Abstract
We show that the difficulties in imaging the dynamics of protein expression in live bacterial cells can be overcome using fluorescent sensors based on Spinach, an RNA that activates the fluorescence of a small-molecule fluorophore. These RNAs selectively bind target proteins, and exhibit fluorescence increases that enable protein expression to be imaged in living cells. These sensors provide a general strategy to image protein expression in single bacteria in real-time.
Protein levels are regulated by complex patterns of synthesis and degradation that may be lost in overexpressed tagged proteins. Therefore, imaging endogenous proteins has the potential to provide insights into physiologic expression patterns.
We recently described a strategy to image metabolites using genetically encoded fluorescent sensors composed of RNA1. This method involves fusing RNA aptamers that bind metabolites to Spinach2, a 98-nt RNA that switches on the fluorescence of 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), an otherwise nonfluorescent small molecule. The metabolite-binding aptamer is fused via a stem required for Spinach fluorescence. The stem is does not form a stable duplex at the imaging temperature. Most aptamers are unstructured before binding their cognate ligand3. After metabolite binding, the aptamer folds, bringing the strands of the stem in proximity, which results in a Spinach structure that can bind DFHBI. The stem sequence that connects the target-binding aptamer to Spinach functions as a “transducer” (Fig. 1a). This transducer module transmits the metabolite-binding event to a fluorescence readout.
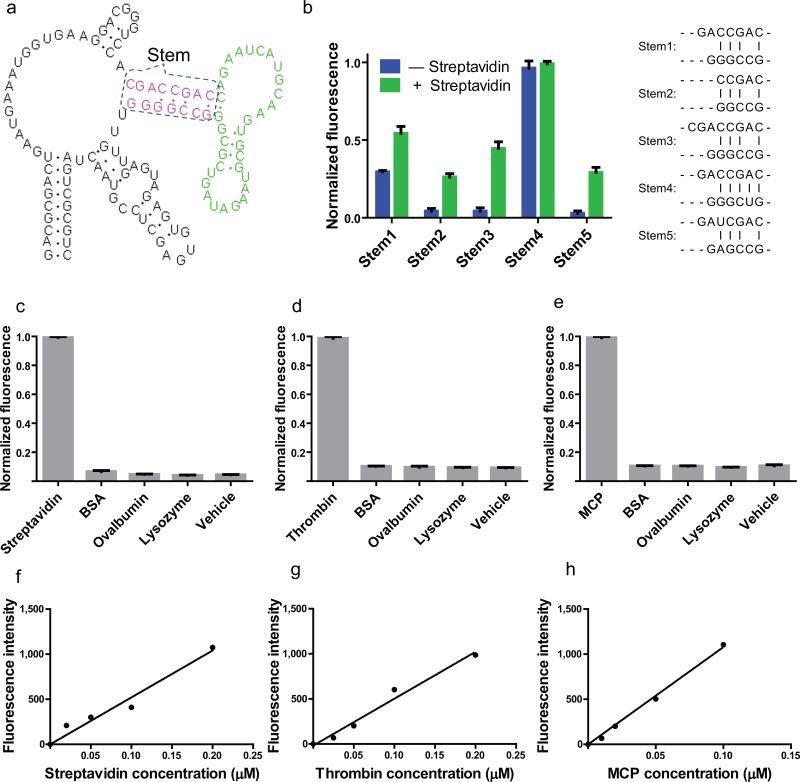
Figure 1. Sensitive and specific detection of proteins in vitro using Spinach-based sensors.
(a) The secondary structure and modular design of streptavidin sensors. The three modular components of the streptavidin sensor are depicted. The recognition module (green) constitutes an aptamer that binds to streptavidin. The transducer module (red) comprises two strands which form a weakly base paired stem. Folding of the recognition domain provides additional stability that facilitates the hybridization of the stem region in the recognition module. The Spinach module (black) binds to and activates the fluorescence of DFHBI, but only when the transducer module forms a stem.
(b) Optimization of stem transducer modules for streptavidin sensors. The streptavidin aptamer was fused to Spinach by one of five different transducer modules. These transducer modules contained different lengths and combination of G-C, A-U and mismatched base pairs, and were chosen because they were predicted to have a very low probability of duplex formation using the prediction software Mfold. Streptavidin sensors containing different stems (stem 1-5) were incubated with 10 μM DFHBI, 0.2 μM RNA in the presence or absence of 100 μg/ml (1.7 μM) streptavidin, and fluorescence emission was measured. The optimal transducer module (stem 3) was chosen because in the context of the sensor it displayed low background fluorescence, with a 10.3-fold increase in fluorescence signal upon incubation with streptavidin. The experiment was replicated three times, an average fluorescence signal and SEM were calculated and show in bar graph.
(c) Emission spectra of the RNA sensor for streptavidin in the presence or absence of streptavidin. Spectra were collected using 0.2 μM RNA, 10 μM DFHBI and 100 μg/ml (1.7 μM) streptavidin. Fluorescence signal is negligible in the absence of streptavidin and increases 10.3-fold in the presence of streptavidin.
(d) Emission spectra of the RNA sensor for human thrombin in the presence or absence of thrombin. Spectra were collected using 0.2 μM RNA, 10 μM DFHBI and 40 μg/mL (1.0 μM) thrombin. Fluorescence signal is negligible in the absence of thrombin and increases 6.9-fold in the presence of thrombin.
(e) Emission spectra of the RNA sensor for MS2 coat protein (MCP) in the presence or absence of MS2. Spectra were collected using 0.5 μM RNA, 10 μM DFHBI and 155 μg/ml (4 μM) MCP. Fluorescence signal is negligible in the absence of MS2 and increases 41.7-fold in the presence of MCP.
(f) Selectivity of streptavidin sensor. 0.2 μM RNA and 10 μM DFHBI were incubated with 0.1 mg/ml (2 μM) streptavidin or 2 μM competing proteins and assayed for fluorescence emission at 500 nm. Only baseline fluorescence is seen in the presence of competing proteins. The experiment was replicated three times, an average fluorescence signal and SEM were calculated and show in bar graph.
(g) Selectivity of human thrombin sensor. 0.2 μM RNA and 10 μM DFHBI were incubated with 0.04 mg/ml (2 μM) thrombin or 2 μM competing proteins and assayed for fluorescence emission at 500 nm. Only baseline fluorescence is seen in the presence of competing proteins. The experiment was replicated three times, an average fluorescence signal and SEM were calculated and show in bar graph.
(h) Selectivity of MS2 coat protein sensor. 0.2 μM RNA and 10 μM DFHBI were incubated with 0.078 mg/ml (2 μM) MS2 or 2 μM competing proteins and assayed for fluorescence emission at 500 nm. Only baseline fluorescence is seen in the presence of competing proteins. The experiment was replicated three times, an average fluorescence signal and SEM were calculated and show in bar graph.
(i) Dose-response curve for fluorescence detection of streptavidin by the RNA-based streptavidin sensor. The fluorescence signal was linear between 0–0.2 μM streptavidin.
(j) Dose-response curve for fluorescence detection of thrombin by the RNA-based thrombin sensor. The fluorescence signal was linear between 0–0.2 μM thrombin.
(k) Dose-response curve for fluorescence detection of MCP by the RNA-based MCP sensor. The fluorescence signal was linear between 0–0.1 μM MCP.
Because this method can detect a variety of small molecules in living cells1, we considered if this approach could be adapted to monitor protein levels in bacteria. To test this idea, we fused a streptavidin-binding aptamer4 to Spinach via different transducer stems, each with different degrees of thermodynamic stability. We tested the ability of each RNA to induce DFHBI fluorescence in a streptavidin-dependent manner (Fig. 1b). Several RNAs were streptavidin sensors based on low fluorescence in the absence of streptavidin and the increase in fluorescence upon streptavidin binding. The optimal sensor exhibited a 10.3-fold increase in fluorescence (Fig. 1c, Supplementary Fig. 1a).
We next fused a thrombin-binding aptamer5 to Spinach, and generated RNAs with transducer stems of various stabilities as described above. Using this approach, we identified a sensor that exhibited 6.9-fold increase following addition of 1 μM thrombin (Fig. 1d, Supplementary Fig. 1b).
We next generated a sensor for the MS2 phage coat protein (MCP). For these sensors we used the natural RNA-binding element of MCP, the MS2 phage coat protein-binding element (MS2E)6. We fused this stem-loop sequence to Spinach with various transducer stems, and generated a sensor that exhibited a 41.7-fold increase in fluorescence upon addition of 4 μM MCP (Fig. 1e, Supplementary Fig. 1c). Each of the sensors is only activated by its cognate protein, but not by other proteins (Fig. 1f,g,h, Supplementary Table 1).
Different transducer modules were found to be optimal for each sensor. The streptavidin sensor contained a transducer domain containing a stem with several mismatches (Fig. 1a; 1b). In the case of the thrombin sensor, a short transducer module with only 2 base pairs resulted in optimal protein-induced fluorescence (Supplementary Fig. 2a). The optimal transducer module for the MCP sensor was composed of a truncated stem found in the MS2E (Supplementary Fig. 2b). These data suggest that different transducer domains should be tested to optimize sensor function.
We next asked if these sensors quantitatively measure protein concentration in vitro. The streptavidin, human thrombin, and MCP sensors all gave linear increases in fluorescence with increasing concentrations of each protein (Fig. 1i,j,k). For example, the thrombin sensor detects human thrombin present in solutions at the nanomolar range (Fig. 1j). Together, these data indicate that sensor fluorescence can be used to measure protein concentrations in vitro.
We used RNA-based sensors to image protein expression in cells. First, we imaged the streptavidin sensor alone or when coexpressed with streptavidin in E. coli BL21 cells. A 10-fold increase in fluorescence signal was observed when streptavidin was coexpressed (Supplementary Fig. 3).
We next monitored MCP expression following infection with MS2 phage. We first tested whether MCP induces MCP sensor fluorescence in vitro on a timescale relevant for monitoring infection. Mixing purified MCP with MCP sensor lead to half the total fluorescence signal in one minute (Supplementary Fig. 4), indicating that the kinetics of fluorescence are rapid.
We monitored fluorescence in individual E. coli following treatment with MS2 phage. F-pilus-bearing E. coli expressing MCP sensor under the control of the IPTG-inducible T7 promoter were induced for 2 h, and then treated with phage at a multiplicity of infection (MOI) of 10. After a short lag of 2-4 minutes, fluorescence increased linearly over time and typically reached a platea ~25 minutes after infection (Fig. 3a). These rates correlate with previous bulk measurements of MCP after infection7.
We observed a linear correlation between MCP sensor signal and MCP levels measured by western blot, indicating that the MCP sensor is useful as direct readout of protein level (Supplementary Fig. 5). The MCP levels were compared to a standard curve of purified MCP (Supplementary Fig. 5a). Based on these measurements, approximately 6 × 104 molecules of MCP were synthesized per cell after infection, for a concentration of approximately 60 ± 4 μM. The linearity of the MCP sensor in sensing 0 to 60 μM of protein in a cell demonstrates that these sensors have the dynamic range necessary to sense most endogenous untagged E. coli proteins8. MCP sensor concentration was 120 ± 10 μM (Supplementary Fig. 5b), which is 2-fold higher than the determined maximal MCP concentration. Since the affinity of MCP to the MCP-binding element is approximately 200 pM6, each synthesized MCP is expected to be bound to a sensor RNA. Conceivably, sensors could be designed with an EC50 in the micromolar range, and therefore the target protein would exhibit a lower fractional occupancy.
We carried out infections under several conditions to ensure that signal increase was specific to MCP synthesis. The sensor fluorescence was blocked by protein synthesis inhibitors and was not seen using control sensors that do not bind MCP (Supplementary Fig. 6a). Taken together, the MCP sensor specifically detects endogenously synthesized MCP.
We next asked whether individual cells exhibited variability in MCP synthesis (Fig. 2b,c). Infections were carried out at a MOI of 0.1 to ensure that each cell was only infected with at most one MS2 phage (Supplementary Fig. 6b). Quantification of the pattern of MCP accumulation in 100 cells showed that 85% of cells exhibited nearly indistinguishable patterns of accumulation of MCP, with near maximal levels of MCP at 20 min. However, 11% of cells displayed significantly faster accumulation of MCP, while 4% exhibited markedly slower rates (Supplementary Fig. 7, Fig. 2c).
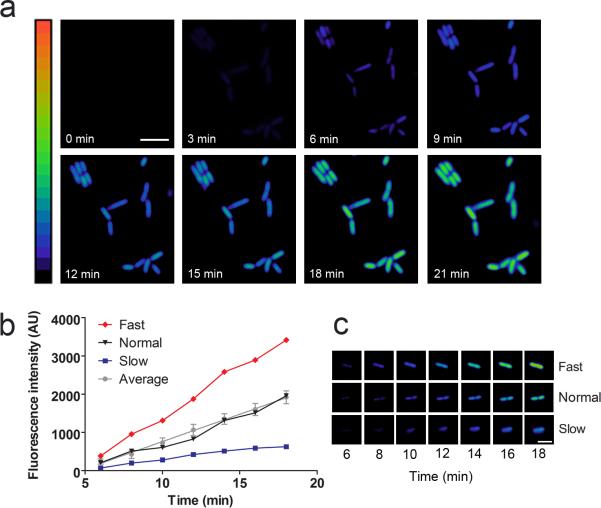
Figure 2. Visualization of MCP synthesis in individual cells after MS2 phage infection.
(a) Kinetics of MCP synthesis after viral infection. E. coli expressing the MCP sensor were infected with MS2 phage at an MOI of 10. MCP synthesis was monitored as an increase in fluorescence signal over time. Images are pseudocolored to show the fold increase in fluorescence at each time point following infection of MS2 phage. Color scale bar represents 0- to 20-fold changes in fluorescence signal. Scale bar, 5 μm.
(b) MCP synthesis kinetics vary in individual cells. E. coli were infected at an MOI of 0.1 and fluorescence was monitored over time. One hundred cells were quantified and an average rate of fluorescence increase was calculated. The average values and SEM are shown (gray line). Based on this average rate, cells were either categorized as Fast (red line), Normal (black line), or Slow (blue line).
(c) Representative examples of Fast and Slow cells. Fast and Slow cells accumulated MCP with a rate greater or less than two standard deviations from the mean rate, respectively. Time points following application of MS2 phage are indicated.
(d) Percentage of cells in fast, average, and low categories for MCP synthesis. The rate of fluorescence increase in 85% of cells was within two standard deviations of the mean value (gray), whereas 4% of cells were faster than average (red), and 11% of cells were slower than average (blue).
In order to confirm that the observed difference in MCP accumulation kinetics was not due to variability in sensor expression, we compared the expression levels of a control protein with the MCP sensor signal after infection. For these experiments, we expressed the blue fluorescent protein (BFP) and the MCP sensor from a single plasmid. In this case, both BFP and the MCP sensor were under the control of separate T7 promoters. After induction, we observed that BFP signal was highly uniform. In contrast, the MCP sensor fluorescence showed more variability than the BFP signal from the same cells (Supplementary Fig. 6b). As an additional control, BFP and Spinach were coexpressed. In this case, BFP and Spinach signals overlapped almost perfectly (Supplementary Fig. 6c), indicating that RNA stability does not play a role in the observed differences in signal from the MCP sensor. These results confirm that cell-to-cell variability exists in MCP synthesis that is independent of the expression level of the MCP sensor.
Unlike the genetically encoded protein-based sensors that are currently available, the sensors described here are composed of RNA and function by allosteric regulation of Spinach fluorescence. These sensors are readily expressed and highly stable in bacteria1, making them useful for probing bacterial signaling pathways. Because RNA aptamers can be readily generated that selectively bind to numerous types of proteins, including those that do not have RNA-binding domains or any known intrinsic RNA-binding capability9, the strategies described here should enable the design of sensors that detect a wide variety of proteins for diverse applications. Strategies to prevent instability of small RNAs10 will be needed for application in eukaryotic cells.
The cell-based approaches described here draw upon previous in vitro technologies for protein detection using aptamer beacons and allosteric ribozymes. Aptamer beacons are aptamers that undergo conformational changes upon protein binding11. Binding changes the distance between a fluorophore/quencher pair, altering the fluorescence signal. Similarly, allosteric ribozymes are activated by protein binding12, and are analogous to the Spinach-based sensors described here. In the case of Spinach-based sensors, protein binding is linked to a fluorescence readout, which makes these sensors useful for real-time imaging in living cells.
Imaging protein expression is important for optimizing production of recombinant proteins13. Optimizing protein yield involves screening mutant or genetically modified bacteria for increased protein expression14-16. However, isolating high-producing cells has been a challenge due to the inability to quantify protein expression in individual cells16. The genetically encoded sensors described a straightforward approach for identifying bacterial cells that exhibit optimized protein expression.
METHODS
Preparation of RNA sensors and mutants
Secondary structure prediction was performed using Mfold online software17. Mutated and truncated RNAs were created by using single stranded DNA templates (Integrated DNA Technologies) with the desired mutations or truncations and PCR amplifying these sequences to create double stranded DNA templates using primers which included a 5’ T7 promoter sequence. PCR products were then purified with PCR purification columns (Qiagen) and used as templates for in vitro T7 transcription reactions (Epicentre) as described previously1.
Linear dose-response curve measurements
Dose-response curves for each sensor in response to the target protein were determined by measuring the increase in fluorescence as a function of increasing target concentration in the presence of a fixed concentration of RNA sensor (200-400 nM) and a fixed concentration of fluorophore (10 μM). Fluorescence measurements were performed using a Perkin Elmer LS-55 fluorescence spectrometer using the following instrument parameters: excitation wavelength, 460 nm; emission wavelength, 501 nm; slit widths, 10 nm. Curves were determined using a linear regression analysis in GraphPad Prism 5 software.
Cloning sensors for expression in E. coli
The MCP and streptavidin sensors were PCR amplified with primers containing either EagI or SacII restriction sites on the 5’ or 3’ ends of the sensor sequence, respectively. The sensors were then cloned into a plasmid containing a chimera of the human tRNALys3 scaffold, which we previously used for Spinach and Spinach-based metabolite sensors, and which has previously been shown to stabilize heterologous expression of RNA aptamers in E. coli18. The entire tRNA-sensor construct was then PCR amplified with a forward primer containing a BglII site (italics) and a T7 promoter sequence (bold): 5’- CAG TAG AGA TCT TAA TAC GAC TCA CTA TAG GCT CGA GGC CCG GAT AGC TCA GTC GG -3’, and a reverse primer containing a BlpI site (italics): 5’- GAT CAG GCT CAG CTG GCG CCC GAA CAG GGAC-3’. This PCR product was then cloned into a pET28c vector directly upstream of the T7 terminator sequence with BglII and BlpI restriction sites.
Live cell imaging of the streptavidin sensor
BL21 E. coli cells were transformed to harbor either the streptavidin sensor alone in the context of a tRNA scaffold in pET28c under the control of a T7 promoter as previously described2 (pET38c-tRNA-Streptavidin sensor) or cotransformed to express both pET28c-tRNA-Streptavidin sensor and pET21a-Streptavidin-Alive19. Cells were grown in LB media supplemented with 50 μg/ml kanamycin and 100 μg/ml ampicillin and 50 μg/ml kanamycin, respectively. Sensor and protein synthesis were induced by addition of IPTG to 1 mM for 2 h. After induction, cells were adhered and imaged as described below.
Live cell imaging of MS2 coat protein synthesis
DH5α Turbo E. coli cells (New England Biolabs) were used for infection by MS2 phage and imaging because they harbor the F plasmid necessary for MS2 infection. Cells were transformed with 40 ng of plasmid DNA expressing the appropriate RNA aptamer in the context of a tRNA scaffold in pET28c under the control of a T7 promoter as previously described2. Additionally, since this strain of DH5α does not already contain a genomically incorporated T7 RNA polymerase, the streptavidin sensor plasmid was cotransformed with a plasmid harboring the T7 polymerase gene under control of a trc promoter20. Cells were grown overnight on LB media supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin to select for those cells that harbored both plasmids. Single colonies were then picked for inoculation in LB supplemented with100 μg/ml carbenicilin and 50 μg/ml kanamycin and grown at 37°C with shaking to OD600 = 0.4, at which point IPTG was added to a final concentration of 1 mM to induce T7 polymerase and MCP aptamer expression. Growth and induction with shaking was continued at 37°C for 2 hours. 100 μL of culture was then removed, spun down to pellet the culture, and resuspended in 2 mL of pH 6.0 M9 minimal media. A 200 μl aliquot of resuspended culture was then plated on PLL-coated 24-well glass-bottom dishes (MatTek) and incubated for 45 minutes at 37°C. Adherent cells were washed twice and then incubated with 50 μM DFHBI in pH 6.0 M9 minimal media for 1 hour at 25°C. Cells were then treated with MS2 phage (ATCC #15597-B1) with an MOI of either 10 or 0.1, as specified. The titer of infection-competent virus was determined by an agar-overlay plaque-formation assay as previously described21. Live fluorescence images were taken with a CoolSnap HQ2 CCD camera through a 60X oil objective mounted on a Nikon TE2000 microscope and analyzed with the NIS-Elements software. The filter set used was a sputter coated filter cube with excitation filter 470/40, dichroic mirror 495 (long pass) and emission filter 525/50 (Chroma Technology). The experiments in which BFP and the MCP sensor were coexpressed were carried out as described above, however, BFP and the MCP sensor were coexpressed from the pET-Duet vector (Novagen). For experiments where tetracycline was used to inhibit protein synthesis, cells were grown, induced, and adhered to slides as described above. Cells were incubated with 10 μg/ml tetracycline for 10 minutes before treatment with MS2 phage. Tetracycline remained in the media throughout the duration of the experiment.
Quantitation of MCP and MCP sensor concentrations
DH5α Turbo E. coli cells were infected with purified MS2 phage at an MOI of 10. At appropriate time points following infection, cells were measured for whole-cell fluorescence using a Tecan SafireII plate reader or collected and lysed by incubation at 95°C for 10 min in SDS sample buffer. For fluorescence measurements, background fluorescence for uninfected cells incubated with 50 μM DFHBI was subtracted from all time points. For analysis by western blot, 0.5 OD units of E. coli in sample buffer per time point was subjected to denaturing polyacrylamide gel electrophoresis. Samples were then transferred to PVDF membrane by electrophoresis. Western blot was carried out using a rabbit polyclonal antibody against MCP (Millipore). An anti-rabbit horse radish peroxidase-conjugated antibody was used to detect signal. Bands from western blot analysis were quantified using Image Lab software version 4.0 (Bio-Rad). Concentration of purified protein was determined by BCA analysis (Pierce). The data presented represent mean and s.e.m. from three independent replicates.
The following calculations were used to measure the approximate concentration of MCP molecules synthesized per cell. Based on quantification, a maximum of 1.3 μg MCP was produced per OD600 unit of infected cells. Use of the conversion 1 OD600 = 109 E. coli cells indicated that 1.3 × 10−15 g of MCP were produced per cell. The molecular weight of MCP is 12.5 kDa, indicating that 6.2 × 104 copies of MCP were synthesized per cell after MS2 infection. Using the general conversion of 1000 copies per cell equals 1 μM, the estimated maximal concentration of MCP produced per E. coli is 60 μM.
The same conversion was applied to determine the total MCP sensor concentration from Northern blot analysis to determine that roughly 120 μM sensor is present under these conditions. Northern blot analysis was carried out as previously described2, and probled using a 5’Cy3-labeled probe against the MCP sensor (5’- GGACCCGTCCTTCACCATTTCATTCAG-3’) (IDT DNA). Bands from northern blot analysis were quantified using Image Lab software version 4.0 (Bio-Rad). The data presented represent mean and s.e.m. from three independent replicates.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. S. Paige for useful comments and suggestions, and F. Dardel (Université Paris Descartes) for providing plasmids containing the tRNA scaffold sequence. This work was supported by NIH NIBIB EB010249 (S.R.J.).
Footnotes
AUTHOR CONTRIBUTIONS
W.S., R.L.S., and S.R.J. conceived and designed the experiments, W.S. and R.L.S. performed experiments and analyzed data, and W.S., R.L.S., and S.R.J. wrote the manuscript.
REFERENCES
- 1.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 4.Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White R, et al. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol Ther. 2001;4:567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- 6.Lowary PT, Uhlenbeck OC. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res. 1987;15:10483–10493. doi: 10.1093/nar/15.24.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viñuela E, Algranati ID, Ochoa S. Synthesis of virus-specific proteins in Escherichia coli infected with the RNA bacteriophage MS2. Eur J Biochem. 1967;1:3–11. doi: 10.1007/978-3-662-25813-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Ishihama Y, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 10.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamaguchi N, Ellington A, Stanton M. Aptamer beacons for the direct detection of proteins. Anal Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 12.Hartig JS, et al. Protein-dependent ribozymes report molecular interactions in real time. Nat Biotechnol. 2002;20:717–722. doi: 10.1038/nbt0702-717. [DOI] [PubMed] [Google Scholar]
- 13.Aucoin MG, et al. Identifying conditions for inducible protein production in E. coli: combining a fed-batch and multiple induction approach. Microb Cell Fact. 2006;5:27. doi: 10.1186/1475-2859-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 15.Huang CJ, Lin H, Yang X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol. 2012;39:383–399. doi: 10.1007/s10295-011-1082-9. [DOI] [PubMed] [Google Scholar]
- 16.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011;10:32. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 19.Howarth M, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosius J, Erfle M, Storella J. Spacing of the −10 and −35 regions in the tac promoter. Effect on its in vivo activity. J Biol Chem. 1985;260:3539–3541. [PubMed] [Google Scholar]
- 21.Adams MH. Bacteriophages. Interscience Publishers; New York: 1959. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.