Abstract
The kidney clears numerous solutes from the plasma; however, retention of these solutes causes uremic illness when the kidneys fail. We know remarkably little about which retained solutes are toxic and this limits our ability to improve dialysis therapies. To explore this we employed untargeted mass spectrometry to identify solutes that are efficiently cleared by the kidney. High resolution mass spectrometry detected 1808 features in the urine and plasma ultrafiltrate of 5 individuals with normal renal function. The estimated clearance rates of 1082 peaks were greater than the creatinine clearance indicating tubular secretion. Further analysis identified 90 features representing solutes with estimated clearance rates greater than the renal plasma flow. Quantitative mass spectrometry with stable isotope dilution confirmed that efficient clearance of these solutes is made possible by the combination of binding to plasma proteins and tubular secretion. Tandem mass spectrometry established the chemical identity of 13 solutes including hippuric acid, indoxyl sulfate, and p-cresol sulfate. These 13 efficiently cleared solutes were found to accumulate in the plasma of hemodialysis patients, with free levels rising to more than 20-fold normal for all but two of them. Thus, further analysis of solutes efficiently cleared by secretion in the native kidney may provide a potential route to the identification of uremic toxins.
Introduction
The kidney removes numerous waste solutes from the blood plasma. When kidney function is lost, these solutes accumulate in the body and cause uremic illness culminating in death unless renal function is partially replaced by dialysis. At present we know remarkably little about which retained solutes are toxic, and this lack of knowledge limits our ability to improve treatment (1, 2). Progress has been slow in part because the number of solutes retained when the kidneys fail is very large (1, 3-6). The current study employed untargeted mass spectrometry to find solutes which are efficiently removed from the plasma by the kidney. It revealed that there are many waste solutes for which renal clearance rates normally exceed the renal plasma flow. Such high clearances require a combination of binding to plasma proteins and active tubular secretion. Concentrations of such solutes can rise to high levels when renal function is replaced by hemodialysis which clears solutes only by diffusion. Presuming that evolution has provided for the kidney to remove toxic substances efficiently, identification of solutes with high renal clearance rates could provide a route to the identification of uremic toxins.
Results
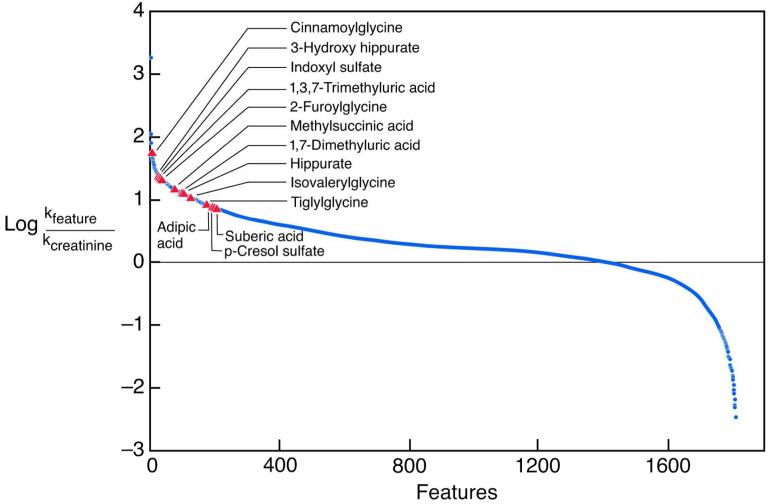
Measurements were made in four men and one woman with normal renal function as reflected by an average creatinine clearance of 142±22 ml/min/1.73 m2. A total of 1808 features were detected in both urine and plasma ultrafiltrate by untargeted high resolution mass spectrometry. Clearance rates for these features were estimated as the urinary excretion rate divided by the concentration in plasma ultrafiltrate. Clearance values are thus expressed in terms of the “free” unbound solute concentration in plasma rather than the total solute concentration. The distribution of estimated clearance rates relative to the creatinine clearance is depicted in Figure 1. For 1082 features, estimated clearance rates were greater than the clearance of creatinine with a false discovery rate of q < 0.05. Because the creatinine clearance is slightly higher than the glomerular filtration rate, these features were considered likely to represent solutes secreted by the renal tubules. There were in contrast only 290 features with estimated clearance rates less than the creatinine clearance with a false discovery rate of q < 0.05.
Figure 1.
The blue line represents the distribution of estimated clearance rates relative to the creatinine clearance for the 1,808 features found in the urine and plasma ultrafiltrate of normal subjects. The red triangles represent the 13 features for which identity was confirmed by analysis of reagent standards.
For 163 features, estimated clearance rates were greater than seven-fold the clearance of creatinine. This suggested that their clearances exceeded the renal plasma flow, which is approximately four-fold the clearance of creatinine. Among these 163 features, 90 were considered to represent unique chemical compounds after elimination of duplicates and features considered to represent dimers, adducts, isotopes, or artifacts on manual review of chromatograms (Supplementary Table 2). Compounds with matching mass values were sought in standard databases and the chemical identities of 13 of these 90 features were established by comparison of their chromatographic retention times and MS/MS spectra with those of reagent standards (Table 1; Supplementary Table 2). For 40 of the 90 features of interest, however, no candidate compounds were found among known human metabolites with mass within 3 parts per million (ppm) (Supplementary Table 2) (7, 8). Clearance values in excess of the renal plasma flow rate were possible because the “free” concentrations of these solutes in plasma ultrafiltrate were lower than their total plasma concentrations, presumably reflecting binding to plasma proteins. While all of the efficiently cleared solutes were protein-bound, the extent of binding varied widely, with the free fraction ranging from 2 to 52 percent of the total plasma concentration. Calculation in terms of total plasma concentration yielded much lower clearance values for the bound solutes (Supplementary Table 2).
Table 1. Solutes Efficiently Cleared by the Native Kidney and Their Accumulation in Hemodialysis Patients.
| Solute | Values in Normal Subjects | Hemodialysis/Normal | |||
|---|---|---|---|---|---|
| Clearance (ml/min/1.73m2) |
Clearance/ Clearancecre |
Free Fraction (%) |
Total | Free | |
| Cinnamoylglycine | 7343 ± 567 | 55 ± 5 | 4 ± 1 | 28b | 123b |
| 3-Hydroxy hippurate | 3617 ± 3618 | 23 ± 19 | 35 ± 6 | 34b | 34b |
| Indoxyl sulfate | 3023 ± 533 | 22 ± 4 | 2 ± 1 | 26b | 77b |
| 1,3,7-Trimethyluric acid | 2896 ± 1117 | 21 ± 7 | 9 ± 3 | 3 | 5b |
| 2-Furoylglycine | 2733 ± 1282 | 21 ± 11 | 49a | 64b | 82b |
| Methylsuccinic acid | 1988 ± 447 | 14 ± 4 | 11 ± 4 | 11b | 91b |
| 1,7-Dimethyluric acid | 1977 ± 2238 | 13 ± 12 | 39 ± 30 | 7b | 8b |
| Hippurate | 1868 ± 1312 | 12 ± 6 | 27 ± 5 | 31b | 53b |
| Isovalerylglycine | 1498 ± 324 | 11 ± 2 | 30 ± 6 | 9b | 47b |
| Tiglylglycine | 1148 ± 235 | 8 ± 2 | 52 ± 19 | 27b | 50b |
| Adipic acid | 1100 ± 304 | 8 ± 2 | 4 ± 1 | 13b | 23b |
| p-Cresol sulfate | 1055 ± 148 | 8 ± 1 | 2 ± 1 | 11b | 34b |
| Suberic acid | 1041 ± 147 | 8 ± 1 | 7 ± 2 | 6b | 30b |
Values are mean±sd.
Clearance/clearancecre is the average ratio of solute clearance to creatinine clearance. Free fraction is the level in plasma ultrafiltrate as a percent of the total plasma level. Hemodialysis/Normal is the ratio of the average pre-treatment concentration in hemodialysis patients to the average concentration in normal subjects.
indicates the free fraction for furoylglycine was calculated in only one subject because peaks in plasma samples from other subjects were too small to quantify.
indicates q < 0.05 for elevation of the solute concentration in hemodialysis patients above the level in normal subjects.
Measurements using LC/MS/MS with isotopically labeled standards confirmed the finding of very high clearance rates for the bound solutes hippurate, indoxyl sulfate, and p-cresol sulfate. Clearance values for these solutes along with urea and creatinine are summarized in Table 2. For comparison, clearance values were also measured for phenylacetylglutamine which previous studies had shown to be secreted by the renal tubules but largely unbound (9). As expected, the clearance of urea was less than the creatinine clearance, reflecting tubular reabsorption after glomerular filtration. The clearance of phenylacetylglutamine in contrast averaged 455±62 ml/min/1.73m2, or approximately three quarters of the estimated renal plasma flow rate. Clearance values for hippurate, indoxyl sulfate, and p-cresol sulfate were much higher. The values obtained by quantitative assay with labeled standards were slightly lower than those estimated from peak areas assessed by untargeted mass spectrometry but still well above the estimated renal plasma flow. Clearance values for these solutes expressed in terms of the total plasma concentration were much lower than those expressed in terms of the free concentration and did not exceed the renal plasma flow.
Table 2. Clearance Values Obtained Using LC/MS/MS Assay.
| Solute | Clearance (ml/min/1.73m2) |
Clearance/ Clearancecre |
Plasma Free Concentration (μM) |
Free Fraction (%) |
Clearancetotal (ml/min/1.73m2) |
|---|---|---|---|---|---|
| Urea | 57 ± 11 | 0.4 ± 0.1 | 2.4 ± 0.6×103 | - | - |
| Creatinine | 142 ± 22 | 1.0 | 74 ± 13 | - | - |
| Phenylacetylglutamine | 455 ± 62 | 3.1 ± 0.1 | 1.8 ± 0.8 | 78 ± 10 | 357 ± 73 |
| Hippurate | 1257 ± 193 | 9 ± 1 | 1.5 ± 0.9 | 41 ± 2 | 518 ± 102 |
| p-Cresol sulfate | 990 ± 151 | 7 ± 2 | 0.46 ± 0.26 | 2.4 ± 0.4 | 22 ± 3 |
| Indoxyl sulfate | 1944 ± 389 | 14 ± 2 | 0.13 ± 0.05 | 2.7 ± 0.4 | 53 ± 7 |
Values are mean±sd.
Clearance/clearancecre is the average ratio of solute clearance to creatinine clearance. Free fraction is the level in plasma ultrafiltrate as a percent of the total plasma level. Clearancetotal is the value that would be obtained if clearance were calculated using the plasma total concentration rather than the plasma free concentration.
Additional studies examined the accumulation in patients with renal failure of solutes found to be efficiently cleared by the native kidney. As summarized in Table 1, free levels of all of the 13 chemically identified solutes with high native kidney clearance rates were elevated in hemodialysis patients. Of note, for all but two of these solutes, the average free concentration in pre-treatment samples from hemodialysis patients were more than 20-fold normal, and the free solute concentrations rose higher than the total solutes concentrations. The great majority of solutes characterized only by exact mass values which have high native kidney clearances were also found to accumulate in hemodialysis patients, as further summarized in Supplementary Table 2.
Discussion
Metabolomic studies have found that the urine contains hundreds of solutes, the majority of which remain to be chemically identified (10-12). Presumably, most of these solutes are cleared by the kidney from the plasma, and their accumulation in the body as uremic solutes may contribute to illness when renal function is reduced. At present, however, we have little knowledge of which of these solutes are clinically important (3, 13).
The current study was designed to identify solutes for which renal clearance rates are normally very high. For any rate of solute production, a high clearance serves to keep the solute level in the body low. We could thus expect to find toxic waste compounds among those solutes with high clearance rates. Analysis of timed urine and plasma ultrafiltrate samples by untargeted mass spectrometry revealed the presence of 90 features considered likely to correspond to solutes with renal clearance rates greater than the renal plasma flow. Of note, compounds with corresponding mass values for many of these features could not be found in standard lists of human metabolites (7, 8). This suggests that many substances for which evolution has provided high clearance rates remain to be identified. The compounds we were able to identify included indoxyl sulfate and p-cresol sulfate. These compounds derive from the action of gut bacteria and are known to be secreted by the renal tubules. They accumulate in the plasma when the kidneys fail and have recently received extensive consideration as uremic toxins (14-17). The other compounds identified comprised three dicarboxylic acids, six acyl glycines, and two substituted purine metabolites. All these substances have previously been found in human urine and hippurate, adipic acid, and cinnamoylglycine have also been reported to accumulate in the plasma of patients with renal failure (http://www.hmdb.ca) (5, 13, 18). Like indoxyl sulfate and p-cresol sulfate, hippurate and 3-hydroxy hippurate have been shown to be secreted by the renal tubules (9, 19). But as far as we can discover, the possibility of a renal clearance exceeding the renal plasma flow has not been considered for any of these solutes.
As described here, the kidney can achieve clearance rates in excess of the renal plasma flow through a combination of tubular secretion and rapidly reversible solute binding to plasma proteins. For solutes which are confined to the plasma and not protein-bound, the maximum clearance is equal to the renal plasma flow rate. But for bound solutes, active secretion lowers the free plasma solute concentration in blood passing through the peritubular capillaries after it leaves the glomeruli so that the bound portion of the solute tends to dissociate from the binding proteins and becomes available for secretion. If the avidity of secretory transport into the tubular lumen is sufficient, the amount of solute secreted is greater than the renal plasma flow multiplied by the free concentration in the plasma entering the peritubular capillaries, and the clearance will rise above the plasma flow rate. The net effect is to reduce the unbound solute concentration in the systemic circulation to a lower level than would be achieved if the solute were completely removed from the plasma passing through the kidneys but were not protein-bound. Since it is the free, unbound concentration of solutes to which tissues throughout the body are exposed, the combination of protein-binding and tubular secretion can provide adaptive advantage in the removal of toxic waste compounds. In essence, the addition of reversible protein-binding to secretion allows the free level of a solute to be reduced without increasing kidney blood flow and size.
The ability of the kidney to reduce the free plasma concentration of bound solutes to very low levels was recognized by E. K. Marshall (20) who provided the first unequivocal demonstration of solute secretion by the renal tubules. Marshall did not measure any natural solutes but hypothesized their potential high clearance based on observation of the renal handling of phenol red, a protein-bound dye. Since then, however, the potential advantage of protein binding has been largely ignored in renal medicine, and clearance values for bound as well as unbound waste solutes have been expressed in terms of the total rather than the more relevant free plasma concentration. Standard practice has been different, however, in the pharmacology literature (21). With pharmaceutical agents as with other compounds, only the free portion of a bound solute is biologically active, and clearance rates for pharmaceuticals have therefore routinely been expressed in terms of their free plasma concentrations. Calculation in terms of the total concentration yields lower values and underestimates the body’s ability to limit the effective, free solute level as revealed in Table 2 and Supplementary Table 2.
An obvious question for renal medicine is whether secretion can persist when glomerular filtration declines. The hope based on early morphologic observations that significant numbers of “aglomerular tubules” continue to operate in patients with glomerular disease was largely disappointed by subsequent analyses (22, 23). More importantly, functional studies have demonstrated repeatedly that secretory clearances decline with the GFR. The evidence is most extensive for the clearance of para-aminohippurate, long used as a measure of renal plasma flow. On average, the clearance of para-aminohippurate declines only slightly less than the GFR, with variation among individual patients and specific diseases (24, 25). A decline in secretory clearance with GFR has also been demonstrated for endogenous organic anion such as hippurate and 5-hydroxyindolacetate and for many pharmaceuticals including protein-bound compounds like furosemide (26-28). Similar reduction in the average secretory clearance of the tightly bound endogenous solutes indoxyl sulfate and p-cresol sulfate in parallel with eGFR have also recently been described in abstract form (Meijers B, Viaene L, Poesen R; Estimated glomerular filtration rate is a good marker of renal clearance for indoxyl sulfate and p–cresyl sulfate. PO384, ASN Kidney Week 2012). Rising plasma concentrations as chronic kidney disease progresses thus do not serve to distinguish compounds which are cleared by secretion from those that are cleared largely by filtration. More subtle questions, such as the extent to which levels of secreted solute are affected by competition for transport molecules and altered expression of transport molecules during renal disease progression, have been less thoroughly studied.
A weakness of untargeted mass spectrometry is that it does not provide precise quantitation. Matrix effects alter the strength of ion signals from individual samples, and particularly from samples of different fluids. The clearance values in Table 1 which are based on relative peak areas in chromatograms of urine and plasma ultrafiltrate must thus be regarded as only approximate. More accurate quantitative LC/MS/MS assays were developed to measure concentrations of selected solutes for which we were able to obtain both unlabeled and isotopically labeled standards. These measurements confirmed that the clearances for hippurate, p-cresol sulfate, and indoxyl sulfate were much higher than the estimated renal plasma flow. Their clearances can be contrasted with that of phenylacetylglutamine, a solute which is actively secreted but largely unbound so that its clearance can only approach the renal plasma flow. Indoxyl sulfate, p-cresol sulfate, 3-hydroxyhippurate, 1,3,7-trimethyluric acid and 1,7-dimethyluric acid are known to be handled by the organic acid transporters OAT1 and/or OAT3 in the renal proximal tubule, and the structures of the other solutes we identified as efficiently cleared make them likely candidates for transport by the same mechanisms (29-33). Identification of which transporters handle which solutes, however, will require studies in cultured cells or animals in which transporter activity has been genetically manipulated.
Confirmation that individual solutes are cleared by the kidney is provided by the finding of high levels in patients with renal failure. Among the compounds we identified as having native kidney clearances greater than the renal plasma flow, the total plasma concentrations of hippurate, p-cresol sulfate, indoxyl sulfate, adipic acid and cinnamoylglycine have previously been reported elevated in patients with renal failure (5, 13, 18). The current study shows further that free levels of bound solutes which are efficiently cleared by the native kidney can rise to high levels in patients maintained on hemodialysis which provides clearance by passive diffusion. Among the 13 compounds we identified as having high native kidney clearances, free levels for all but two were more than 20-fold normal in hemodialysis patients. The exceptions, 1,3,7-trimethyluric acid and 1,7-dimethyluric acid are caffeine metabolites (34). All of our normal subjects were coffee or tea drinkers, while intake among the hemodialysis patients was restricted to a single daily cup of tea in one individual. In addition to differences in production, the non-renal clearance and volume of distribution of individual solutes can influence the degree to which their levels are elevated in dialysis patients.
Several limitations should be acknowledged. First, our list of features corresponding to solutes with high renal clearance rates is undoubtedly incomplete. Previous studies have shown that the number of features identified by mass spectrometry increases when samples are assayed using multiple chromatographic and ionization methods (11, 12, 35, 36). Second, the current approach would fail to identify compounds that are efficiently removed from the plasma but then either degraded in the kidney or altered before excretion into the urine. Finally, we may have calculated falsely high clearance values for substances that are excreted in the urine following production in the kidney.
In summary, the current study shows that the combination of protein-binding with tubular secretion allows the kidney to clear waste solutes at rates exceeding the renal plasma flow. And it suggests that such high clearance rates are achieved for a large number of natural solutes, many of which remain to be chemically identified. Further analysis of the group of solutes efficiently cleared by the kidney could provide a route to identification of these uremic toxins.
Methods
Blood samples were obtained at the midpoint of timed urine collections following an overnight fast in five subjects with normal renal function whose characteristics are noted in Supplementary Table 1. Studies were performed in accord with the Declaration of Helsinki. Plasma was deproteinized with methanol (1:3 vol:vol), dried, and reconstituted in 90:10 vol:vol water:acetonitrile to half the original concentration. Ultrafiltrate was obtained using Nanosep 30K Omega separators (Pall), dried, and reconstituted in 90:10 water:acetonitrile to five times the original concentration. Urine was diluted with water to provide solute concentrations which would be found in a urine flow of 100 ml/min, dried, and reconstituted in 90:10 water:acetonitrile to twice the concentration of the diluted sample.
Chromatography was performed on an ACQUITY UPLC system (Waters). Ten Kl of each sample was loaded to a Kinetex XB-C18 150 × 2.1 mm, 1.7 Km particle size column (Phenomenex) maintained at 40 °C. Mobile phase flow was 0.4 ml/min using 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with gradient from 3% B to 25% B over 9 minutes, from 25% B to 100% B to 15 minutes, remaining at 100% B to 19 minutes and returning to 3% B to 21 minutes. MS was performed on an Exactive orbitrap mass spectrometer (Thermo Fisher) with data collected over the range m/z 70 to 800 at 50,000 FWHM resolution using electrospray ionization with heated probe (400 °C). The MS was calibrated in positive mode using a standard LTQ ESI positive ion calibration solution (Pierce) in m/z 138-1922 mass range. The negative ion calibration was extended down to m/z 97 from the m/z 265-1980 mass range of a standard LTQ ESI negative ion calibration solution by spiking it with p-cresol sulfate and indoxyl sulfate at 3.3 ug/mL, which generated additional calibration ions of m/z 96.9601 (sulfate fragment), m/z 187.0070 (p-cresol sulfate), and m/z 212.0023 (indoxyl sulfate). Modification of the calibration mix was intended to allow sub 1 ppm accuracies in the mass range of the majority of solutes excreted in the urine and thereby decrease the number of possible elemental compositions. To minimize effects of instrument drift, samples from each subject were run together in triplicate and samples from all subjects were run first in negative and then positive mode.
MZmine software v2.2 (Okinawa Institute of Science and Technology) was used to identify features characterized by retention time and m/z from each LC/MS run and to assign amplitudes to these features based on integration of the ion current values (37). Because our goal was to identify solutes cleared by the kidney, analysis was further restricted to the 754 negative ion and 1054 positive ion features with average peak area greater than 4,000 in the triplicate runs in the plasma ultrafiltrate of at least three of five subjects and also in the urine of at least three of five subjects. Clearance rates for these 1808 features were estimated by comparing the concentrations in urine and plasma ultrafiltrate to those of creatinine. For 1082 of the 1808 features the estimated clearance rates were greater than the creatinine clearance with a false discovery rate of q < 0.05. This was considered evidence of tubular secretion. Chromatograms for the 163 of the 1082 secreted features with clearance values more than seven-fold the creatinine clearance were examined manually. The cut-off of seven fold was chosen to be well above the renal plasma flow, which is about four-fold the creatinine clearance, and to include the feature identified as p-cresol sulfate for which a deuterated standard was available allowing its high renal clearance to be confirmed by a more quantitative assay (below). Elimination of duplicates when features appeared in both positive and negative mode and of features considered to represent dimers, adducts, isotopes, or artifacts on manual review reduced the total number of features with estimated clearance rates greater than seven-fold the creatinine clearance to 90 as summarized in Supplementary Table 2. Compounds with m/z values corresponding to these features were identified using the Human Metabolome and Metlin Databases (http://www.hmdb.ca, http://metlin.scripps.edu/) and chemical standards were obtained as further summarized in Supplementary Table 2. To confirm chemical identities, selected urine samples and chemical standards were run using the same LC method coupled to an LTQ Orbitrap Velos (Thermo Fisher) for features appearing with negative mode and to an LTQ XL ion trap (Thermo Fisher) for features appearing with positive mode. Chemical identify was established by match of retention time, principle ion mass, and MS/MS spectrum (Supplementary Table 3). Among the 13 chemically identified features, the average magnitude of the mass error assigned to peaks extracted by MZmine was 0.5±0.5 ppm, encouraging confidence that correct mass values were assigned to the unidentified features listed in Supplementary Table 2.
Pretreatment plasma from 6 hemodialysis patients and from 6 normal subjects whose characteristics are summarized in Supplementary Table 1 was also analyzed by high resolution mass spectrometry as described above. Plasma was deproteinized, dried, and reconstituted in 90:10 vol:vol water:acetonitrile to one fourth the original concentration for dialysis patients and to the original concentration for normal subjects. Ultrafiltrate was reconstituted in 90:10 water:acetonitrile to the original concentration for dialysis patients and ten fold the original concentration for normal subjects. Peaks areas for plasma and ultrafiltrate obtained with MZmine software were averaged excluding values of less than 4,000 in duplicate runs of each sample. Peaks corresponding to solutes previously identified as having very high renal clearance values were identified by matching retention time and mass. Peaks corresponding to the 13 chemically identified features were reintegrated manually using Xcalibur Software (ThermoFisher).
Concentrations of p-cresol sulfate, indoxyl sulfate, hippurate, and phenylacetylglutamine in the normal samples were further assayed by stable isotope dilution LC/MC/MS using p-cresol-d8-sulfate (synthesized from p-cresol d8 (38) (Cambridge Isotopes)), indoxyl-2,4,5,6,7-d5-sulfate (Isosciences), and N-benzoyl-d5-glycine and Nα-(phenyl-d5-acetyl)-L-glutamine (both C/D/N Isotopes) as internal standards. Preparation was the same except that dried samples were reconstituted in water to twice the concentration used for untargeted analysis. Ten Kl of each sample was loaded to a Kinetex C18 150 mm × 2.1 mm, 1.7 μm particle size column maintained at 30 °C. Buffer flow was set at 0.35 ml/min using 10 mM ammonium formate in water (A) and 10 mM ammonium formate in methanol (B) with 5% B for 1 minute, from 5% B to 30% B to 6 minutes, rapidly to 90% B to 8.5 minutes, and then down to 5% B to 13.5 minutes. MS was performed on an Agilent 6430 Triple Quadrupole mass spectrometer with electrospray ionization in the negative mode (Agilent Technologies). Solute concentrations were calculated using manufacturer’s software (MassHunter Quant). Ion transitions used for quantitation were m/z 187.0 → 107.0 for p-cresol sulfate, m/z 212.0 → 80.0 for indoxyl sulfate, m/z 178.1 → 134.2 for hippurate, and m/z 263.2 → 145.1 for phenylacetylglutamine with corresponding transitions for the deuterated internal standards. Recoveries for p-cresol sulfate, indoxyl sulfate, hippurate, and phenylacetylglutamine were 100 ± 4 %, 114 ± 6 %, 93 ± 3 %, and 105 ± 3 % for reagents added to plasma ultrafiltrate, 93 ± 7 %, 108 ± 25 %, 107 ± 6 % f, and 95 ± 4 % for reagents added to plasma, and 91 ± 17 %, 105 ± 12 %, 95 ± 15 %, and 102 ± 14 % for reagents added to urine to achieve concentrations similar to those found in experimental subjects. Creatinine and urea were assayed in the clinical laboratory.
Statistics
Clearance rates for the 1808 features detected in urine and in plasma ultrafiltrate were compared to creatinine clearance rates in each subject by the Wilcoxon signed-rank test using SAS Enterprise Guide 4.3 and unpaired comparisons between values for hemodialysis patients and normal subjects were performed using the Mann-Whitney U test using SPSS V20. False discovery rates were then calculated using Q-VALUE (http://genomics.princeton.edu/storeylab/qvalue/).
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Andres Martinez of the California Polytechnic State University, San Luis Obispo, CA for the synthesis of deuterated p-cresol sulfate, the staff of the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University for assisting with analyses, and Dr. Jenny Shen of Stanford University for help with statistics.
Support: TLS was supported by the Mitsubishi Tanabe Pharma Corporation NKF Fellowship for the Study of Uremia. Other support was provided by the NIH (R21DK84439 and RO1 DK80123 to TWM and RO1DK80123 to THH).
Footnotes
Disclosure
The authors have no financial interests to disclose.
References
- 1.Vanholder R, Abou-Deif O, Argiles A, et al. The role of EUTox in uremic toxin research. Semin Dial. 2009;22:323–328. doi: 10.1111/j.1525-139X.2009.00574.x. [DOI] [PubMed] [Google Scholar]
- 2.Meyer TW, Sirich TL, Hostetter TH. Dialysis cannot be dosed. Semin Dial. 2011;24:471–479. doi: 10.1111/j.1525-139X.2011.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer TW, Hostetter TH. Uremia. N Engl J Med. 2007;357:1316–1325. doi: 10.1056/NEJMra071313. [DOI] [PubMed] [Google Scholar]
- 4.Niwa T. Update of uremic toxin research by mass spectrometry. Mass Spectrom Rev. 2011;30:510–521. doi: 10.1002/mas.20323. [DOI] [PubMed] [Google Scholar]
- 5.Rhee EP, Souza A, Farrell L, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato E, Kohno M, Yamamoto M, et al. Metabolomic analysis of human plasma from haemodialysis patients. Eur J Clin Invest. 2011;41:241–255. doi: 10.1111/j.1365-2362.2010.02398.x. [DOI] [PubMed] [Google Scholar]
- 7.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith CA, O’Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman L, Jornvall H, Bergstrom J. Phenylacetylglutamine and hippuric acid in uremic and healthy subjects. Nephron. 1990;55:265–271. doi: 10.1159/000185973. [DOI] [PubMed] [Google Scholar]
- 10.Guo K, Peng J, Zhou R, et al. Ion-pairing reversed-phase liquid chromatography fractionation in combination with isotope labeling reversed-phase liquid chromatography-mass spectrometry for comprehensive metabolome profiling. Journal of chromatography A. 2011;1218:3689–3694. doi: 10.1016/j.chroma.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 11.Roux A, Xu Y, Heilier JF, et al. Annotation of the human adult urinary metabolome and metabolite identification using ultra high performance liquid chromatography coupled to a linear quadrupole ion trap-Orbitrap mass spectrometer. Anal Chem. 2012;84:6429–6437. doi: 10.1021/ac300829f. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Creek DJ, Barrett MP, et al. Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal Chem. 2012;84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- 13.Duranton F, Cohen G, De Smet R, et al. Normal and pathologic concentrations of uremic toxins. Journal of the American Society of Nephrology: JASN. 2012;23:1258–1270. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evenepoel P, Meijers BK, Bammens BR, et al. Uremic toxins originating from colonic microbial metabolism. Kidney Int Suppl. 2009:S12–19. doi: 10.1038/ki.2009.402. [DOI] [PubMed] [Google Scholar]
- 16.Schepers E, Glorieux G, Vanholder R. The gut: the forgotten organ in uremia? Blood Purif. 2010;29:130–136. doi: 10.1159/000245639. [DOI] [PubMed] [Google Scholar]
- 17.Meyer TW, Hostetter TH. Uremic solutes from colon microbes. Kidney Int. 2012;81:949–954. doi: 10.1038/ki.2011.504. [DOI] [PubMed] [Google Scholar]
- 18.Aronov PA, Luo FJ, Plummer NS, et al. Colonic contribution to uremic solutes. Journal of the American Society of Nephrology: JASN. 2011;22:1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith HW, Finkelstein N, Aliminosa L, et al. The Renal Clearances of Substituted Hippuric Acid Derivatives and Other Aromatic Acids in Dog and Man. The Journal of clinical investigation. 1945;24:388–404. doi: 10.1172/JCI101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall EK., Jr. Two lectures on renal physiology. The Physiologist. 1966;9:367–384. [PubMed] [Google Scholar]
- 21.Benet LZ, Hoener BA. Changes in plasma protein binding have little clinical relevance. Clin Pharmacol Ther. 2002;71:115–121. doi: 10.1067/mcp.2002.121829. [DOI] [PubMed] [Google Scholar]
- 22.Oliver j. Architecture of the Kidney in Chronic Brith’s Disease. Paul B. Hoeber; New York: 1939. [Google Scholar]
- 23.Meyer TW. Tubular injury in glomerular disease. Kidney Int. 2003;63:774–787. doi: 10.1046/j.1523-1755.2003.00795.x. [DOI] [PubMed] [Google Scholar]
- 24.Bradley SE, Bradley GP, Tyson CJ, et al. Renal function in renal diseases. The American journal of medicine. 1950;9:766–798. doi: 10.1016/0002-9343(50)90292-0. [DOI] [PubMed] [Google Scholar]
- 25.Bricker NS, Klahr S, Lubowitz H, et al. Renal Function in Chronic Renal Disease. Medicine (Baltimore) 1965;44:263–288. doi: 10.1097/00005792-196507000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi P, Gulyassy P, Stanfel L, et al. Plasma hippurate in renal failure: high-performance liquid chromatography method and clinical application. Nephron. 1987;47:290–294. doi: 10.1159/000184526. [DOI] [PubMed] [Google Scholar]
- 27.Hannedouche T, Laude D, Dechaux M, et al. Plasma 5-hydroxyindoleacetic acid as an endogenous index of renal plasma flow. Kidney Int. 1989;35:95–98. doi: 10.1038/ki.1989.13. [DOI] [PubMed] [Google Scholar]
- 28.Voelker JR, Cartwright-Brown D, Anderson S, et al. Comparison of loop diuretics in patients with chronic renal insufficiency. Kidney Int. 1987;32:572–578. doi: 10.1038/ki.1987.246. [DOI] [PubMed] [Google Scholar]
- 29.Enomoto A, Takeda M, Taki K, et al. Interactions of human organic anion as well as cation transporters with indoxyl sulfate. Eur J Pharmacol. 2003;466:13–20. doi: 10.1016/s0014-2999(03)01530-9. [DOI] [PubMed] [Google Scholar]
- 30.Sugawara M, Mochizuki T, Takekuma Y, et al. Structure-affinity relationship in the interactions of human organic anion transporter 1 with caffeine, theophylline, theobromine and their metabolites. Biochim Biophys Acta. 2005;1714:85–92. doi: 10.1016/j.bbamem.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Anzai N, Kanai Y, Endou H. Organic anion transporter family: current knowledge. J Pharmacol Sci. 2006;100:411–426. doi: 10.1254/jphs.crj06006x. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto Y, Watanabe H, Noguchi T, et al. Organic anion transporters play an important role in the uptake of p-cresyl sulfate, a uremic toxin, in the kidney. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2011 doi: 10.1093/ndt/gfq785. [DOI] [PubMed] [Google Scholar]
- 33.Wikoff WR, Nagle MA, Kouznetsova VL, et al. Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1) J Proteome Res. 2011;10:2842–2851. doi: 10.1021/pr200093w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang-Liu DD, Williams RL, Riegelman S. Disposition of caffeine and its metabolites in man. The Journal of pharmacology and experimental therapeutics. 1983;224:180–185. [PubMed] [Google Scholar]
- 35.Psychogios N, Hau DD, Peng J, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordstrom A, Want E, Northen T, et al. Multiple ionization mass spectrometry strategy used to reveal the complexity of metabolomics. Anal Chem. 2008;80:421–429. doi: 10.1021/ac701982e. [DOI] [PubMed] [Google Scholar]
- 37.Pluskal T, Castillo S, Villar-Briones A, et al. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feigenbaum J, Neuberg C. Simplified method for the preparation of aromatic sulfuric acid esters. J Am Chem Soc. 1941;63:3529–3530. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.