Abstract
The complex nature of cell migration highlights the power and challenges of applying mathematics to biological studies. Mathematics may be used to create model equations that recapitulate migration, which can predict phenomena not easily uncovered by experiments or intuition alone. Alternatively, mathematics may be applied to interpreting complex data sets with better resolution—potentially empowering scientists to discern subtle patterns amid the noise and heterogeneity typical of migrating cells. Iteration between these two methods is necessary in order to reveal connections within the cell migration signaling network, as well as to understand the behavior that arises from those connections. Here, we review recent quantitative analysis and mathematical modeling approaches to the cell migration problem.
In parallel rather than in series: mathematics in experimental biological methods
Complex biochemical networks responsible for the process of cell migration in different cell types have been identified through traditional biological assays, such as Western blots and morphological studies. From the vantage point of this tremendous body of work, a current challenge is to understand how these many signaling components receive and coordinate signals to produce productive migration. Mathematics has often been viewed as an endpoint for experimental studies: experimentally measured biochemical constants for known interactions are incorporated into equations, which can then be tested for their ability to recapitulate observed behavior. Recent advances have, however, allowed the emergence of a complementary approach: mathematics may be used to enhance experimental resolution and analysis (Figure 1). While traditional biological assays will continue to reveal important facets of cell migration, cell migration is also a particularly suitable subject for mathematically fine-tuned experimental methods, for reasons which we discuss below.
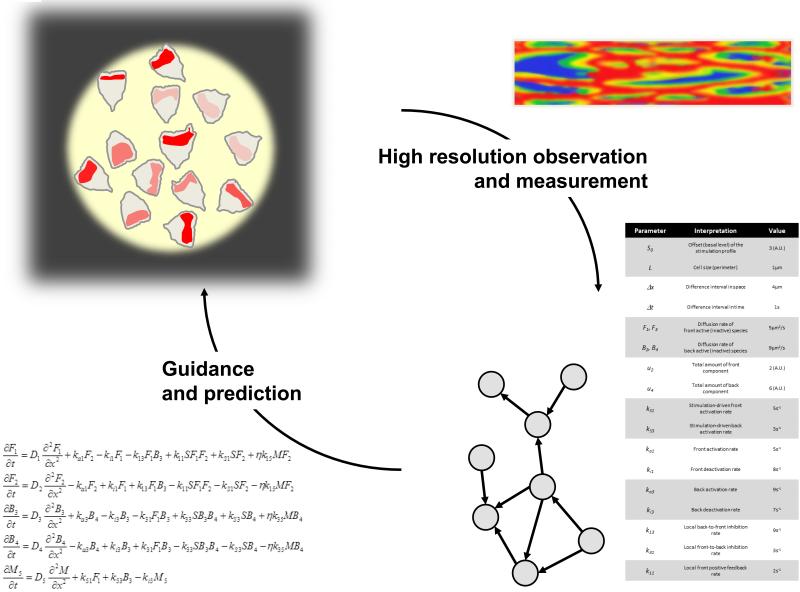
Figure 1.
Envisioning migration. Mathematically fine-tuned experimental analysis enhances the resolution with which behaviors can be observed, while mathematical models provide guidance and prediction of behaviors and networks.
Spatial coordination
The process of cell migration begins with polarization, in which cells generate a front and a back in order to achieve movement in one direction [1-4]. This spatial segregation of signaling components is critical for migration, but is difficult to analyze; traditional methods such as Western blots [5], light scattering [6], and standard flow cytometry cannot provide spatial information. Recent complementary approaches allow quantitative characterization of this spatial information. Machacek et al. performed cross-correlative analyses to track the activity of GTPases with respect to the protruding edge of a cell [7]; this allowed measurement of the distance from the cell edge at which GTPase activity peaked. Welf et al.’s cross-correlative analysis of PI3K signaling [8] showed that PI3K reinforces rather than produces protrusions in fibroblasts, while Galic et al. performed spatial cross-correlation analysis on cells adhered to surfaces patterned with nanocones to demonstrate that N-BAR proteins are directly recruited to the plasma membrane by membrane-curving forces [9]. Analysis of the spatial distributions of cytoskeletal readouts of polarizing primary neutrophils identified differential paths of information flow [10] and the insulation of the back signaling from the front signaling by the microtubules [11]. Quantifying spatial parameters allows trends in probe localization to be not only identified, but also tested for statistical significance, thus opening a new arena of study.
Meanwhile, clever image analysis has further been used to extract much more quantitative spatial information from the cell shape itself. Driscoll et al. used kymographs to study the evolution of cell shape, and found for Dictyostelium discoideum that cells change shape via traveling curvature waves, possibly due to actomyosin dynamics [12]. Barnhart et al. [13] tracked contours and created edge velocity maps of keratocytes on substrates of different adhesion strengths. They found that keratocyte speed and shape have a biphasic dependence on adhesion strength, and that adhesion strength (without long-term adaptation) is sufficient for switching the migration behavior of cells. These authors’ methods transform cell shape itself into a rich resource for the study of cell migration.
Timescales and cell-to-cell asynchrony
An additional difficulty in studying cell migration is achieving adequate temporal resolution, as the characteristic timescales of cell migration are very short. For example, neutrophils can rapidly transform extracellular cues into protrusive changes, creating actin ruffles within 20 seconds of initial chemoattractant exposure [14], and undergoing shape oscillations with a period of roughly 8 seconds [15]. This is in stark contrast to studies of other periodic behavior such as circadian clocks or stages of the cell cycle, where timescales may be on the order of hours to days. An offset of seconds between two migrating cells can be equivalent to a half-period shift. Readouts across a population of cells will thus be heterogeneous and hard to interpret in absolute time. Further, unlike cell cycle studies, in which cells may be synchronized with methods such as serum starvation, the protrusion-retraction cycles of separately migrating cells are not synchronized with one another. Traditional biochemical assays may demonstrate whether one protein activates, inhibits, or does not affect another protein. However, to further explore the signaling behavior and functional implications of a biochemical circuit, the temporal coordination of the components must be studied. The study of migrating cells thus requires high temporal resolution and resourceful computational methods that circumvent the difficulty of interpretation caused by cell-to-cell asynchrony within a population.
Recent studies have addressed the issue of asynchrony with an elegant computational multiplexing approach: the activity of a probe and the edge movement of the membrane are read out from the same cell, and the relative time and distance between probe activation and cell edge movement is recorded. This approach thus allows one readout from many individual cells to be pooled, and pools may be compared for multiple probes to uncover the coordination of the proteins represented by the probes. Using this approach, Machacek et al. uncovered the relative timing of GTPases within protrusions of fibroblasts [7], Tkachenko et al. demonstrated RhoGDI’s role in the temporal regulation of RhoA [16], and Ji et al. predicted intracellular forces from the F-actin network [17]. Meanwhile, Marco et al. [18] developed a math model to characterize the relations of several parameters of cell polarization, and then designed an experiment that tied in closely with their model. This model-directed experiment allowed extraction of different parameters, which are experimentally difficult to measure independently, and preserved context by allowing simultaneous measurement from the same timepoints and individual cells. Specialized application of mathematics can permit both computational alignment of asynchronous events and synchronous extraction of parameters, and is thus especially powerful for the study of cell migration.
Ab initio ad finem: the spectrum of uses for math modeling
Mathematical models vary widely in their levels of abstraction and biological detail [19]. At the one extreme are conceptual models, which may seek to identify minimal circuits and ab initio mechanisms underlying observed phenomena. At the other extreme are data-driven models, which incorporate experimental data in order to ask whether the resultant equations recapitulate, ad finem, experimentally observed behavior. What kind of models do we need to build to learn more about a behavior as complex as cell migration? Here, we argue that the path toward greater understanding of cell migration is not a straight shot through increasingly mechanistic territory.
Conceptual modeling is commonly used when a circuit is poorly characterized. However, conceptual models can also be applied to well characterized systems to replace the biological circuit’s details with simpler functional units–much like the creation of Thévenin equivalent circuits [20] for electronic circuits. Meanwhile, data-driven models are often applied to well-known systems to identify missing pieces. For example, in the case of bacterial chemotaxis, ultrasensitivity of the system could not be explained by known components, and data-driven modeling was used to demonstrate that the motor itself adapts at the level of switch component subunit clusters [21]. However, a data-driven approach may also be used to guide our knowledge of which pieces of a poorly-characterized system are most important to its operation. Iteration between conceptual and data-driven models can identify design principles and constraints which may guide future experimental and modeling endeavors in cell migration (Figure 1).
Bird’s eye view: conceptual modeling
Conceptual models can be used to capture the essence of what is currently known about a biological system. For instance, Ofer et al. [22] studied the simple migration system of keratocyte fragments, which lack cell bodies. The authors created force-balance equations to demonstrate the emergence of global shape and speed from underlying actin dynamics and membrane tension. This study elegantly highlighted the minimum requirements for coordinating retraction of the rear with protrusion at the front. Similarly, Neilson et al. [23] [24] studied pseudopod formation in Dictyostelium discoideum. The authors modified Meinhardt’s discrete model of chemotactic orientation [25] to demonstrate that a cyclical internal process can be used to recapitulate both pseudopod formation and orientation bias. This simple conceptual model suggested a pseudopod-centered mechanism of chemotaxis. Conceptual models can identify minimal circuits, and thus reveal general principles at the heart of more complex circuits.
Conceptual models for zooming out and narrowing in
Conceptual modeling can also be used to survey minimal circuits capable of creating observed phenomena, which in turn can guide future studies—that is, zooming out can allow the field to narrow in. For example, perfect or near-perfect adaptation enables cells to respond to the gradient, rather than the average value, of a signal [26]. Ma et al. explored this property not by modeling the known signaling components of these networks, but by generalizing chemotactic and other biological networks into abstract three-node networks, and performing a topology search over all of the possible relations of the three nodes [27]. This search revealed that only two network designs, the negative feedback loop and the incoherent feed forward loop, are capable of achieving perfect adaptation.
This general finding may then guide future mechanistic studies of cell migration. A recent cross-correlative study of GTPases, actin, phosphoinositides, and edge velocity by Kunida et al. indicated adaptation of Rac1 activity in response to PI3K inhibition [28]. To search for a source of this behavior, Kunida et al. looked to the abstract, conceptual search performed by Ma et al. for networks capable of adaptation. Of the two candidate topologies that Ma et al. identified, the negative feedback topology recapitulated Kunida et al.’s experimental findings. Kunida et al. thus searched for a negative regulator of actin and Rac1, and found that myosin light chain kinase acts as a node in the causative negative feedback loop for the observed adaptation. Although negative feedback between frontness signals such as Rac1 and backness signals such as myosin light chain kinase has long been established in migrating cells [5], the relevance of such feedback to adaptation is an unexplored area. Thus, mathematical modeling and constraints from an abstract topology search were used in this mechanistic study to find a new behavioral implication of a known biochemical link. More recent topology searches for networks capable of polarization [1] or for networks capable of front-back buffering [11] may provide similar guidance for future mechanistic efforts.
Data-driven models as predictors
Data-driven models may be used not only as final tests of understanding, but also as ways of picking up in silico where observation is limited in vitro. For instance, Shibata et al. [29] created kinetic equations to describe the reactions of phosphatidylinositol lipids in chemotactic cells. Their simulations and experiments both showed two types of behaviors for the localization of PtdIns(3,4,5)P3: traveling waves and the formation of transient domains. The authors explored ranges of variables in their simulations in a systematic manner that is not possible in vitro, and found that the traveling waves are induced by an instability of the stationary uniform state, while stochastic noise was important for transient domain-formation.
Similarly, Marée et al. [30] created a data-driven model to explore the effects of both biophysical and chemical feedback on cell polarization and motility. Their model suggested that cell shape is not just a downstream readout, but also feeds back by directly affecting the internal distribution of GTPases. Meanwhile, their model allowed them to test the effect of different values of phosphoinositide feedback; this allowed the authors to note that an intermediate level of phosphoinositide feedback creates normal migration. Lin et al. [31] expanded this model and predicted that a gradient of Rac activation can create a strong cell polarization response, that the timing of this polarization depended strongly on the gradient of Rac, and that antagonism between Rac and Rho could amplify polarization; in vitro creation of such a gradient with a rapamycin stimulation system confirmed their in silico prediction.
Finally, data-driven models have been used to predict not only outcomes, but also new hypotheses altogether. Wu et al. [32] created data-driven decision-tree models to analyze a ‘cue-signal-response’ data set from multipotent stromal cells. Similar to their work in fibroblasts [33, 34], the modeling classified responses to combinations of signals and conditions. Their decision trees revealed a non-intuitive prediction that decreasing ERK would promote cell migration, which they then confirmed in vitro.
Modeling new realms
How will we begin to combine the many models of different subprocesses of migration into a multiscale model? As the field progresses, previously coarse-grained models are becoming finer-grained as experimental findings provide more detailed metrics [4]. In addition, the field is growing wider with new models of subprocesses of migration that were not built in original models, including: social migration [35, 36], integrin-clustering [37], interactions with the extracellular matrix [38], haptotaxis [39, 40], and N-BAR domain proteins [41]. Iteration between conceptual and data-driven modeling will be required to navigate the future challenge of weaving together these subprocesses (some of which will be dependent on others).
Outlook
Cell migration is a multifaceted process, many pieces of which remain poorly elucidated. While the utility of mathematics in biology is often thought to be the creation of detailed models, the path to the elusive end-to-end, whole-cell model is not a straight line through increasingly mechanistic territory. Instead, the path to this distant goal requires feedback between experimental analysis, conceptual modeling, and data-driven modeling. Beyond the power of testing experimentally-derived models, mathematics may be incorporated throughout the iterative cycle of experiment and theory: to increase the resolution with which behaviors are observed and to predictively associate behaviors and networks. Importantly, these applications of mathematics may not only push the field closer to a whole-cell understanding of migration, but also reveal general principles utilized in cell migration and perhaps across many other cell signaling processes.
ACKNOWLEDGEMENTS
We thank Kevin Thurley for critical feedback. LFW and SJA were supported by the NIH (R01CA133253, R01 GM071794) and the Welch Foundation (I-1644, I-1619).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Chau AH, et al. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell. 2012;151(2):320–32. doi: 10.1016/j.cell.2012.08.040. In this study, Chau et al. systematically searched over 81 simple network topologies to find core topologies capable of spontaneous polarity. Eight core topologies were identified, all of which contained one or more minimal motifs: positive feedback, mutual inhibition, and/or inhibitor with positive feedback.
- 2.Swaney KF, Huang CH, Devreotes PN. Eukaryotic Chemotaxis: A Network of Signaling Pathways Controls Motility, Directional Sensing, and Polarity. Annual Review of Biophysics. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F. The Signaling Mechanisms Underlying Cell Polarity and Chemotaxis. Cold Spring Harbor Perspectives in Biology. 2009;1(4) doi: 10.1101/cshperspect.a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welf ES, Haugh JM. Signaling pathways that control cell migration: models and analysis. Wiley Interdisciplinary Reviews-Systems Biology and Medicine. 2011;3(2):231–240. doi: 10.1002/wsbm.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, et al. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114(2):201–14. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 6.Omann GM, Sklar LA. Response of neutrophils to stimulus infusion: differential sensitivity of cytoskeletal activation and oxidant production. J Cell Biol. 1988;107(3):951–8. doi: 10.1083/jcb.107.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. Here, Machacek et al. spatially and temporally characterized GTPase activitiy in fibroblasts by tracking signaling events relative to cell protrusion. RhoA was found to activate close to the cell edge and temporally near the moment of protrusion initiation, while Cdc42 and Rac1 were implicated in the reinforcement of protrusions.
- 8*.Welf ES, et al. Migrating fibroblasts reorient directionality by a metastable, PI3K-dependent mechanism. J Cell Biol. 2012;197(1):105–14. doi: 10.1083/jcb.201108152. In this study, Welf et al. spatiotemporally mapped PI3K activity and cell protrusion/retraction in randomly migrating fibroblasts. PI3K was shown to reinforce, not initiate, protrusions.
- 9.Galic M, et al. External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat Cell Biol. 2012;14(8):874–81. doi: 10.1038/ncb2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ku CJ, et al. Network crosstalk dynamically changes during neutrophil polarization. Cell. 2012;149(5):1073–83. doi: 10.1016/j.cell.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Wang Y, et al. Cell Reports. Identifying network motifs that buffer front-to-back signaling in polarized neutrophils. In Press. This study combined studied natural variation in control and microtubule-inhibited cells. Experiments, mathematical modeling, and a systematic topology search demonstrated that microtubules insulate back signaling from variations in the front signaling by acting as a long-range, positive link from front to back.
- 12.Driscoll MK, et al. Cell shape dynamics: from waves to migration. Plos Computational Biology. 2012;8(3):e1002392. doi: 10.1371/journal.pcbi.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Barnhart EL, et al. An adhesion-dependent switch between mechanisms that determine motile cell shape. PLoS Biol. 2011;9(5):e1001059. doi: 10.1371/journal.pbio.1001059. Here, Barnhart et al. created edge-velocity maps of keratocytes on substrates of different adhesion strengths in order to characterize the dependence of motility properties on adhesion. Cell shape and speed were shown to have a biphasic dependence on adhesion strength, and adhesion strength was found to be sufficient for causing cells to switch between types of migration behavior.
- 14.Zigmond SH, Sullivan SJ. Sensory adaptation of leukocytes to chemotactic peptides. J Cell Biol. 1979;82(2):517–27. doi: 10.1083/jcb.82.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrengruber MU, Deranleau DA, Coates TD. Shape oscillations of human neutrophil leukocytes: characterization and relationship to cell motility. J Exp Biol. 1996;199(Pt 4):741–7. doi: 10.1242/jeb.199.4.741. [DOI] [PubMed] [Google Scholar]
- 16.Tkachenko E, et al. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell Biol. 2011;13(6):660–7. doi: 10.1038/ncb2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10(12):1393–400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marco E, et al. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129(2):411–22. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogilner A, Allard J, Wollman R. Cell polarity: quantitative modeling as a tool in cell biology. Science. 2012;336(6078):175–9. doi: 10.1126/science.1216380. [DOI] [PubMed] [Google Scholar]
- 20.Brittain JE. Thevenin Theorem. Ieee Spectrum. 1990;27(3):42–42. [Google Scholar]
- 21.Yuan JH, et al. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484(7393):233–U115. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Ofer N, Mogilner A, Keren K. Actin disassembly clock determines shape and speed of lamellipodial fragments. Proc Natl Acad Sci U S A. 2011;108(51):20394–9. doi: 10.1073/pnas.1105333108. Ofer et al. quantitatively analyzed shape and speed of fish keratocyte fragments to guide mathematical modeling. Force-balance equations demonstrated that actin dynamics and membrane tension were sufficient to produce global shape and speed.
- 23*.Neilson MP, et al. Chemotaxis: a feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol. 2011;9(5):e1000618. doi: 10.1371/journal.pbio.1000618. In this study, Neilson et al. modeled internal cell signaling to demonstrate the feasibility of a pseudopod-centered mechanism of chemotaxis, in which the cell creates pseudopods by an intrinsic cycle and is only biased, not driven, by external cues.
- 24.Neilson MP, et al. Modeling cell movement and chemotaxis using pseudopod-based feedback. SIAM Journal on Scientific Computing. 2011;33(3):1035–1057. [Google Scholar]
- 25.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112(Pt 17):2867–74. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 26.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82(1 Pt 1):50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Ma W, et al. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138(4):760–73. doi: 10.1016/j.cell.2009.06.013. Here, Ma et al. systematically searched over abstract three-node networks to identify core topologies capable of perfect and near-perfect adaptation. Only two network topologies, the negative feedback loop and the incoherent feedforward loop, were shown to achieve perfect adaptation.
- 28.Kunida K, Matsuda M, Aoki K. FRET imaging and statistical signal processing reveal positive and negative feedback loops regulating the morphology of randomly migrating HT-1080 cells. J Cell Sci. 2012;125(Pt 10):2381–92. doi: 10.1242/jcs.096859. [DOI] [PubMed] [Google Scholar]
- 29.Shibata T, et al. Modeling the self-organized phosphatidylinositol lipid signaling system in chemotactic cells using quantitative image analysis. J Cell Sci. 2012;125(Pt 21):5138–50. doi: 10.1242/jcs.108373. [DOI] [PubMed] [Google Scholar]
- 30*.Maree AF, Grieneisen VA, Edelstein-Keshet L. How cells integrate complex stimuli: the effect of feedback from phosphoinositides and cell shape on cell polarization and motility. Plos Computational Biology. 2012;8(3):e1002402. doi: 10.1371/journal.pcbi.1002402. This study’s model of biophysical and chemical feedback showed that cell shape may not just be a downstream readout, but also a source of feedback. In silico testing demonstrated that an intermediate level of phosphoinositide feedback creates normal migration.
- 31.Lin B, et al. Synthetic spatially graded Rac activation drives cell polarization and movement. Proc Natl Acad Sci U S A. 2012;109(52):E3668–77. doi: 10.1073/pnas.1210295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Wu S, et al. Controlling multipotent stromal cell migration by integrating “course-graining” materials and “fine-tuning” small molecules via decision tree signal-response modeling. Biomaterials. 2011;32(30):7524–31. doi: 10.1016/j.biomaterials.2011.06.050. This study classified migrating multipotent stromal cell responses to combinations of signals and conditions to create decision-tree models. This modeling method predicted and in vitro experiments confirmed that decreasing ERK would promote cell migration.
- 33.Hautaniemi S, et al. Modeling of signal-response cascades using decision tree analysis. Bioinformatics. 2005;21(9):2027–35. doi: 10.1093/bioinformatics/bti278. [DOI] [PubMed] [Google Scholar]
- 34.Kharait S, et al. Decision tree modeling predicts effects of inhibiting contractility signaling on cell motility. BMC Syst Biol. 2007;1:9. doi: 10.1186/1752-0509-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vedel S, et al. Migration of cells in a social context. Proc Natl Acad Sci U S A. 2013;110(1):129–34. doi: 10.1073/pnas.1204291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szabo B, et al. Phase transition in the collective migration of tissue cells: Experiment and model. Physical Review E. 2006;74(6) doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
- 37.Welf ES, Naik UP, Ogunnaike BA. A spatial model for integrin clustering as a result of feedback between integrin activation and integrin binding. Biophys J. 2012;103(6):1379–89. doi: 10.1016/j.bpj.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schluter DK, Ramis-Conde I, Chaplain MA. Computational modeling of single-cell migration: the leading role of extracellular matrix fibers. Biophys J. 2012;103(6):1141–51. doi: 10.1016/j.bpj.2012.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pohlmeyer JV, Waters SL, Cummings LJ. Mathematical model of growth factor driven haptotaxis and proliferation in a tissue engineering scaffold. Bull Math Biol. 2013;75(3):393–427. doi: 10.1007/s11538-013-9810-0. [DOI] [PubMed] [Google Scholar]
- 40.Mallet DG, Pettet GJ. A mathematical model of integrin-mediated haptotactic cell migration. Bull Math Biol. 2006;68(2):231–253. doi: 10.1007/s11538-005-9032-1. [DOI] [PubMed] [Google Scholar]
- 41.Kabaso D, et al. Theoretical Model for Cellular Shapes Driven by Protrusive and Adhesive Forces. Plos Computational Biology. 2011;7(5) doi: 10.1371/journal.pcbi.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]