative real time RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). cDNAs were synthesized by iSCRIPT cDNA Synthesis Kit (Bio-Rad). Quantitative real time RT-PCR amplifications were performed in the iCycler (Bio-Rad) real time PCR machine using iQ SYBR Green supermix (Bio-Rad) according to the manufacturer’s instruction. gapdh was amplified on the same plates and used to normalize the data. Each sample was prepared in triplicate and each experiment was repeated at least three times. The relative abundance of each gene was calculated by subtracting the CT value of each sample for an individual gene from the corresponding CT value of gapdh (ΔCT). ΔΔCT were obtained by subtracting the ΔCT of the reference point. These values were then raised to the power of 2 (2ΔΔCT) to yield fold-expression relative to the reference point. Representative data are presented as means + SD of the triplicates or of four wells of cell culture. The sequences of primer sets for ALP, osteocalcin (OCN), PPARgama, C/EBPalpha, C/EBPbeta and gapdh mRNAs are shown in Table.
Table.
| Name | F/R | Sequences |
|---|---|---|
| Coll | F | TCTCCACTCTTCTAGTTCCT |
| R | TTGGGTCATTTCCACATGC | |
| ALP | F | CTTGCTGGTGGAAGGAGGCAGG |
| R | CACGTCTTCTCCACCGTGGGTC | |
| OCN | F | CAAGTCCCACACAGCAGCTT |
| R | AAAGCCGAGCTGCCAGAGTT | |
| PPARγ | F | TCGCTGATGCACTGCCTATG |
| R | GAGAGGTCCACAGAGCTGATT | |
| C/EBPα | F | CAAGAACAGCAACGAGTACCG |
| R | GTCACTGGTCAACTCCAGCAC | |
| C/EBPβ | F | CGCAGACAGTGGTGAGCTT |
| R | CTTCTGCTGCATCTCCTGGT | |
| C/EBPδ | F | CGACTTCAGCGCCTACATTGA |
| R | CTAGCGACAGACCCCACAC | |
| GAPDH | F | GGTCGGTGTGAACGGATTTG |
| R | ATGAGCCCTTCCACAATG |
Western blot analysis and ubiquitination assay
For Western blot analysis, whole-cell lysates were prepared from B-MPCs. Whole cell lysates (10 μg protein/lane) were loaded in 10% SDS-PAGE gels and immunoblotted with antibodies to Runx2, JunB, and β-actin. For ubiquitination assay, B-MPCs were treated with the proteasome inhibitor MG 132 (10 μM) for 4 hours. Whole cell lysates (200μg protein/sample) were incubated with UbiQapture-Q Matrix (Biomol) by gentle agitation at 4°C overnight to pull down all ubiquitinated proteins according to the manufacturer’s instructions. After washing three times, captured proteins were eluted with 2× SDS-PAGE loading buffer and analyzed by Western blotting using anti-JunB antibody, as described previously 18.
T cell proliferation assay
The effect of mesenchymal cells on T cell proliferation was examined according to a published protocol 20. In brief, mononuclear cells were obtained from the spleen and incubated with 1 μM Carboxyfluorescein succinimidyl ester (CFSE, Molecular Probes) for 8 min at 20°C. Cells were quenched with fetal calf serum, washed twice with media, and then suspended in complete RPMI medium containing 2 mM glutamine (Sigma-Aldrich), 100 IU/mL penicillin, 100 μg/mL streptomycin. CFSE labeled spleen cells (2× 105 in 200 μL) were seeded into 96-well plate at a final cell density of 1× 106/mL along with 5× 104 B-MPCs, in the presence of 10 μg/mL anti-CD3 antibody (R&D Systems, Minneapolis, MN). After 3 days, cells were stained with CD4-PE and were analyzed by FACS. The percentage of CD4+ T cells and distribution of CFSE-labeled CD4+ T cells were determined.
Statistics
All results are given as mean ± SD. Comparisons between 2 groups were analyzed using the 2-tailed unpaired Student’s t test. One way ANOVA and Dunnett’s post-hoc multiple comparisons were used for comparisons among 3 or more groups. P values less than 0.05 were considered statistically significant.
RESULTS
Itch−/− mice have increased bone volume and osteoblastic bone formation
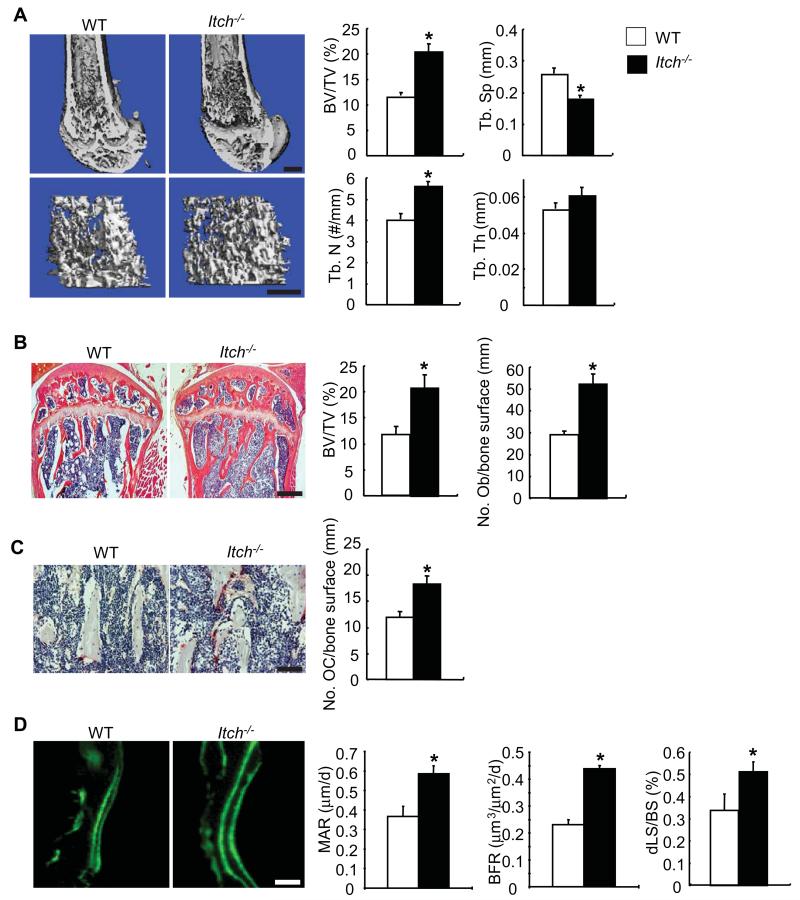
To determine the role of Itch in bone mass, we examined bone phenotype of 1-month-old Itch−/− mice and WT littermates. μCT of femoral bones showed that Itch−/− mice have increased bone volume, trabecular numbers and decreased trabecular separation compared with WT littermates (Figure 1A). Histomorphometric analysis of H&E-stained bone sections revealed 45-50% increased bone volume and osteoblast numbers in Itch−/− mice (BV/TV: 20.6±2.6% vs 11.6±1.6% in WT mice; and No. osteoblasts/mm bone surface: 52±5 vs 29±2 in WT mice)(Figure 1B). TRAP stained sections showed 30-35% increased osteoclasts on trabecular bone surface in Itch−/− mice (No. osteoclasts/mm bone surface: 18±2 vs 12±1.5 in WT mice)(Figure 1C). Double calcein labeling indicated increased MAR (0.59±0.04 vs 0.37±0.05 μm/day), BFR (0.44±0.01 vs 0.23±0.02 μm3/μm2/day), and dLS/BS (0.51±0.05 vs 0.34±0.08 %) in bone sections from Itch−/− mice compared to bone sections from WT mice. These data suggested that the increased bone volume in Itch−/− mice is caused by increased osteoblastic bone formation, although the increased osteoclastic formation is also observed.
Figure 1. Itch−/− mice have increased bone volume.
Bones from 1-month-old Itch−/− mice and WT littermates were analyzed. (A) Representative image of μCT scanning of femur and μCT data analyses. Bar = 300μm. (B) H&E-stained tibial sections and histomorphometric analyses of the percentage of BV/TV and the number of osteoblast (Ob)/mm bone surface. Bar = 250μm. (C) TRAP-stained sections and the number of TRAP+ osteoclasts (OC)/mm bone surface. Bar = 50μm. (D) Double calcein labeling of femoral sections and bone formation parameters. Bar = 25μm. Values are mean ± SD of 5 mice/group. *p<0.05 vs WT mice.
Mesenchymal progenitor cells from Itch−/− mice have increased CFU colony formation and osteoblast differentiation, and decreased adipocyte differentiation
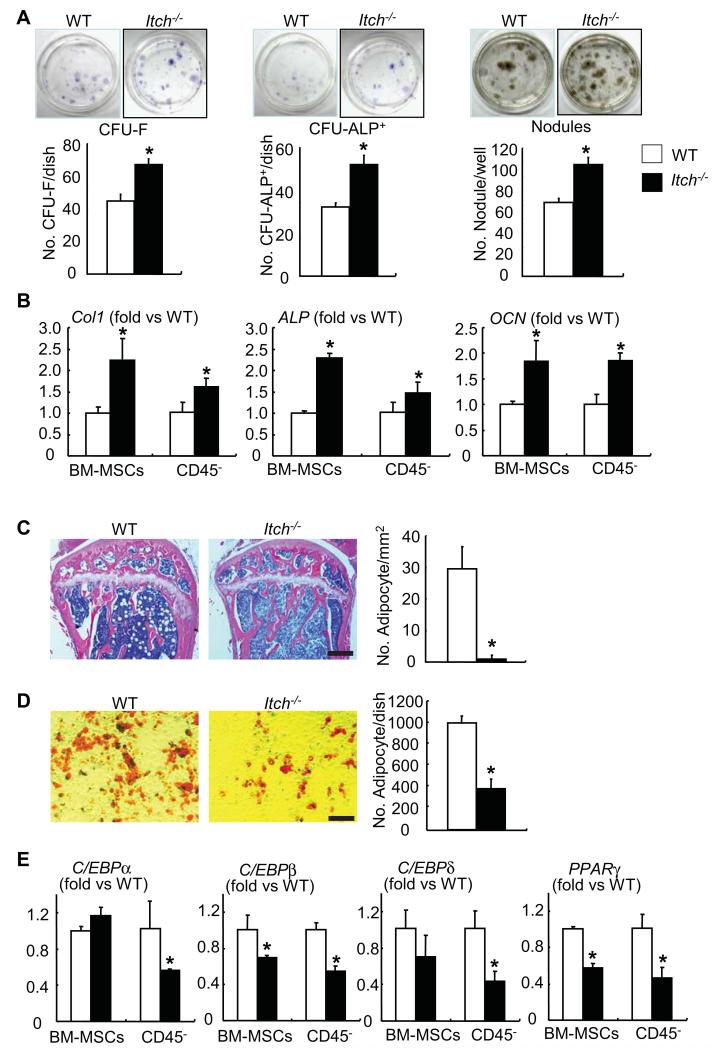
To determine if increased osteoblastic bone formation in Itch−/− mice is due to the alteration of osteoblast differentiation from bone marrow mesenchymal progenitor cells (BM-MPCs), we first examined if Itch−/− bone marrow cells could give rise more mesenchymal colonies when they are cultured in basal medium and more ALP+ colonies when they are cultured in the osteoblast inducing medium. Itch−/− cells formed significantly increased number of CFU-F and CFU-ALP+ colonies compared to WT cells (CFU-F colony No./well: 79±4 vs 52±3; CFU-ALP+ colony No./well: 63±5 vs 38±2). To examine osteoblast function, we examined mineralized nodule formation using Itch−/− BM-MPCs. Itch−/− BM-MPCsformed more mineralized nodules than those from WT littermates (Mineralized nodule No./well: 105±7 vs 68±8)(Figure 2A). Consistently, the expression levels of osteoblast marker genes, Col1, ALP and OCN, were all increased in BM-MPCs and CD45− MPC-enriched cells isolated from Itch−/− mice (Figure 2B).
Figure 2. Itch−/− bone marrow mesenchymal progenitors have increased osteoblast differentiation and decreased adipocyte differentiation.
(A) Bone marrow mesenchymal progenitor cells (BM-MPCs) from Itch−/− mice and WT littermates were cultured in the osteoblast-inducing medium for 7-21 days. (A) Representative image of CFU-F, CFU-ALP+ colonies or mineralized nodules (upper panels) and the number of CFU-F and CFU-ALP+ colonies or mineralized nodules (lower panels). Values are mean ± of 3 dishes. (B) The expression levels of osteoblast marker genes were examined by qPCR in B-MPCs and CD45− MPC-enriched cells. (C) H&E-stained sections from tibial bone of 3-month-old Itch−/− mice and WT littermates were analyzed. The number of adipocytes/mm bone surface was counted. Bar = 250μm. Values are mean ± SD of 5 mice/group. (D) BM-MPCs were cultured in the adipocyte inducing medium for 21 days and the number of adipocytes was counted. Bar = 100μm. Values are mean ± SD of 3 dishes. (E) The expression levels of adipocyte master genes were examined by qPCR in BM-MPCs and CD45− MPC-enriched cells. Values are mean ± SD of 5 mice. *p<0.05 vs WT cells.
Bone marrow progenitor cells can differentiate into osteoblasts and adipocytes. These reciprocal differentiations often happen in bone disease, such as osteoporosis 17. The adipocyte differentiation was examined in Itch−/− mice. H&E-stained bone sections from 3-month-old Itch−/− mice and WT littermates showed numerous adipocytes in bone marrow of WT mice, however, few adipocytes were observed in Itch−/− mice (Figure 2C). BM-MPCs were cultured and induced into adipocytes using the adipocyte inducing medium. Cells from Itch−/− mice formed fewer adipocytes than WT cells (Oil red O+ cells No./well: 390±83 vs 1005±71)(Figure 2D). Similarly, the expression levels of adipocyte marker genes, C/EBPα, C/EBPβ, C/EBPδ and PPARγ were all decreased in BM-MPCs and CD45− MPC-enriched cells isolated from Itch−/− mice compared to WT littermates. (Figure 2E), indicating decreased adipocyte differentiation in cells from Itch−/− mice.
Itch−/− mesenchymal progenitor cells have increased bone formation potential in recipient mice
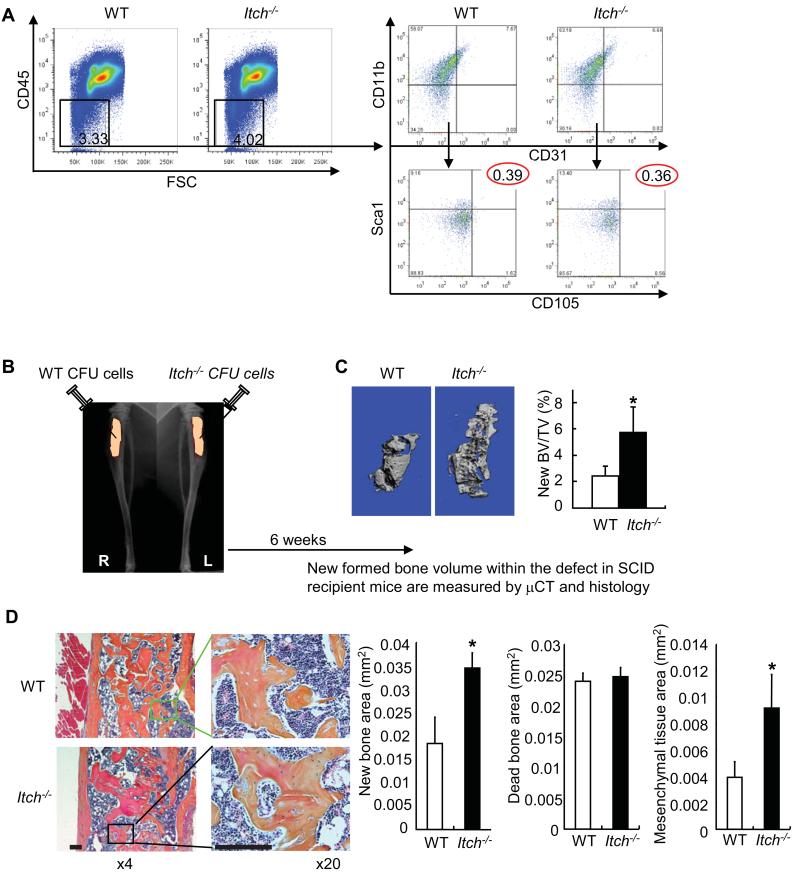
To elucidate whether increased osteoblast differentiation in Itch−/− cells is due to increased number of mesenchymal stem cells, we examine the frequency of cells that express surface markers for mesenchymal stem cells. Although there are no standard mesenchymal stem cell surface markers, murine mesenchymal stem/progenitor cells have been identified as CD45−/CD11b−/CD31−/CD105+/Sca1+ cells 21. We assessed the percentage of CD45−/CD11b−/CD31−/CD105+/Sca1+ cells in bone marrow cells from Itch−/− mice and WT littermates by FACS analysis. We first demonstrated that bone marrow cells from Itch−/− mice and WT littermates have a similar percentage of CD45− cells, which are gated off CD45+ hematopoietic lineage cells and contain enriched MPCs 18. We then indicated that they have same percentage of CD11b−/CD31−/CD105+/Sca1+ cells, cells that are often used as mesenchymal progenitor cells 22(Figure 3A). To directly test the effect of Itch depletion on osteoblast differentiation potential in the absence of its effect on other cell types, we implanted Itch−/− or WT CFU cells which are composed of MPCs and decalcified bone matrix into tibial defects of SCID mice (Figure 3B). Itch−/− cells formed significantly more new bone in defects of recipient mice compared with WT cells examined by μCT (new bone BV/TV: 5.8±1.9 vs 2.4±0.7%)(Figure 3C) and histomorphemetic analysis in H&E stained sections new bone area: 0.035±0.003 vs 0.018±0.006 mm2) and mesenchymal tissue area: 0.009±0.003 vs 0.004±0.001 mm2) (Figure 3D). Thus MPCs from Itch−/− mice have great bone formation potential in vivo.
Figure 3. Mesenchymal colony cells from Itch−/− mice form increased new bones in recipient mice.
(A) The % of non-hematopoietic lineage cells (CD45−) and the % cells that express MPC surface markers (CD11b−/CD31−/Sca1+/CD105+) in primary bone marrow cells by FACS analysis. (B) CFU cells from Itch−/− and WT littermates were implanted into tibial defects of SCID mice with decalcified bone matrix as scaffold. Mice were sacrificed 6 weeks post-implantation. (C) μCT images and μCT data of the % of new formed bone volume vs total tissue volume. (D) Histology and histomorphometric data of the area of new bone, dead bone and mesenchymal tissue containing spindle-shaped fibroblasts observed in decalcified H&E-stained sections of the bones Bar = 125μm. Values are mean ± SD of 5 mice. *p<0.05 vs WT cells.
Decreased ubiquitination and proteasome degradation of JunB protein in the absence of Itch
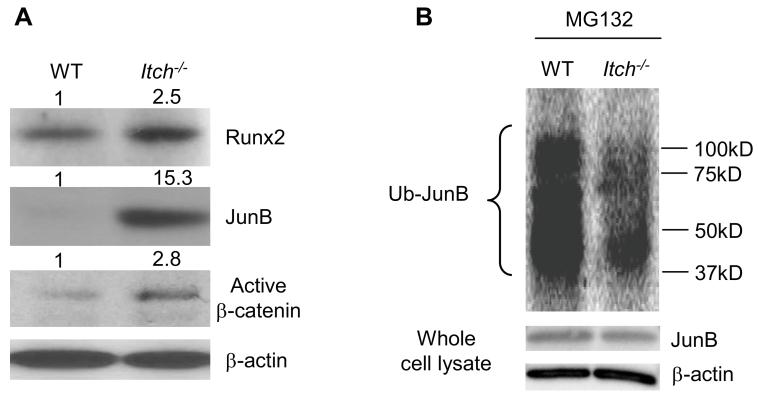
Itch functions as E3 ligase to promote ubiquitination of targeting proteins. JunB and Runx2 are two positive osteoblast regulators which are regulated partially through ubiquitination and proteasome degradation in osteoblasts 6, 23. In vitro studies reported the over-expression of E3 ligase Smurf1 or Wwp1 promote the ubiquitination and proteasome degradation of Runx2 protein in osteoblast cell lines 23, 24. A recent report indicated that disheveled protein, a negative regular of Wnt/β-catenin signaling pathway is regulated by Itch in HEK293T cells 25. We examined the expression levels of Runx2, JunB and activated form of β-catenin to determine which one of them is a potential endogenous target of Itch in MPCs to account for altered osteoblast differentiation. We used the expression level of activated form of β-catenin as readout for disheveled protein. Interestingly, we found dramatically increased JunB, moderate increased activated form of β-catenin, and slightly increased Runx2 in Itch−/− BM-MPCs (Figure 4A). These data indicate that under condition where Itch−/− cells have normal proteasome function, the degradation of JunB and perhaps disheveled protein was reduced. Because of markedly accumulated JunB, we decided to determine whether the increased JunB protein in Itch−/− BM-MPCs is due to abnormal ubiquitination by comparing the amount of ubiquitinated JunB protein between Itch−/− BM-MPCs and WT cells in the presence of proteasome inhibitor MG132. Itch−/− BM-MPCs had a significantly reduced JunB ubiquitination with a similar level of total JunB protein compared to WT cells (Figure 4B).
Figure 4. Decreased JunB ubiquitination in Itch−/− bone marrow mesenchymal progenitors.
(A) Protein expression levels of Runx2, JunB and active β-catenin proteins in BM-MPCs from WT and Itch−/− mice by Western blot analysis. The fold changes were calculated from the intensity of bands on Western blot image using Scion Image Beta 4.02 (Scion Corporation, NIH). (B) Itch−/− and WT BM-MPCs were treated with MG132 for 4 hrs and whole cell lysates were incubated with UbiQapture-Q matrix to pull down ubiquitinated proteins. Ubiquitinated proteins were blotted with anti-JunB antibody. Expression level of JunB and β-actin was measured in the same whole cell lysates.
Itch-inhibited osteoblast differentiation requires the E3 ligase activity
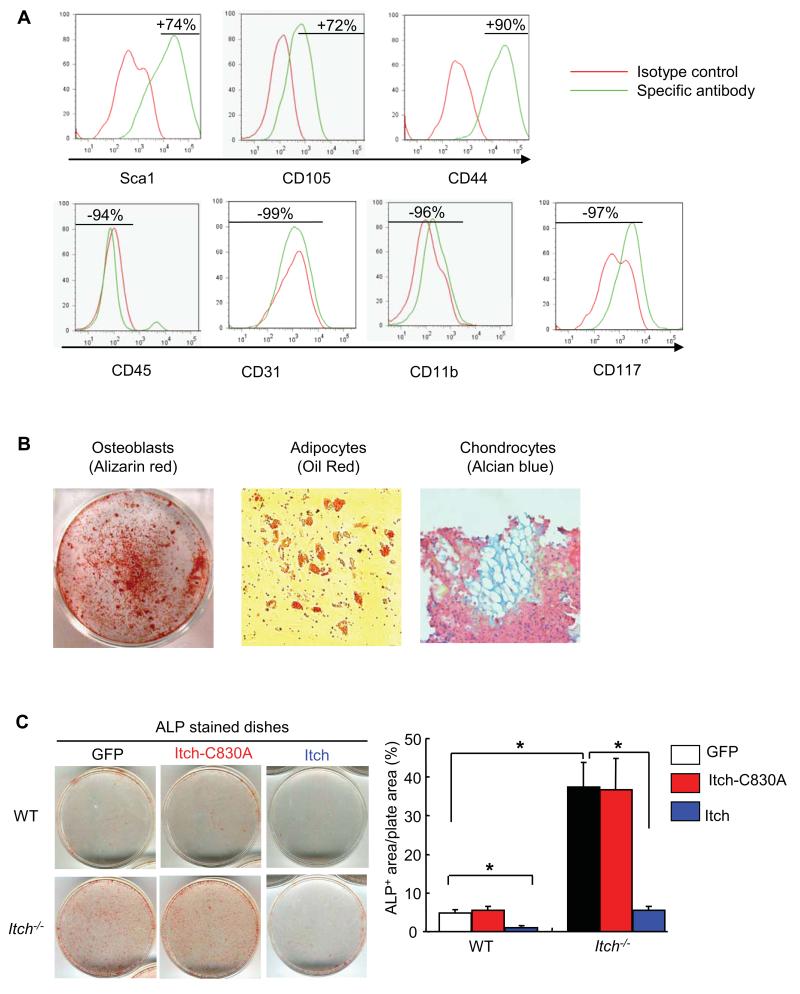
Ubiquitin E3 ligases promote ubiquitination of targeting proteins by facilitating the transfer ubiquitin moiety from E2 protein substrate via its ligase activity 1. To elucidate whether Itch ligase activity is required for regulating osteoblast differentiation by Itch, we need to demonstration the WT Itch, but not ligase inactive Itch mutant (Itch-C830A), could rescue the osteoblast defect of Itch−/− cells. Because it is very difficult to infect bone marrow progenitor cells, we used bone-derived MPCs (B-MPCs) according to a recent published protocol with minor modification 19. We first characterized immunophenotype of B-MPCs by FACS (Figure 5A) and confirmed that these cells had capacity to differentiate to osteoblast, adipocytes and chondrocyte lineages (Figure 5B). We then infected B-MPCs with retroviral supernatant containing GFP, Itch-C830A or WT Itch virus for 4 days, changed to osteoblast-inducing medium for another 5 days, and examined the ALP+ area as an indicator of osteoblast differentiation. Similar to BM-MPCs, Itch−/− B-MPCs gave rise to more ALP+ osteoblasts than WT cells. Increased ALP+ area in Itch−/− cells was completely reversed in cells that were infected by WT Itch virus, but not in cells that were infected by Itch-C830A virus. Consistently, WT Itch virus reduced ALP+ area in WT cells while Itch-C830A virus had no effect (ALP+ area/plate area: 6.5±1 in GFP- vs 0.5±0.1 in WT Itch- vs 6±0.8 % in Itch-C830A-infected WT cells)(Figure 5C).
Figure 5. Itch ligase activity is required for increased osteoblast differentiation of Itch−/− cells.
The bone-derived cells from WT mice were cultured in αMEM medium and passaged for 3 times as bone-derived mesenchymal progenitor cells (B-MPCs). (A) Expression profile of surface markers for mesenchymal stem cells in B-MPCs was examined by FACS. MPC surface markers are CD45−/Sca1+/CD105+/CD44+/CD31−/CD11b−/CD117−. (B) The differentiation potential to osteoblasts, adipocytes and chondrocytes were examined by culturing cells in the appropriate inducing media for 2-4 weeks. (C) Bone-MPCs were infected with retroviral supernatant containing GFP, Itch-C830A mutant or WT Itch virus. Cells were cultured in the osteoblast-inducing medium for 5 days and stained for ALP activity. The % of ALP+ area/total plate area was measured. Values are mean ± SD of 3 plates. *P<0.05 vs different groups as indicated.
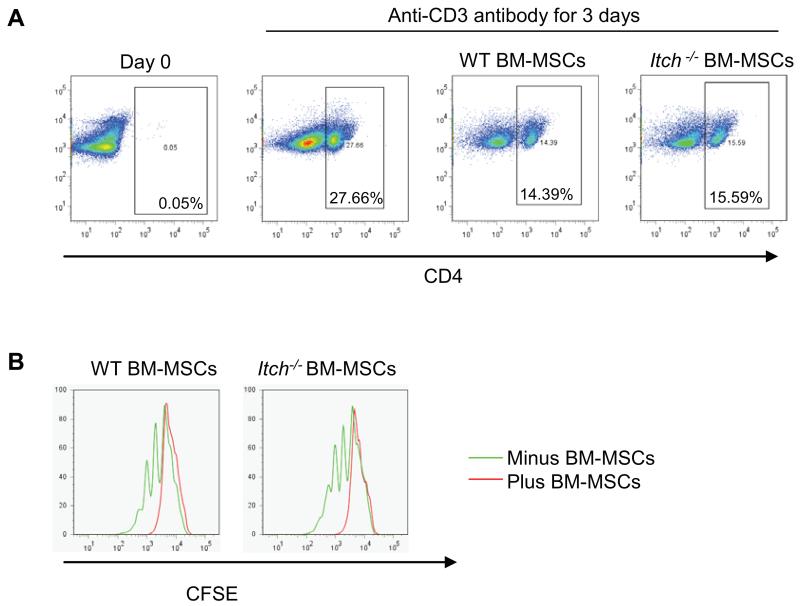
As mesenchymal stem cells are known to have a role in immune modulation 26 and Itch−/− mice have impaired T cell function 10 we performed a T cell proliferation assay by co-culturing T cells with or without addition of BM-MPCs from Itch−/− mice or WT littermates. Results showed both WT and Itch−/− BM-MPCs could decrease the number (Figure 6A) and division (Figure 6B) of T cells at a compatible level, indicating that Itch−/− BM-MPCs have a similar immune modulation effect as WT cells.
Figure 6. Itch−/− mesenchymal progenitors have a comparable inhibitory effect on T cell proliferation.
WT splenocytes were labeled with CFSE. Labeled splenocytes were then co-cultured with B-MPCs from Itch−/− mice and WT littermates in the presence of anti-CD3 antibody for 3 days. Cells were stained with anti-CD4 antibody. The percentage of CD4+ T cells (A) and distribution of CFSE-labeled CD4+ T cells (B) were determined by FACS analysis.
DISCUSSION
In this study, we used global Itch knockout mice and demonstrated that Itch−/− mice have high bone volumes with increased osteoblast-mediated bone formation. Itch depletion does not affect the number of cells that express mesenchymal stem cell markers within bone marrow. However, mesenchymal progenitor cells (MPCs) from bone marrow Itch−/− mice have increased osteoblast differentiation in vitro and new bone formation in vivo. Itch−/− MPCs expressed very high levels of osteoblast positive regulator JunB with significantly reduced amount of ubiquitinated JunB protein. Furthermore, increased osteoblast differentiation of Itch−/− MPCs requires Itch ligase activity. Thus, Itch is a new E3 ligase that negatively regulates osteoblast function at the progenitor levels.
Itch belongs to the family of WW domain-containing ubiquitin ligases. Among them, Smurf1 and Wwp1 have been reported to have negative effects on osteoblast function. Smurf1 promotes the degradation of Smad1/5 3, Runx2 23, JunB 6 and MEKK2 27 proteins. Smurf1−/− mice develop age-related bone loss. Wwp1 plays an important role in chronic inflammation-induced osteoblast inhibition and regulates JunB degradation 18. How does Itch deficiency-associated bone and osteoblast defects differ from these already described bone abnormalities in Smurf1−/− or Wwp1−/− mice? First, increased bone volume occurs at a young age (1-month-old) in Itch−/− mice before skeleton maturation while Smurf1−/− mice have detectable increased bone mass until 6-months of age 6. Secondly, Itch−/− mice develop multiple- organ inflammation with age, a phenotype similar to patients with the Itch gene mutation 9 while Smurf1−/− or Wwp1−/− mice have relatively normal immune system. Finally, Itch−/− mice have increased osteoclast formation while Smurf1−/− or Wwp1−/− mice have normal osteoclastogenesis. Thus Itch likely regulates cell function at different stages or/and in different cell types from Smurf1 and Wwp1.
Like other members of WW domain-containing ubiquitin ligases, Itch regulates the stability of multiple proteins via ubiquitination and subsequent proteasomal or lysosomal degradation 28. Among multiple Itch target proteins, we found that JunB is a major endogenous substrate of Itch in BM-MPCs. JunB is a positive regulator of osteoblast differentiation. Elevated total JunB protein when the proteasome function is intact and decreased amount of ubiquitinated JunB when the proteasome function is blocked in Itch−/− BM-MPCs indicate that JunB is a major endogenous substrate of Itch in these cells. Previous studies propose that ubiquitination and proteasome degradation is one of regulatory mechanisms of Runx2 29. However, we found that Runx2 protein levels are only slightly changed, suggesting that Runx2 is not the major target of Itch in BM-MPCs. JunB is a member of AP-1 transcription factor. We reported that abnormal JunB degradation is responsible for the negative effect of Smurf1 on osteoblast proliferation and differentiation Smurf1−/− BM-MPCs express elevated levels of JunB protein and JunB depletion abolishes increased osteoblast marker gene expression in Smurf1−/− BM-MPCs 6. Compared Smurf1−/− BM-MPCs, Itch−/− cells had much higher level of JunB protein and osteoblast differentiation potential. Itch−/− mice develop high bone mass at young age (1-month-old) while Smurf1−/− mice have increased bone volume when they become at least 6-8-month-old. Thus, Itch may be a stronger osteoblast negative regulator than Smurf1.
Itch deficiency causes progressive autoimmune diseases 8, 9 and increased osteoclastogenesis 30. We found that Itch−/− mice at a young age (1-month-old) have high bone mass while at an old age (1-year-old) they develop osteoporosis. This is not due to changes of osteoblast and osteoclast differentiation potential of Itch−/− cells at different ages because increased osteoblast and osteoclast numbers were detected in young and old Itch−/− mice. It is possible that the aging process potentates the catabolic effect of Itch depletion on osteoclast-mediated bone resorption, which overrides the anabolic effect of Itch depletion on osteoblasts. Because the potential adverse effects of Itch inhibition on immune system and osteoclasts, Itch inhibitors may be more suitable for local administration in a short duration to treat patients with local bone damage. Administration of Itch depleted mesenchymal stem and progenitor cells may be used in facilitating bone repairs given the fact that Itch−/− MPCs formed much more new bone in bone defects in recipient mice, and Itch is stronger osteoblast inhibitor than Smurf1 and Wwp1 based on bone phenotype analysis of knockout mice6. Another use of Itch−/− mesenchymal stem and progenitor cells is to search for new potential bone anabolic proteins that are also endogenous substrates of Itch by comparing proteomic information among Itch−/− cells, Itch−/− cells infected with WT Itch, and Itch−/− cells infected with ligase-inactive mutant of Itch mutant.
In summary, we found that ubiquitin E3 ligase Itch is a negative regulator of osteoblast differentiation from mesenchymal stem and progenitor cells. We found increased osteoblast numbers and bone formation rate in Itch−/− mice. Itch−/− bone marrow progenitor cells formed more mesenchymal colonies that have increased capacity to differentiate to osteoblasts. More importantly, Itch−/− mesenchymal colony cells formed much more new bone in recipient mice, indicating that Itch depletion has a strong positive effect on osteoblast differentiation in a cell autonomous fashion. Thus, Itch is a potential new target for development of bone anabolic drug, which could accelerate local bone repair in patients with bone disorders such as bone fracture and defects.
Acknowledgments
The authors thank Dr. Lydia E. Matesic (University of South Carolina) for providing Itch−/− mice, Dr. Abbott DW (Case Western Reserve University School of Medicine) for Flag-Itch and Itch-C830A mutant expression vectors and Dr. Igor Kuzin (University of Rochester) for helping T cell proliferation test. This work was supported by research grants from National Institute of Health PHS awards (AR48697 to L. Xing). MicroCT was supported by P30AR0613007 to Edward M. Schwarz.
This work was supported by Grants R01 AR48697 to LX from the National Institute of Health.
Footnotes
Author Contribution:
Zhang, Hengwei: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
Xing, Lianping: conception and design, financial support, data analysis and interpretation manuscript writing, final approval of manuscript
No Disclosure of Potential Conflicts of Interest
REFERENCES
- 1.Wullaert A, Heyninck K, Janssens S, et al. Ubiquitin: tool and target for intracellular NF-kappaB inhibitors. Trends in immunology. 2006;27:533–540. doi: 10.1016/j.it.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Durrington HJ, Upton PD, Hoer S, et al. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. The Journal of biological chemistry. 2010;285:37641–37649. doi: 10.1074/jbc.M110.132415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo R, Yamashita M, Zhang Q, et al. Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins. The Journal of biological chemistry. 2008;283:23084–23092. doi: 10.1074/jbc.M709848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneki H, Guo R, Chen D, et al. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. The Journal of biological chemistry. 2006;281:4326–4333. doi: 10.1074/jbc.M509430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Q, Chen D, Zuscik MJ, et al. Overexpression of Smurf2 stimulates endochondral ossification through upregulation of beta-catenin. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2008;23:552–563. doi: 10.1359/JBMR.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L, Huang J, Guo R, et al. Smurf1 inhibits mesenchymal stem cell proliferation and differentiation into osteoblasts through JunB degradation. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25:1246–1256. doi: 10.1002/jbmr.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou W, Chen X, Shim JH, et al. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nature cell biology. 2011;13:59–65. doi: 10.1038/ncb2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry WL, Hustad CM, Swing DA, et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nature genetics. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 9.Lohr NJ, Molleston JP, Strauss KA, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet. 86:447–453. doi: 10.1016/j.ajhg.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang D, Elly C, Gao B, et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nature immunology. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- 11.Gao M, Labuda T, Xia Y, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 12.Tao M, Scacheri PC, Marinis JM, et al. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Current biology : CB. 2009;19:1255–1263. doi: 10.1016/j.cub.2009.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shembade N, Harhaj NS, Parvatiyar K, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nature immunology. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 14.Shembade N, Parvatiyar K, Harhaj NS, et al. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. The EMBO journal. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita T, Yao Z, Li F, et al. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. The Journal of biological chemistry. 2007;282:18245–18253. doi: 10.1074/jbc.M610701200. [DOI] [PubMed] [Google Scholar]
- 16.Darzynkiewick, Roederer, Tanke . Cytometry. 4th Edition 2004. New Developments; p. 75. [Google Scholar]
- 17.Zhang HW, Ding J, Jin JL, et al. Defects in mesenchymal stem cell self-renewal and cell fate determination lead to an osteopenic phenotype in Bmi-1 null mice. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2010;25:640–652. doi: 10.1359/jbmr.090812. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Huang J, Zhang H, et al. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells. 2011;29:1601–1610. doi: 10.1002/stem.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H, Guo ZK, Jiang XX, et al. A protocol for isolation and culture of mesenchymal stem cells from mouse compact bone. Nature protocols. 2010;5:550–560. doi: 10.1038/nprot.2009.238. [DOI] [PubMed] [Google Scholar]
- 20.Zappia E, Casazza S, Pedemonte E, et al. Mesenchymal stem cells ameliorate experimental autoimmune encephalomyelitis inducing T-cell anergy. Blood. 2005;106:1755–1761. doi: 10.1182/blood-2005-04-1496. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Qu J, Cao L, et al. Mesenchymal stem cell-based angiopoietin-1 gene therapy for acute lung injury induced by lipopolysaccharide in mice. The Journal of pathology. 2008;214:472–481. doi: 10.1002/path.2302. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto N, Akamatsu H, Hasegawa S, et al. Isolation of multipotent stem cells from mouse adipose tissue. Journal of dermatological science. 2007;48:43–52. doi: 10.1016/j.jdermsci.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M, Qiao M, Oyajobi BO, et al. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. The Journal of biological chemistry. 2003;278:27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- 24.Jones DC, Wein MN, Oukka M, et al. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 25.Wei W, Li M, Wang J, et al. The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol Cell Biol. 2012;32:3903–3912. doi: 10.1128/MCB.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartholomew A, Sturgeon C, Siatskas M, et al. Mesenchymal stem cells suppress lymphocyte proliferation in vitro and prolong skin graft survival in vivo. Experimental hematology. 2002;30:42–48. doi: 10.1016/s0301-472x(01)00769-x. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita M, Ying SX, Zhang GM, et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heissmeyer V, Rao A. Itching to end NF-kappaB. Nature immunology. 2008;9:227–229. doi: 10.1038/ni0308-227. [DOI] [PubMed] [Google Scholar]
- 29.Xing L, Zhang M, Chen D. Smurf control in bone cells. Journal of cellular biochemistry. 2010;110:554–563. doi: 10.1002/jcb.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Wu C, Matesic L, et al. Itch, an E3 ligase, negatively regulates osteoclastogenesis by promoting deubiquitination of TRAF6 through interaction with the deubiquitinating enzyme, cylindromatosis. ASBMR Annual meeting. 2010 Abstract 1081. [Google Scholar]